Abstract
Xanthomonas axonopodis pv. citrumelo is a citrus pathogen causing citrus bacterial spot disease that is geographically restricted within the state of Florida. Illumina, 454 sequencing, and optical mapping were used to obtain a complete genome sequence of X. axonopodis pv. citrumelo strain F1, 4.9 Mb in size. The strain lacks plasmids, in contrast to other citrus Xanthomonas pathogens. Phylogenetic analysis revealed that this pathogen is very close to the tomato bacterial spot pathogen X. campestris pv. vesicatoria 85-10, with a completely different host range. We also compared X. axonopodis pv. citrumelo to the genome of citrus canker pathogen X. axonopodis pv. citri 306. Comparative genomic analysis showed differences in several gene clusters, like those for type III effectors, the type IV secretion system, lipopolysaccharide synthesis, and others. In addition to pthA, effectors such as xopE3, xopAI, and hrpW were absent from X. axonopodis pv. citrumelo while present in X. axonopodis pv. citri. These effectors might be responsible for survival and the low virulence of this pathogen on citrus compared to that of X. axonopodis pv. citri. We also identified unique effectors in X. axonopodis pv. citrumelo that may be related to the different host range as compared to that of X. axonopodis pv. citri. X. axonopodis pv. citrumelo also lacks various genes, such as syrE1, syrE2, and RTX toxin family genes, which were present in X. axonopodis pv. citri. These may be associated with the distinct virulences of X. axonopodis pv. citrumelo and X. axonopodis pv. citri. Comparison of the complete genome sequence of X. axonopodis pv. citrumelo to those of X. axonopodis pv. citri and X. campestris pv. vesicatoria provides valuable insights into the mechanism of bacterial virulence and host specificity.
INTRODUCTION
Xanthomonas is an important genus of plant pathogenic bacteria (87). These Gram-negative rod-shaped pathogens belong to the class Gammaproteobacteria and can infect over 350 species of plants (12). Among the diseases on citrus, citrus bacterial canker (CBC) and citrus bacterial spot (CBS) are caused by distinct pathovars of Xanthomonas species. Citrus canker is caused by several pathogenic variants of X. axonopodis pv. citri (synonyms Xanthomonas citri, Xanthomonas campestris pv. citri, or Xanthomonas citri subsp. citri) (90, 109), whereas CBS is caused by X. axonopodis pv. citrumelo. X. axonopodis pv. citri strain 306 with a suspected origin in southeastern Asia causes Asiatic type A canker and is the most widespread and virulent form of CBC. It produces hyperplasic and hypertrophic (corky) lesions surrounded by oily or water-soaked margins and a yellow halo on leaves, stems, and fruits.
In 1984, a disease similar to citrus canker was discovered in citrus nurseries in central Florida, leading to the destruction of millions of seedlings (98). This new form of citrus canker was described as the “E-strain” group, also known as nursery strain canker. Leaf spots of this strain are irregular to round, 3 to 5 mm in diameter, flat, water soaked, often necrotic in the center, and usually surrounded by a chlorotic halo. Water-soaked elongate lesions with necrotic centers are also observed on twigs but not on fruits (16). Extensive efforts were put forth to eradicate this outbreak, resulting in the destruction of 20 million citrus plants at the cost of $94 million (91).
Strains in this group do not cause hyperplasia and the lesions continue to be flat with time, unlike CBC, which results in raised callus-like lesions. Further research revealed that the CBS pathogen is variable but widely distributed in the state and does not have the same host range as the Asiatic strain (34). CBS bacteria are most aggressive on trifoliate orange hybrids, including Swingle citrumelo (36). Populations of CBS bacteria decreased or varied in leaves of grapefruit (92). The origin of the strain remains unknown as it is not found outside Florida, and it is speculated to have moved to citrus from existing populations of Xanthomonas in Florida (33). Further research showed that these strains are serologically, genetically, and physiologically distinct from the previously known citrus canker pathogenic groups (35) and are not susceptible to any of the phages commonly used to differentiate these groups (37). Because of the canker-like symptoms it caused, the disease was wrongly termed as E-strain citrus canker (32). The disease is now recognized as distinct from citrus canker and was named citrus bacterial spot, caused by Xanthomonas axonopodis pv. citrumelo (25). Other names associated with the bacteria include X. campestris pv. citrumelo and X. alfalfae pv. citrumelonis (31, 90). The nomenclature and classification for the strains of Xanthomonas that infect citrus have undergone extensive taxonomic revision in recent years and are still under debate (90, 110). Hence, in this report we chose to use classical nomenclature and address the CBS pathogen as Xanthomonas axonopodis pv. citrumelo, as approved by Bergey's Manual of Systematic Bacteriology (88).
Compared to X. axonopodis pv. citri, X. axonopodis pv. citrumelo has much reduced pathogenicity with limited host range. The host range of X. axonopodis pv. citri is broad, including most commercial citrus varieties, while X. axonopodis pv. citrumelo does not infect any commercial citrus varieties and is limited primarily to trifoliate orange, its hybrids, and a few other individual species (37). Citrus canker is endemic in Florida in most citrus-producing areas, while CBS occurs almost exclusively in nurseries, where young, susceptible tissue is abundant and irrigation is frequent (107). In field, greenhouse, and growth chamber, X. axonopodis pv. citrumelo generally does not cause disease when applied as a spray. Bacterial populations of CBS strains within lesions on most hosts except Swingle citrumelo decline rapidly with time (21).
In comparison with X. axonopodis pv. citri, the X. axonopodis pv. citrumelo strains isolated from Florida nurseries were found to vary widely in aggressiveness from each other (36). Restriction fragment length polymorphism analysis by Hartung and Civerolo (45) showed that X. axonopodis pv. citrumelo strains are not very closely related to X. axonopodis pv. citri and that they are not a form of canker. This was further corroborated by comparison of X. axonopodis pv. citrumelo strains with other xanthomonads with DNA-DNA hybridization, which showed that X. axonopodis pv. citrumelo is only about 60% similar to X. axonopodis pv. citri (21). Later, Cubero and Graham deduced that X. axonopodis pv. citrumelo is much more closely related to X. campestris pv. vesicatoria than to X. axonopodis pv. citri by using 16S rRNA gene analysis (16) as well as leucine-responsive regulatory protein gene analysis (15).
The mechanism accounting for the reduced pathogenicity and limited host range of X. axonopodis pv. citrumelo compared to X. axonopodis pv. citri remains unknown. To address this question, comparative genomic study was conducted in this present research by completing the genome sequence of X. axonopodis pv. citrumelo strain F1 (36). In comparison with X. axonopodis pv. citri, X. axonopodis pv. citrumelo is a genetically, pathogenically, and serologically diverse pathogen (4, 29, 36, 45). We decided to sequence X. axonopodis pv. citrumelo F1, which is a highly aggressive citrus bacterial spot bacterial strain and thus is more likely to infect citrus nurseries as a pathogen. To gain a better understanding of the ecological and evolutionary relationships between strains and species of Xanthomonas, we also compared X. axonopodis pv. citrumelo with the closely related X. campestris pv. vesicatoria strain 85-10 (syn. X. euvesicatoria or X. axonopodis pv. vesicatoria), which causes bacterial spot on tomato and pepper (64, 87, 109).
MATERIALS AND METHODS
Bacterial strain and DNA sequencing.
The X. axonopodis pv. citrumelo strain F1 sequenced in this study was isolated from Avon Park, FL, in 1984. Genomic DNA was extracted from bacterial culture grown overnight at 28°C in nutrient broth medium by using a Wizard DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. Quantity and quality of the DNA were measured spectrophotometrically (Nanodrop ND-1000; NanoDrop Tech. Inc., Wilmington, DE). Whole-genome sequencing was performed using two high-throughput sequencing techniques, 454 pyrosequencing and Illumina Solexa GA sequencing. Single and paired-end reads were generated on a 454 GS-FLX Titanium sequencer (454 Life Sciences, Branford, CT) in accordance with the manufacturer's protocol at the Interdisciplinary Center for Biotechnology Research (ICBR), University of Florida. Paired-end Illumina sequence reads were obtained using Illumina genome analyzer IIx (Illumina, Hayward, CA) at the Yale University Center for Genomics and Proteomics.
A total of 367,109 high-quality sequences with an average read length of 332 bp, representing more than 21-fold genome coverage, were obtained by 454 FLX sequencing. These sequences were assembled into contigs and scaffolds using the 454 de novo assembler Newbler 2.0. In total, 72 contigs were generated, of which 61 contigs were larger than 500 bp. The average size of the large contigs was 81 kb. These contigs were further grouped into five scaffolds based on paired-end reads. The maximum size of the scaffolds was 2,559,303 bases with an average of 990,948 bp. Solexa sequencing generated a total of 37,695,118 high-quality filtered sequence reads with an average read length of 74 bp. Average coverage was more than 400-fold. All reads were de novo assembled using CLCbio Genomics Workbench version 4.0, length fraction and similarity were set at 0.9, and all the other parameters were set as default values. This yielded 1,350 contigs (length weighted median N50 = 8,322; maximum length = 36,202; minimum length = 102).
Gap closure and assembly validation.
The 1,350 contigs obtained from Illumina were used to confirm the assembly of 454 scaffolds. The Illumina contigs were aligned against the 454 scaffolds by using BLASTn to confirm the orientations and integrity of the assembled sequences and to close gaps and link contigs together within the scaffold. A de novo BamHI optical map of the genome of X. axonopodis pv. citrumelo was generated by OpGen technologies (Madison, WI). In silico BamHI restriction maps of the 5 scaffolds were constructed and aligned to the optical map according to their restriction fragment pattern by using MapSolver v.3.1 software (OpGen Technologies, Inc.). PCR primers (available upon request) were designed and Sanger sequences of these PCR products were used to close the gaps between the scaffolds. Final assembly was correlated with the optical map for further validation.
Annotation and curation.
Coding genes were identified using Softberry's FgenesB suite of bacterial operon- and gene-finding programs, based on the Markov chain model prediction algorithm at ICBR, UF (108). Predicted proteins were annotated by similarity searches against the NCBI nonredundant (nr) protein database (http://ncbi.nlm.nih.gov) and the clusters of orthologous groups (COG) database. A function was assigned to a predicted gene if it met the criteria of a minimum cutoff of 50% identity and 80% coverage of the gene length. In a few cases, additional putative protein-coding genes were annotated by direct homology search at the nr protein database using BLASTp. Each gene was also functionally classified by assigning a COG number. The rRNA genes were annotated by the FgenesB tool based on sequence conservation, while tRNA genes were detected with the tRNAscan-SE program (63). Insertion sequences were identified by submitting the whole genome to the IS Finder website (95). The CGView Server was used to generate graphical views of the genome (38). The results of the automated annotation were examined and curated manually using the JGI GenePRIMP pipeline (77).
Phylogenetic analysis.
To determine the position of X. axonopodis pv. citrumelo within the evolutionary precinct of Xanthomonas pathovars, we used protein sequences of nine housekeeping genes, uvrD, secA, carA, recA, groEL, dnaK, atpD, gyrB, and infB, from 10 completely sequenced Xanthomonas spp. We also added sequences from three Xylella fastidiosa strains and three Pseudomonas spp. as well as from two Stenotrophomonas maltophilia strains. The sequences of Ralstonia solanacearum strains GMI1000 and PSI07 and Burkholderia cenocepacia strain NCTC 10247 were used as the outgroup species. Amino acid sequences of nine proteins from the above genomes were aligned using Clustal W (59), and the resulting alignments were concatenated. A phylogenic tree from concatenated genes was constructed using PAUP 4.0 (100) by the maximum likelihood method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches in the tree.
Comparative analysis.
For comparative analyses, the sequences of X. campestris pv. vesicatoria strain 85-10 (GenBank accession no. NC_007508) and X. axonopodis pv. citri strain 306 (GenBank accession no. NC_003919), which were determined as the closest relatives to X. campestris pv. citrumelo F1 in BLAST analyses as well as phylogenetic analysis, were retrieved from GenBank. Complete genome sequences of all the three Xanthomonas spp. and also specific regions were aligned and visualized in progressive mode using MAUVE (17). Pangenome analysis that included the core genome shared by all the three strains was done by an “all-against-all” BLAST of the protein sequences of the above genomes. The genes aligned based on amino acid sequence were considered orthologous if reciprocal BLASTp hits were found between two genes with e values of ≤10−20 and alignments exceeding 80% sequence identity and 80% query gene length. A gene was considered a singleton or unique to each strain if it had no hits with an e value of ≤10−5.
Additional sequence analysis.
Candidate type III secretion system (T3SS) effectors were identified using both nucleotide and protein blasts by comparison to the Xanthomonas effector database (http://www.xanthomonas.org). Putative perfect and imperfect PIP box sequences TTCGC-N15-TTCGC and TTCGC-N16-TTCG, respectively, were identified using custom scripts (23). Genomic regions with atypical G+C content were identified using the web-based software GC-profile. It uses a suite of segmentation programs to identify regions with differential G+C content in the genome (26).
Pectate lyase assay.
Cultures were grown on nutrient agar at 28°C and then suspended in sterile deionized water to an optical density at 540 nm of 0.3. Hildebrand's media A, B, and C were used to test for pectolytic activity (47). In short, the medium contained bromothymol blue dye, calcium chloride, 2% sodium polypectate, and 0.4% agar. The pH was adjusted to 4.5, 7.0, and 8.5 for media A, B, and C. One-microliter portions of the cultures were inoculated onto the plates and incubated at 28°C for 6 days before pitting was confirmed due to pectate lyase production.
Nucleotide sequence accession number.
The complete genome of Xanthomonas axonopodis pv. citrumelo strain F1 has been deposited at EMBL/DDBJ/GenBank under the accession number XACM_F1.sqn XACM CP002914.
RESULTS AND DISCUSSION
Sequencing and general features of the genome.
X. axonopodis pv. citrumelo was sequenced using 454 GS-FLX pyrosequencing (both unpaired and paired end) (65) and paired-end Illumina/Solexa sequencing (8). The reads from 454 and Illumina were assembled into contigs separately, an overview of which is presented in Table S1 in the supplemental material. Although the genome coverage obtained through Illumina was much higher than that obtained through 454, the longer GS-FLX reads resulted in a much better assembly of contigs. The 72 contigs obtained were aligned in the right order to obtain 5 scaffolds by using the paired-end reads. Aligning 1,350 Illumina contigs and using the ones overlapping the 454 contigs solved most gaps within scaffolds. Pyrosequencing has a higher error rate around homopolymers (48), resulting in insertion-deletion errors in assembly and thus in-frame stop codons in genes. Illumina data, on the other hand, have errors mainly due to mismatches (19). Hence, they were used to correct the errors in the scaffold sequences by mapping the Illumina reads against the 454 consensus sequences using CLCbio Genomics Workbench version 4.0 (6).
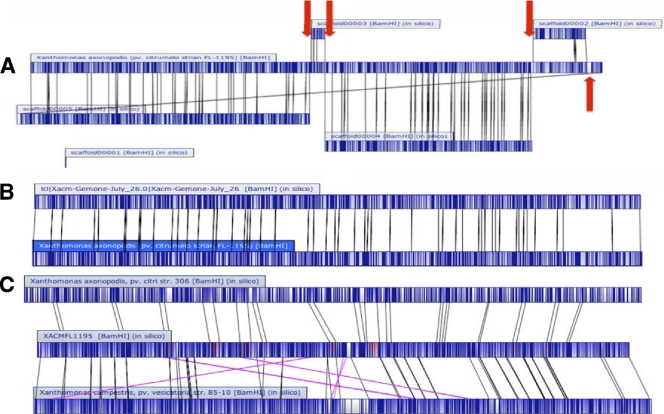
After all these intensive and time-consuming efforts, the assembly still contained 5 scaffolds with internal gaps, which were difficult to resolve due to repeat regions. Thus, optical mapping was used to obtain a de novo BamHI restriction map with no requirement for previous sequence information (60). The in silico restriction maps of scaffolds were aligned to this structural map to reveal the correct alignment and orientation of all the contigs as shown in Fig. 1 A. The genome was completely closed by primer walking and validated by manually inspecting all areas of imperfect match between the optical map and the sequence assembly (Fig. 1B). The genome sequence was further corroborated either by high coverage with the 454 and Illumina data or by resequencing of the region (data not shown).
Fig. 1.
Alignments between the whole-genome optical maps and the in silico genome sequence assemblies at various stages of the project. Dark blue represents cut sites, light blue regions indicate alignment, and white regions indicate no alignment. (A) Early comparison of an optical map derived from BamHI digestion of the X. axonopodis pv. citrumelo F1 genome to the assembled scaffolds generated by sequencing. The X. axonopodis pv. citrumelo optical map derived from BamHI digestion of the chromosome is presented as a single contig in the center. The sequenced genome contains five scaffolds that have a corresponding match to the optical map. Scaffold 1 is too small to be mapped using current optical map technology. However, during gap closure it was placed between contigs 3 and 4. The finishing strategy including gap closure was simplified using the optical map as an assembly model. Red arrows indicate where PCR gap closure was done. (B) Comparison of the final assembly of the X. axonopodis pv. citrumelo genome (top) to the optical map (bottom) for the BamHI digestion. (C) Comparison of the finished sequence of X. axonopodis pv. citrumelo (center) to the BamHI optical map of X. axonopodis pv. citri strain 306 (top) and X. campestris pv. vesicatoria strain 85-10 (bottom). Dark blue represents cut sites, light blue represents aligned regions, red represents regions aligning to both sequences, and white represents unaligned regions. Alignment lines for inversions and translocations are highlighted in pink. Inverted and translocated regions are highlighted in yellow.
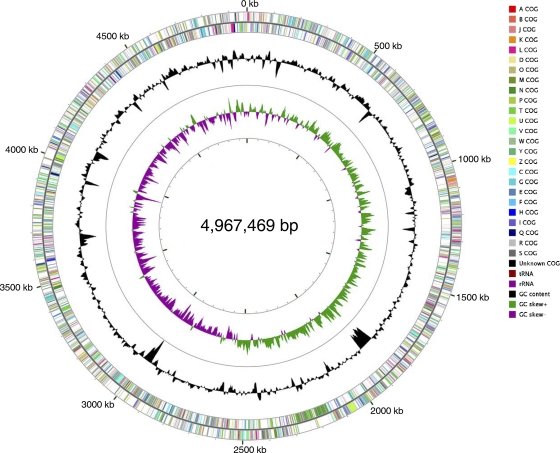
X. axonopodis pv. citrumelo strain F1 has a single, circular chromosome of 4,967,469 bp (Fig. 2) with no plasmids. Details of the general features of the genome are shown in Table 1. The G+C content of the chromosome averages 64.92%, which is similar to that of other Xanthomonas genomes. The chromosome displays a clear GC skew transition typical of prokaryotic genomes, indicative of a bidirectional replication mechanism (82). GC skew analysis and blast comparison were used to locate the origin of replication at the point with an excess of G over C corresponding to the beginning of the leading strand, and dnaA was the first of the coding sequences (CDSs) of the genome.
Fig. 2.
Circular representation of X. axonopodis pv. citrumelo F1. Circles from outside to inside: first, scale bar in kilobases; second and third, predicted coding sequences of chromosome on leading and lagging strands, respectively (colors according to COGs); fourth, G+C content; fifth, G+C skew.
Table 1.
General features of X. axonopodis pv. citrumelo strain F1 genome
| Chromosome feature | Value |
|---|---|
| Genome size (bp) | 4,967,469 |
| GC content (%) | 64.92 |
| Plasmids | 0 |
| Protein-coding region (%) | 86.53 |
| Predicted CDSs | |
| Protein-coding genes | 4,202 |
| With COGs | 3,087 |
| With Pfam | 3,293 |
| With TIGRfam | 1,314 |
| Connected to KEGG pathways | 1,189 |
| rRNA | 6 |
| rRNA operons | 2 |
| tRNA | 54 |
The X. axonopodis pv. citrumelo genome contains 4,202 putative CDSs and 60 structural RNAs (Table 1). The genome shows a coding density of 86.53%, characteristic of most xanthomonads. There is no asymmetry in the distribution of the CDSs on the chromosome between the leading-strand 2,131 (50%) and the lagging-strand 2,131 (50%). After FgenesB annotation and manual curation, 3,481 CDSs (82.42%) could be assigned to one or more COG functional classes (see Table S2 in the supplemental material), whereas there was not enough evidence for 721 CDSs to be assigned to any COG category. Two sets of 5S-16S-23S rRNA, clustered in operons, were found located in a region of approximately 500 kb (between bp 4379256 and bp 4847563) on the left replichore. A total of 54 tRNA genes with specificities for all 20 amino acids were also identified.
Phylogenetic relatedness of X. axonopodis pv. citrumelo to other xanthomonads.
To establish the phylogenetic relationship of X. axonopodis pv. citrumelo strain F1 with respect to other selected members of completely sequenced Xanthomonas, we compared a set of nine housekeeping genes. These genes are highly conserved and show no evidence of horizontal transfer among the 10 xanthomonads as well as the other plant pathogens sequenced. These genes have provided robust analysis and resolved evolutionary relationships reliably in other studies (14). For this analysis, we focused on bacteria with complete genomes and excluded draft genomes due to their limitations (75). We created an alignment of the nine proteins, concatenated the sequences, and reconstructed the phylogenetic tree by using the maximum likelihood method (Fig. 3). The phylogenetic tree indicates that X. axonopodis pv. citrumelo strain F1 groups most closely with X. campestris pv. vesicatoria and Xanthomonas axonopodis pv. citri, forming a clade distinct from other xanthomonads. The closest relative of X. axonopodis pv. citrumelo is X. campestris pv. vesicatoria 85-10, which causes bacterial spot disease in tomato and pepper (105). This is consistent with the results obtained by comparison of optical maps of the three chromosomes (Fig. 1C). X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria appear to be separated from X. axonopodis pv. citri, which is included in this cluster; this is supported by a good bootstrap value of 100 at this node. However, the relationship with X. axonopodis pv. citri is sufficiently close that they share nucleotide sequence identity of over 98% in most conserved regions. Interestingly, X. axonopodis pv. citrumelo F1 has been shown to be 56% and 58% similar to X. axonopodis pv. citri A strain 9771 and X. campestris pv. vesicatoria strain 58, respectively, by DNA-DNA hybridization analysis (21). It is noteworthy that other strains of X. axonopodis pv. citri and X. campestris pv. vesicatoria were used in that comparison rather than the sequenced X. axonopodis pv. citri A strain 306 and X. campestris pv. vesicatoria strain 85-10. Interestingly, Gent and colleagues (29) had shown that the pathovars of citrumelo are indistinguishable from a few other X. axonopodis pathovars and that they do not form a monophyletic cluster by repetitive element PCR. However, X. axonopodis pv. citrumelo F1 sequenced here is an aggressive strain which was not included in the previous study.
Fig. 3.
Maximum likelihood tree of the genome of X. axonopodis pv. citrumelo F1 showing the relationship to other fully sequenced xanthomonads and related species. The tree was constructed using concatenated protein sequences of nine housekeeping genes (uvrD, secA, carA, recA, groEL, dnaK, atpD, gyrB, and infB) aligned using Clustal W. A phylogenic tree was constructed in PAUP (version 4.0) from concatenated sequences by using the maximum likelihood method. The sequences of Ralstonia solanacearum strains GMI1000 and PSI07 and Burkholderia cenocepacia strain NCTC 10247 were used as outgroup species. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The bar (0.1) at the bottom represents the number of amino acid substitutions per site.
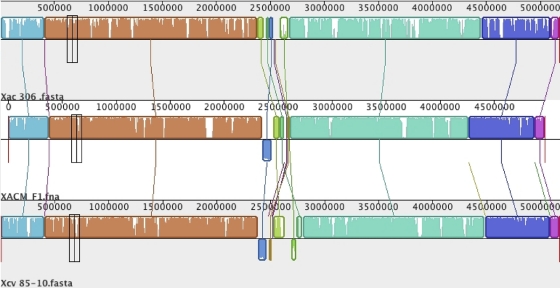
Comparison of chromosome organization of X. axonopodis pv. citrumelo to that of X. axonopodis pv. citri and X. campestris pv. vesicatoria.
The chromosome organization of X. axonopodis pv. citrumelo was compared with that of two closely related strains, X. axonopodis pv. citri and X. campestris pv. vesicatoria, by using MAUVE in progressive mode. Though most of the genome is colinear, X. axonopodis pv. citrumelo harbors some translocations and inversions around the replication terminus of the chromosome (Fig. 4). The unequal replichores might have been due to this reorganization. The X. axonopodis pv. citrumelo genome has one major inversion with a translocation and two major deletions compared to X. axonopodis pv. citri, whereas there are three inversions with translocations and two major deletions compared to X. campestris pv. vesicatoria. Many of the rearranged and deleted blocks were flanked by transposons and/or integrases, indicating that this rearrangement may be a result of horizontal gene transfer (HGT).
Fig. 4.
MAUVE alignment of the genome sequences of X. axonopodis pv. citri (Xac) strain 306, X. axonopodis pv. citrumelo (XACM) strain F1 and X. campestris pv. vesicatoria (Xcv) strain 85-10. Conserved and highly related regions are colored, and low-identity unique regions are in white (colorless).
The genome of X. axonopodis pv. citrumelo does not harbor any plasmid. In contrast, X. axonopodis pv. citri contains two (pXAC33 and pXAC64) and X. campestris pv. vesicatoria contains four (pXCV2, pXCV19, pXCV38, and pXCV183) plasmids. Plasmids of xanthomonads have been reported to play important roles in pathogenicity. Plasmid pXAC64 of X. axonopodis pv. citri carries the pthA4 gene, a homolog of pthA, which is capable of conferring the ability to cause canker-like symptoms to strains of X. axonopodis pv. citrumelo (99). On the other hand, plasmids pXCV38 and pXCV183 carry putative vir/tra- and icm/dot-like type IV secretion systems, respectively (105). The absence of plasmids from X. axonopodis pv. citrumelo may have contributed to the reduced virulence of the CBS strain.
HGT and genome plasticity.
Horizontal gene transfer (HGT) is recognized as one of the major mechanisms for genome plasticity leading to diversification and speciation of the bacteria (74). A simple method to identify potential horizontally transferred genes is to look for regions having atypical G+C content in the genome. The G+C content of X. axonopodis pv. citrumelo genome ranged from 48.90% to 68.41%, with an average of 64.92%. The segmentation results predicted eight regions of low GC content, which are recognized as genomic islands (see Table S3 in the supplemental material). The negative cumulative GC profile of these regions is also different in comparison to the whole genome (see Fig. S1A). A sharp drop in the G+C content of these regions distinctly separates them from the rest of the genome (see Fig. S1B).
These regions vary in size from approximately 3 kb to 64 kb. It was noteworthy that one of the genomic islands, from bp 2970113 to bp 3004362, codes for virB4-, virB11-, virB9-, and virD4-expressed proteins, which are part of the type IV secretion system, and components of type IV pilus like fimT, pilE, and pilus tip-associated proteins. The presence of such genes in genomic regions indicative of horizontal gene transfer is in agreement with earlier reports (105). It was also observed that about 50% of open reading frames (ORFs) in the two biggest regions, from bp 1827507 to bp 1891340 and from bp 3664590 to bp 3686175, were determined to be orphan genes. Orphan genes have a very limited phylogenetic distribution and have no recognizable homologs. A recent study in Escherichia coli demonstrated that most orphan genes encode functional proteins (18). Thus, orphan genes may encode functional proteins in X. axonopodis pv. citrumelo and might contribute to virulence or differential host range of the strain.
In addition, these regions have a high number of integrase and transposase genes, which is a conserved feature of genomic islands. The genome of X. axonopodis pv. citrumelo includes 41 insertion sequence (IS) elements. A majority of these transposases belong to the IS3 family, including Ixac2 and IS1404. We also found 2 IS elements each from the IS5 and IS1595 families and 3 elements belonging to the Tn3 family.
Comparison of proteins encoded by X. axonopodis pv. citrumelo to those encoded by X. axonopodis pv. citri and X. campestris pv. vesicatoria.
We compared the proteomes of the above three Xanthomonas spp. by using reciprocal BLASTp. A Venn diagram representing the pangenome of all three genomes is shown in Fig. 5. The comparison of the predicted protein sequences revealed that 3,292 CDSs are shared by all three genomes. These genes represent about three-quarters of the genome, forming the core set that includes conserved housekeeping and virulence genes essential for plant infection. Of the remaining 910 predicted genes in X. axonopodis pv. citrumelo, 119 have homologs only in X. axonopodis pv. citri and are absent from X. campestris pv. vesicatoria. These genes may include virulence factors necessary for infecting the common citrus host. The number of homologs in X. campestris pv. vesicatoria, at 385, is much higher than that in X. axonopodis pv. citri, further confirming that the CBS strain is much closer to X. campestris pv. vesicatoria than it is to X. axonopodis pv. citri. A total of 406 protein coding genes are unique to X. axonopodis pv. citrumelo, of which 298 are hypothetical genes, 26 are mobile genetic elements, and 82 are singletons with predicted functions. Moreover, 174 genes show homologs in distinctly related Xanthomonas or other highly related bacteria, suggesting their acquisition by horizontal gene transfer. The significant features shared between the genomes as well as the differences between the genomes are discussed in detail below.
Fig. 5.
Venn diagram representing the pangenome of X. axonopodis pv. citrumelo F1 (XACM), X. campestris pv. vesicatoria strain 85-10 (XCV), and X. axonopodis pv. citri strain 306 (XAC). Numbers in parentheses represent the protein coding genes on the chromosome in each species; numbers of shared genes are also shown within the diagram.
T3SS gene clusters.
Gram-negative bacteria use T3SS to translocate virulence factors into the host cell. In X. axonopodis pv. citrumelo, the T3SS is encoded by 27 genes, the organization of which is in close synteny with the hrp cluster of X. campestris pv. vesicatoria and X. axonopodis pv. citri (see Fig. S2 in the supplemental material). The cluster includes all nine hrc (hypersensitive response conserved) genes, which encode T3SS structural components, and all 9 hrp genes, some of which encode components of the hrp pilus. These genes, present only in phytopathogenic bacteria, are associated with the rapid programmed death of plant cells at the site of infection in most nonhost or resistant host plants and for pathogenesis in susceptible hosts (10). The major difference from X. axonopodis pv. citri is that the X. axonopodis pv. citrumelo cluster lacks the hypothetical gene upstream of hrpF and instead has three additional genes in the same locus. These 3 genes consist of XACM_0383, which encodes a hypothetical protein with 97% amino acid identity to a hypothetical protein XPE_2921 in X. perforans, a pathogen of tomato causing bacterial spot (51); XACM_0384, which is 99% identical to outer membrane protein F1 (XopF1) from X. campestris pv. vesicatoria; and XACM_0385, with no obvious homologs. The final gene in this cluster, XACM_0407, shares 98% similarity to putative transglycosylase gene hpaH from X. perforans. As compared to that of X. campestris pv. vesicatoria, the T3SS of X. axonopodis pv. citrumelo lacks 2 hrp-associated genes, hpaG and hpaE; 2 outer protein genes, xopD and xopA; and 5 hypothetical genes, XCV_0410, XCV_0412, XCV_0436, XCV_0438, and XCV_0439, with no known functions. The loss of these genes from X. axonopodis pv. citrumelo might have contributed to host range and virulence variation.
Repertoire of T3SS effectors of X. axonopodis pv. citrumelo in comparison to those of X. axonopodis pv. citri and X. campestris pv. vesicatoria.
T3SS effectors were identified in the X. axonopodis pv. citrumelo genome and compared with those of X. axonopodis pv. citri and X. campestris pv. vesicatoria. Considerable differences were observed in the effector repertoires present in these three strains (Table 2). Twenty-two effectors were identified in X. axonopodis pv. citrumelo, whereas 25 and 30 effectors were identified in X. axonopodis pv. citri and X. campestris pv. vesicatoria, respectively. We subdivided them into three groups—core, partially shared, and species specific—depending on their presence in the three strains. The core effectors shared by all the three strains consist of 17 effector genes. Of this core set, 9 effector genes (avrBs2, xopK, xopL, xopN, xopP, xopQ, xopR, xopX, and xopZ) are present in the genomes of all sequenced Xanthomonas spp. with the exception of X. albilineans and X. campestris pv. armoraciae, the latter of which has only xopP and xopR. These genes might be essential effector genes required for the pathogenicity of xanthomonads in plant hosts. The effector avrBs2 belongs to a family known for effector-triggered immunity in plants (101). It elicits hypersensitive reaction (HR) in plants carrying the Bs2 resistance gene (67) and is needed for the full virulence of the pathogen on susceptible hosts. Effector xopQ, which belongs to the hopQ1 family from Pseudomonas, has also been known as an avirulence determinant in Nicotiana benthamiana, since the Pseudomonas syringae pv. tomato DC3000 deletion mutant (deletion of hopQ1-1) acquired the ability to grow to high levels and produce bacterial speck lesions in nonhost N. benthamiana (111). XopN has been shown to interact with TARK1 and TFT1 proteins from tomato, thus repressing pathogen-associated molecular pattern-triggered immunity (55). The homologs of effectors xopL, xopP, and xopQ have been shown to contribute to pathogenicity in X. campestris pv. campestris (49). Both xopX and xopZ potentially interfere with host innate immunity, thus making the plant more susceptible. The remaining 8 core effectors (xopA, xopE1, xopF2, xopI, xopV, xopAD, xopAE, and xopAK) are not present in all xanthomonads and might be responsible for pathogenicity in some plant hosts while inducing resistance in others (113). It is likely that none of the effectors belonging to the core group are responsible for the difference in virulence and host range of X. axonopodis pv. citrumelo, X. axonopodis pv. citri, and X. campestris pv. vesicatoria.
Table 2.
Effector repertoire of X. axonopodis pv. citrumelo strain F1, X. axonopodis pv. citri strain 306, and X. campestris pv. vesicatoria strain 85-10
| Effector class | Designation(s) in: |
Pfam domains | Reference(s) | ||
|---|---|---|---|---|---|
| X. axonopodis pv. citrumelo strain F1 | X. axonopodis pv. citri strain 306 | X. campestris pv. vesicatoria strain 85-10 | |||
| Core effectors present in all three strains | |||||
| AvrBs2 | XACM_0049 | XAC0076 | XCV0052 | Glycerophosphoryl diester phosphodiesterase | 53 |
| XopA (HpaI/HpaG) | XACM_0406 | XAC0416 | XCV0440 | 73 | |
| XopE1 (AvrXacE1) | XACM_0271 | XAC0286 | XCV0294 | Putative transglutaminase | 106 |
| XopF2 | XACM_2726 | XAC2785a | XCV2942 | 86 | |
| XopI | XACM_0750 | XAC0754 | XCV0806 | F-box protein | 104 |
| XopK | XACM_3001 | XAC3085 | XCV3215 | 24 | |
| XopL | XACM_3007 | XAC3090 | XCV3220 | LRR protein | 50 |
| XopN | XACM_2728 | XAC2786 | XCV2944 | ARM/HEAT repeat | 55 |
| XopP | XACM_1178 | XAC1208 | XCV1236 | 86 | |
| XopQ | XACM_4215 | XAC4333 | XCV4438 | Inosine uridine nucleoside N-ribohydrolase | 86 |
| XopR | XACM_0263 | XAC0277 | XCV0285 | 24 | |
| XopV | XACM_0604 | XAC0601 | XCV0657 | 24 | |
| XopX | XACM_0532 | XAC0543 | XCV0572 | 66 | |
| XopZ | XACM_2036 | XAC2009 | XCV2059 | 24 | |
| XopAD | XACM_4086 | XAC4213 | XCV4315a XCV4314a XCV4313a | SKWP repeat protein | 40,79 |
| XopAE (HpaF/HpaG) | XACM_0381 | XAC0393 | XCV0409a XCV0408a | LRR protein | 113 |
| XopAK | XACM_3563 | XAC3666 | XCV3786 | 79 | |
| Effectors shared by X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria but not present in X. axonopodis pv. citri | |||||
| XopC1 | XACM_2129b XACM_2132b XACM_2248b | XCV2435 | Phosphoribosyltransferase domain and haloacid dehalogenase-like hydrolase | 86 | |
| XopF1 (Hpa4) | XACM_0384 | XCV0414 | 86 | ||
| XopAJ (AvrRxo1) | XACM_4204 | XCV4428 | Zeta toxin | 117 | |
| Effectors shared by X. axonopodis pv. citri and X. campestris pv. vesicatoria but not present in X. axonopodis pv. citrumelo | |||||
| XopE2 (AvrXacE3, AvrXccE1) | XACb0011 | XCV2280 | Putative transglutaminase | 106 | |
| Effectors unique to X. axonopodis pv. citri | |||||
| PthA (AvrBs3, TAL) | XACa0022(PthA1) XACa0039(PthA2) XACb0015(PthA4) XACb0065(PthA4) | Transcriptional activator, nuclear localization | 2 | ||
| XopE3 (AvrXacE2) | XAC3224 | Putative transglutaminase | 71 | ||
| XopAI | XAC3230 | Putative ADP- ribosyltransferase | 105 | ||
| HrpW (PopW) | XAC2922 | Pectate lyase | 76 | ||
| Effectors unique to X. campestris pv. vesicatoria | |||||
| AvrBs1 | XCVd0104 | 105 | |||
| XopB | XCV0581 | 72 | |||
| XopD | XCV0437 | C48-family SUMO cysteine protease (Ulp1 protease family), EAR motif | 86 | ||
| XopG | XCV1298 | M27 family peptidase clostridium toxin | 105 | ||
| AvrBs1.1 (XopH) | XCVd0105 | Putative tyrosine phosphatase | 105 | ||
| XopJ1 | XCV2156 | C55-family cysteine protease or Ser/Thr acetyltransferase | 86 | ||
| XopJ3 (AvrRxv) | XCV0471 | C55-family cysteine protease or Ser/Thr acetyltransferase | 105 | ||
| XopO | XCV1055 | 105 | |||
| XopAA | XCV3785 | Early chlorosis factor, proteasome/cyclosome repeat | 105 | ||
| Effectors unique to X. axonopodis pv. citrumelo | |||||
| XopC2 | XACM_1180 | XAC1209a XAC1210a | XCV1238a XCV1237a | Haloacid dehalogenase-like hydrolase | 113 |
| XopW | XACM_0435 | 24 | |||
Inactive/pseudogene.
Partial sequences due to interruption by IS elements during HGT.
The partially shared effectors are present in only two of the three strains. This group consists of xopC1, xopF1, and xopAJ, shared by X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria only, as well as xopE2, shared by X. axonopodis pv. citri and X. campestris pv. vesicatoria but absent from X. axonopodis pv. citrumelo. xopF1 is found in all Xanthomonas species except X. axonopodis pv. citri, which encodes a truncated version of the same. In X. axonopodis pv. citrumelo, a homolog of xopF1 is present, which shares 99% similarity to that gene in X. campestris pv. vesicatoria. The xopAJ homolog of X. axonopodis pv. citrumelo shares 99% similarity with xopAJ-avrRxo1 of X. campestris pv. vesicatoria. This gene is truncated in X. axonopodis pv. citrumelo due to a deletion mutation at bp 1056 in the gene that resulted in early termination of the protein at 379 amino acids as opposed to its homolog of 450 amino acids in X. campestris pv. vesicatoria. The xopC1 effector gene encodes a haloacid dehalogenase-like hydrolase with several phosphoribosyltransferase domains. This gene is present in X. axonopodis pv. citrumelo but is fragmented across the genome (XACM_2129, XACM_2132, and XACM_2248) due to genome rearrangement and transposon insertion and thus is likely to be nonfunctional. xopE2, which has been identified in various xanthomonads, has recently been shown to be involved in the virulence of X. campestris pv. vesicatoria group B strains on tomato but not in that of group A strains (62). It has also been related to suppression of HR, indicating that it plays a dual role in different host plants (62).
Two species-specific effectors, xopC2 and xopW, were found in X. axonopodis pv. citrumelo. Though homologs of xopC2 are found in both X. axonopodis pv. citri and X. campestris pv. vesicatoria, they might be nonfunctional. X. axonopodis pv. citrumelo consists of a xopC2 gene with its closest homolog in X. perforans, which causes bacterial spot only on tomato (80). X. axonopodis pv. citrumelo also has a truncated gene, XACM_0435, which is a homolog of xopW from X. oryzae pv. oryzicola and might be nonfunctional. X. campestris pv. vesicatoria has at least 9 unique effectors, as listed in Table 2. Some effectors, such as avrBs1, avrBs1.1, and xopJ3, are known avirulence factors. These effectors might be important for pathogenicity in tomato and pepper. X. axonopodis pv. citri possesses four unique effectors, avrBs3-pthA, xopE3, xopAI, and hrpW, which are absent from X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria (Table 2).
The differences in the repertoires of T3SS effectors in X. axonopodis pv. citrumelo and X. axonopodis pv. citri might contribute to their differences in virulence and host range. T3SS effectors have been known to contribute to pathogenicity and multiplication of pathogens in planta (42). T3SS effectors benefit the pathogens by altering the physiology of the host cell and suppressing plant defenses (39). T3SS effectors might contribute to host range by suppressing host defenses as virulence factors or narrowing the host range when certain effectors are specifically recognized by the plant as avirulence factors (43). Importantly, avrBs3-pthA is present in X. axonopodis pv. citri A strain 306 while absent from X. axonopodis pv. citrumelo F1 and X. campestris pv. vesicatoria 85-10. However, it is noteworthy that many other X. campestris pv. vesicatoria strains contain avrBs3 and homologs (103). In X. axonopodis pv. citri strain 306, there are 4 copies of pthA, pthA1, pthA2, pthA3, and pthA4, on two plasmids, which are all absent from X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria. PthA4 with 17.5 repeats, which is same as PthA, is known to play an important role in citrus canker as knockout of pthA4 abolished the development of citrus canker symptom development (115). PthA is responsible for the development of hypertrophic and hyperplasic symptoms and cell death, and its mutation leads to a reduction in the ability of bacteria to disseminate from infected lesions (116). PthA also contributes to epidermal rupture and necrosis, which promotes the exudation and dissemination of X. axonopodis pv. citri. Interestingly, the pthA gene from the X. axonopodis pv. citri strain, when introduced to X. axonopodis pv. citrumelo, conferred the ability to cause raised pustules (99). PthA and its homologs do not determine host range according to a previous study (3), indicating that neither of the complementing homologs nor any of the noncomplementing paralogs of pthA suppresses the avirulence of the X. axonopodis pv. citri A* strain on grapefruit. However, hssB3.0, a homolog of pthA, was shown to be responsible for the host-specific suppression of virulence of X. axonopodis pv. citri A strain KC21 on Citrus grandis cultivars but not on other Citrus species, such as Citrus sinensis (93). This suppression led to reduced aggressiveness rather than a change in host range, since X. axonopodis pv. citri A strain KC21 still causes citrus canker symptoms on Citrus grandis.
Other X. axonopodis pv. citrumelo-specific effectors might contribute to the broad host range of X. axonopodis pv. citri compared to that of X. axonopodis pv. citrumelo. xopE3 (avrXacA2) is a putative transglutaminase enzyme gene that belongs to the hopX (avrPphE) family and is widespread among phytopathogenic bacteria (71). XopAI is putative effector protein reported only in the three canker-causing strains X. axonopodis pv. citri 306, X. aurantifolii strain B, and X. aurantifolii strain C as well as in one X. vesicatoria strain, namely, 1111 (68, 80). The role of XopAI in the virulence of Xanthomonas remains to be characterized. HrpW is not known to be associated with virulence, although it contains domains resembling hairpins and pectate lyases. It may not function as an intracellular effector but is secreted by the T3SS. HrpW in several other phytopathogens is known to elicit an HR in nonhost plants (54). Alternately, the limited host range of X. axonopodis pv. citrumelo might result from the presence of the X. axonopodis pv. citrumelo-specific xopC2 and xopW serving as avirulence factors. The presence of all the species-specific effectors in X. axonopodis pv. citri and X. axonopodis pv. citrumelo may be the main factors determining the host range of the pathogens. Further study is needed to understand their contribution to X. axonopodis pv. citri and X. axonopodis pv. citrumelo for infecting different hosts.
The difference in effector repertoires of X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria might contribute to their different host specificities, with X. axonopodis pv. citrumelo infecting citrus seedlings and X. campestris pv. vesicatoria 85-10 causing bacterial spot disease on both pepper and tomato plants (80, 87). It has been suggested that the specific effector set of a given bacterial strain is the potential determinant of host range (105). Compared to X. campestris pv. vesicatoria, X. axonopodis pv. citrumelo contains xopC2 and xopW, (which are absent from X. campestris pv. vesicatoria), while X. axonopodis pv. citrumelo lacks avrBs1, xopB, xopD, xopG, xopH (avrBs1), xopJ1, xopJ3 (avrRxv), xopO, and xopAA (which are present in X. campestris pv. vesicatoria) (Table 2). avrBs1 is known to encode a 50-kDa protein with homology to AvrA of Pseudomonas syringae pv. glycinea. This protein specifies avirulence on pepper cultivars containing the resistance gene Bs1 (69). XopD is known to alter host transcription, promote pathogen growth, and delay the development of disease symptoms (56). XopB attenuated cell proliferation when expressed in yeast and also caused cell death in N. benthamiana leaves but not in tomato (89). XopJ homologs are known to inhibit host protein secretion and interfere with defense responses (7). Other effectors including XopG, XopO, and XopAA in X. campestris pv. vesicatoria have not been studied in detail. How these effectors contribute to the different host ranges and different virulences of X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria needs further characterization.
Other secretion systems associated with virulence.
Xanthomonads have at least five more protein secretion systems other than the T3SS, including types I, II, IV, V, and VI. Genes involved in all the secretion systems were identified in the X. axonopodis pv. citrumelo F1 genome. Secretion systems are of fundamental importance for the translocation of proteins and other molecules. They play important roles in the virulence of different bacterial pathogens. The relevant features of these virulence-associated secretion systems of X. axonopodis pv. citrumelo shared with X. axonopodis pv. citri and X. campestris pv. vesicatoria are presented below.
(i) T1SS.
The type I secretion system (T1SS) has not been shown to contribute to virulence in Xanthomonas spp. (11). Instead, T1SS is required for Xa21-mediated immunity in rice against X. oryzae pv. oryzae PXO99. Ax21 (activator of Xa21-mediated immunity) is highly conserved in Xanthomonas spp. and secreted by T1SS. In X. axonopodis pv. citrumelo, the Ax21 protein (XACM_0208) is 100% identical with the X. axonopodis pv. citri and X. campestris pv. vesicatoria proteins and 93% identical with the X. oryzae pv. oryzae PXO99 protein. In addition, RaxST is required for sulfation, and three genes, raxA, raxB, and raxC, are required for the secretion of Ax21 (44). The gene raxA in X. axonopodis pv. citrumelo (XACM_1188) may be nonfunctional due to a frameshift mutation, whereas the proteins encoded by raxB (XACM_1189), raxC (XACM_3355), and raxST (XACM_1187) are 99 to 100% identical to X. campestris pv. vesicatoria proteins. X. axonopodis pv. citri, on the other hand, contains only the raxC gene.
(ii) T2SS.
X. axonopodis pv. citrumelo is also equipped with the xps and xcs type II secretion system (T2SS) clusters, which secrete toxins and degradative enzymes. The T2SS clusters in X. axonopodis pv. citrumelo are very conserved compared to those identified in X. campestris pv. vesicatoria and X. axonopodis pv. citri, with xps being composed of 11 genes and xcs of 12 genes (see Fig S3 in the supplemental material). The xps T2SS, which is found in all the sequenced xanthomonads, is known to affect virulence in X. campestris pv. vesicatoria and also enhance translocation by T3SS. The xcs T2SS, on other hand, is restricted to only some Xanthomonas spp. and has no obvious virulence function (102). The T2SSs are known to secrete many plant cell wall-degrading enzymes like cellulases, xylanases, lipases, and proteases, among others. Each species has its unique set of enzymes, which helps degrade components of the plant cell wall, thus assisting in pathogenesis. The range of these enzymes in X. axonopodis pv. citrumelo was compared to the ones found in X. campestris pv. vesicatoria and X. axonopodis pv. citri (see Table S4 in the supplemental material). Enzymes such as cellulase, protease, and pectate lyase have been known to promote bacterial nutrition and also virulence (20, 83). XynC, an endoxylanase in X. campestris pv. vesicatoria, is known to be secreted by the xps T2SS under the control of hrpG and hrpX and contribute to virulence (102). X. axonopodis pv. citrumelo contains a homolog of this gene, XACM_0913, that may play a similar role. Most of the enzymes show functional redundancy and hence the loss of one gene might not affect virulence. Rajeshwari et al. (81) showed that double mutants, with mutations of both lipase and xylanase, show much reduced virulence as compared to the single mutants in X. oryzae pv. oryzae.
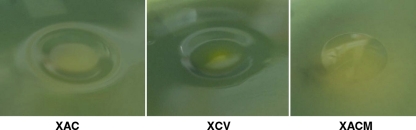
It is interesting that X. axonopodis pv. citrumelo is deficient in pectate lyase function. In contrast with the four genes in X. campestris pv. vesicatoria and the three in X. axonopodis pv. citri, X. axonopodis pv. citrumelo shows the presence of only two genes. However, both the genes, XACM_2919 and XACM_3456, are pseudogenes that have stop codons, resulting in truncated protein. Thus, these proteins may be nonfunctional. This was confirmed by inoculating the strains onto Hildebrand's medium. Both X. axonopodis pv. citri and X. campestris pv. vesicatoria produced pitting in the agar at pH 8.5, confirming pectate lyase activity. X. axonopodis pv. citrumelo did not produce any pitting, as seen in Fig. 6, supporting the hypothesis that it is pectate lyase deficient. A pectate lyase gene homolog, xagP, has been shown to induce an HR in tobacco and pepper in X. axonopodis pv. glycines (52). The role of pectate lyase in citrus pathogens remains to be determined. Several T2SS substrates have been shown not only to affect virulence but also to induce plant defense responses. T2SS and its substrates are also controlled by T3SS regulators (41).
Fig. 6.
Comparison of the pecate lyase production by X. axonopodis pv. citri strain 306 (XAC), X. axonopodis pv. citrumelo strain F1 (XACM), and X. campestris pv. vesicatoria strain 85-10 (XCV). Pitting can be seen in Hildebrand's agar medium at pH 8.5 when pectate lyase-positive strains X. axonopodis pv. citri 306 and X. campestris pv. vesicatoria 85-10 were inoculated. No pitting is seen for X. axonopodis pv. citrumelo strain F1. All strains were incubated in Hildebrand's agar medium at 28°C for 6 days.
(iii) T4SS.
In bacteria, the type IV secretion system (T4SS) is known to contribute to virulence. Two T4SS clusters are present in both X. axonopodis pv. citri and X. campestris pv. vesicatoria. Both the clusters are of the Vir type in X. axonopodis pv. citri (see Fig. S4 in the supplemental material), where one is located on the chromosome and the other is on plasmid (1). X. campestris pv. vesicatoria, on the other hand, codes for one Vir and the other Dot/Icm-type cluster (see Fig. S4), both on plasmids, and a partial Vir cluster on the chromosome (105). X. axonopodis pv. citrumelo codes for only one Vir-type T4SS cluster on the chromosome. The cluster in X. axonopodis pv. citrumelo does not show high similarity to Vir-type T4SS of either X. axonopodis pv. citri or X. campestris pv. vesicatoria. With the exception of virD4, which shows homologs in both X. axonopodis pv. citri and X. campestris pv. vesicatoria, most of the predicted T4SS genes in X. axonopodis pv. citrumelo share sequence similarity with genes in strains of Stenotrophomonas maltophilia, which is an aerobic Gram-negative environmental bacterium commonly found in soil, water, and animals (46). X. axonopodis pv. citrumelo also codes for a VirK and two VirJ-like proteins outside the T4SS cluster. The function of the virK protein is unknown and it has been linked to T2SS substrates instead of T4SS (41). VirJ is a perisplasmic chaperone believed to mediate the association between the T4S pilus and substrate proteins (13).
(iv) T5SS.
Both X. axonopodis pv. citri and X. campestris pv. vesicatoria encode a two-partner secretion system, which belongs to the type V secretion system (T5SS). It translocates large proteins such as adhesins and has been identified in many bacterial pathogens. X. axonopodis pv. citrumelo codes for a filamentous hemagglutinin-like protein, FhaB. A complete homolog of the corresponding gene can be found in X. axonopodis pv. citri but is inactivated in X. campestris pv. vesicatoria by an internal stop codon-inducing mutation. In X. axonopodis pv. citrumelo, the gene is interrupted due to genomic rearrangements, which indicates that it might be inactive. The rearrangement has caused the gene to split, with insertion of 2 hypothetical genes, XACM_1838 and XACM_1839, between the two fhaB fragments. Transposon genes in the vicinity may have instigated this change. fhaB is involved in attachment and biofilm formation in X. axonopodis pv. citri, and its loss affects the virulence of the bacterium (30). The fhaB gene in X. axonopodis pv. citrumelo is likely to be inactive due to the insertion, and the lack of the functional gene could contribute to the low virulence of X. axonopodis pv. citrumelo compared to that of X. axonopodis pv. citri.
(v) T6SS.
Recently, a new secretion system was identified in Vibrio cholerae and Pseudomonas aeruginosa and was named the type VI secretion system (T6SS). T6SS is evolutionarily related to bacteriophage, likely a remnant of phage injection machinery (84). T6SS has diverse roles in virulence, symbiosis, interbacterial interactions, and antipathogenesis in different bacteria (84). X. campestris pv. vesicatoria is found to have two clusters of T6SS: cluster type I and cluster type II, of which the latter is split into two locations. X. axonopodis pv. citri has only cluster type II T6SS, which like that of X. campestris pv. vesicatoria is split into two locations (9). The distribution of the T6SS in all three Xanthomonas is compiled in Table S5 in the supplemental material. X. axonopodis pv. citrumelo codes for two clusters, cluster type I from XACM_2098 to XACM_2121 and cluster type II from XACM_4015 to XACM_3979. Both of the clusters are homologs to the ones found in X. campestris pv. vesicatoria. X. axonopodis pv. citri, on the other hand, has only T6SS cluster type II, encoded from XAC4147 to XAC4112.
Bacterial surface structures. (i) LPS.
Lipopolysaccharide (LPS) serves a dual role as a physical barrier by protecting bacteria from antibacterial substances produced by plants and also as an inducer of plant defense-related genes (70). Flanked by highly conserved housekeeping genes for cystathionine gamma lyase (met) at one end and electron transport flavoprotein (etf) at the other end, the genome of X. axonopodis pv. citrumelo contains a cluster of 22 genes involved in LPS biosynthesis. The LPS gene cluster in a 24.5-kb region in X. axonopodis pv. citrumelo is markedly different in both number and composition from the 17-gene cluster in X. axonopodis pv. citri and more so from the 16-gene cluster in X. campestris pv. vesicatoria (Fig. 7). The flanking genes of etfB, etfA, and metB-metC are conserved in all three genomes. The LPS locus of X. axonopodis pv. citrumelo has at least six homologs to X. axonopodis pv. citri and only one to X. campestris pv. vesicatoria (Fig. 7). The LPS cluster is involved in synthesis of O-antigen polysaccharide. It is known to be important for biofilm formation on the host and contributes to virulence. Two such loci, XAC3586 and rfbC, have been experimentally shown to contribute to the virulence of X. axonopodis pv. citri on grapefruit (Citrus paradisi cv. Duncan grapefruit) (61). X. axonopodis pv. citrumelo does not have homologs for either of these genes. This may contribute to the poor survival of X. axonopodis pv. citrumelo in planta and hence the low virulence compared to X. axonopodis pv. citri. Interestingly, the wzt mutants of X. axonopodis pv. citri showed more water soaking on citrus plants (58). XACM_3499 in X. axonopodis pv. citrumelo is the closest homolog to wzt, with a low protein identity of 34% and 41% to its orthologs in X. axonopodis pv. citri and X. campestris pv. vesicatoria. Also, the X. axonopodis pv. citrumelo gene codes for a truncated protein with half missing from the C-terminal region as compared to its orthologs; thus, it might be nonfunctional. This may have led to variation in the symptoms of X. axonopodis pv. citrumelo, which shows pronounced water soaking compared to X. axonopodis pv. citri. It was also suggested that there is no obvious correlation of the content of the LPS gene cluster with host specificity (64). Thus, the variation in the LPS gene cluster among X. axonopodis pv. citrumelo, X. axonopodis pv. citri, and X. campestris pv. vesicatoria might contribute to their differences in virulence or symptom development in plant hosts rather than serving as a determinant of their differential host range. In either case, these differences are consistent with the hypothesis of strong diversifying selection on this locus put forward by Patil et al. (78).
Fig. 7.
Relative organization of the LPS gene cluster in the genomes of X. axonopodis pv. citrumelo strain F1 (XACM), X. axonopodis pv. citri strain 306 (XAC), and X. campestris pv. vesicatoria strain 85-10 (XCV). Not to scale. Conserved and highly related genes are colored and low-identity or unique genes are white.
(ii) Extracellular polysaccharides.
Extracellular polysaccharides (EPS, called xanthan gum in Xanthomonas) are an important component of a biofilm and contribute to the epiphytic fitness of Xanthomonas spp. (85). They are postulated to promote colonization of plant tissues by protecting the pathogens from harsh environmental conditions and to contribute to occlusion of vascular tissues in wilts and blights (57). X. axonopodis pv. citrumelo contains the complete gum gene cluster from gumA to gumP, which is syntenic to those found in X. axonopodis pv. citri and X. campestris pv. vesicatoria. The identity of gum genes ranges from 88 to 100% among the three Xanthomonas species. Thus, it is unlikely that EPS plays any role in the differences in virulence and host range of X. axonopodis pv. citrumelo, X. axonopodis pv. citri, and X. campestris pv. vesicatoria.
(iii) Flagella.
Xanthomonas is known to contain all the genes for flagellum synthesis and motility. Flagella are encoded by various genes located in 4 clusters, which are characteristically flanked by transposase genes. X. axonopodis pv. citrumelo contains complete flagellar structure and motility genes in four similar clusters. The flagellar genes in X. axonopodis pv. citrumelo are mostly organized in an order similar to those in X. campestris pv. vesicatoria and X. axonopodis pv. citri (see Fig. S5 in the supplemental material). Cluster 1 consists of motA (XACM_3590) and motB (XACM_3592) and cluster 2 of motB (XACM_1939) and motC (XACM_1940). These genes encode flagellar motor proteins required for the rotation of flagella. Cluster 3 in X. axonopodis pv. citrumelo consists of 24 genes from XACM_1954 to XACM_1977, involved in synthesis and regulation of flagella that is syntenic with X. axonopodis pv. citri and X. campestris pv. vesicatoria. The gene fliK in this cluster may be a pseudogene, which is nonfunctional due to frameshift mutations. Mutations in fliK affect flagellar hook length in animal pathogenic bacteria (114). However, no detectable difference was observed between the motilities of X. axonopodis pv. citri and X. axonopodis pv. citrumelo (data not shown). X. axonopodis pv. citrumelo has a cluster comprising 24 genes from XACM_1991 to XACM_2014, which is highly conserved as compared to both X. axonopodis pv. citri and X. campestris pv. vesicatoria.
Interestingly, the genes that lie between these clusters are different in X. axonopodis pv. citrumelo and X. axonopodis pv. citri. A notable difference includes the absence of a homolog of XAC1927 from X. axonopodis pv. citrumelo. This gene, encoding an Fe-S oxidoreductase and located on a probable pathogenicity island, has been linked to virulence in X. axonopodis pv. citri (58). The absence of this gene from X. axonopodis pv. citrumelo could possibly contribute to the low virulence of X. axonopodis pv. citrumelo compared to X. axonopodis pv. citri on citrus.
(iv) rpf cluster.
The regulation of pathogenicity factors (rpf) genes control the synthesis of diffusible signal factor (DSF), which plays a major role in quorum sensing, thus controlling various virulence factors in X. axonopodis pv. citri (94). Three core genes, rpfF, rpfC, and rpfG, control the synthesis of the DSF molecule and signal transduction. rpfF is responsible for the production of DSF, whereas rpfC-rpfG are two-component signaling factors. RpfC is a sensor protein and RpfG is a response regulator. RpfG has a HD-GYP domain that regulates the amount of cyclic di-GMP. Furthermore, this is involved in the regulation of the DSF regulon, thus affecting the virulence of the pathogen X. axonopodis pv. citri (5). In addition to all the rpf genes found in X. axonopodis pv. citri, the CBS pathogen X. axonopodis pv. citrumelo contains a functional rpfH, which lies nestled between rpfC and rpfG. This gene encodes a protein which is structurally similar to the sensory domain of RpfC. rpfH is also present in X. campestris pv. vesicatoria and X. campestris pv. campestris but absent from X. axonopodis pv. citri. A study by Slater et al. (97) showed that mutation in the rpfH gene in X. campestris pv. campestris did not affect the DSF pathway, and thus its role in this operon is unclear. It would be interesting to study whether its presence affects the virulence of X. axonopodis pv. citrumelo on citrus.
Other strain-specific genes that might contribute to the distinct virulences of X. axonopodis pv. citri and X. axonopodis pv. citrumelo on citrus.
Overall, 807 X. axonopodis pv. citri-specific genes were missing in X. axonopodis pv. citrumelo and X. campestris pv. vesicatoria. Besides the genes discussed above, X. axonopodis pv. citri also contains other genes missing from X. axonopodis pv. citrumelo that may be responsible for its higher virulence. A plant-like natriuretic peptide (PNP) gene (XacPNP), which is expressed specifically during the infection process in X. axonopodis pv. citri, is one such gene. XAC2654 encodes this plant-like hormone that induces changes in host photosynthetic efficiency, thereby weakening host defense (28). It has been shown that X. axonopodis pv. citri PNP mimics host PNP and results in improved host tissue health and consequently better pathogen survival in the lesions (27). The X. axonopodis pv. citrumelo genome was found to have neither a homolog of XAC2654 nor the surrounding region in its genome. Interestingly, X. axonopodis pv. citri also contains genes with putative toxin-producing functions. The genes syrE1 and syrE2 are similar to those found in Pseudomonas syringae that encode the phytotoxin syringomycin (22). These nonribosomal peptide synthetases, which might produce toxins, are absent from the X. axonopodis pv. citrumelo genome. X. axonopodis pv. citri also codes for hemolysin-type calcium-binding proteins XAC2197 and XAC2198 and also contains potential secretion genes hlyB and hlyD. These genes are also found in the citrus pathogen Xylella fastidiosa, and their products belong to the RTX toxin family (96). They are known to be pore-forming cytotoxins which act as virulence factors, and individual toxins often exhibit host specificity in eukaryotes (112). The region containing the toxin genes is absent from X. axonopodis pv. citrumelo. Another region of 20 kb, approximately from Mb 1.72 to 1.74, is specific to X. axonopodis pv. citri and is not found in X. axonopodis pv. citrumelo. It contains at least two genes, XAC1496 and XAC1507 (mobL), which are involved in the virulence of the canker pathogen (115). This region has a very low G+C content of 50% and is surrounded by integrase genes, suggesting that X. axonopodis pv. citri might have acquired it through recent HGT events. These might be potential genes contributing to the high aggression shown by X. axonopodis pv. citri on citrus as compared to X. axonopodis pv. citrumelo.
In conclusion, we completed the genome sequencing of X. axonopodis pv. citrumelo F1, combining 454 GS-FLX pyrosequencing (both unpaired and paired-end) and paired-end Illumina/Solexa sequencing and closing the gap using Sanger sequencing. Comparison of the finished genome sequence of X. axonopodis pv. citrumelo to those of X. axonopodis pv. citri and X. campestris pv. vesicatoria provides valuable insights into the emergence of new virulent strains with different host ranges and distinct virulences. Such knowledge contributes to our understanding of bacterial evolution and the role of various systems in virulence and host range of the pathogen. These strain-specific genes need to be functionally characterized to understand their roles in virulence and host specificity.
Supplementary Material
ACKNOWLEDGMENTS
This work has been supported by the Florida Citrus Research and Development Foundation.
We thank N. Potnis for her critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Alegria M., et al. 2005. Identification of new protein-protein interactions involving the products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J. Bacteriol. 187:2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Saadi A. 2005. Phenotypic characterization and sequence analysis of pthA homologs from five pathogenic variant groups of Xanthomonas citri. Ph.D. dissertation. University of Florida, Gainesville, FL [Google Scholar]
- 3. Al-Saadi A., et al. 2007. All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host-range variation. Mol. Plant Microbe Interact. 20:934–943 [DOI] [PubMed] [Google Scholar]
- 4. Alvarez A. M., Benedict A. A., Mizumoto C. Y., Pollard L. W., Civerolo E. L. 1991. Analysis of Xanthomonas campestris pv. citri and X. c. citrumelo with monoclonal antibodies. Phytopathology 81:9 [Google Scholar]
- 5. Andrade M., et al. 2006. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol. Microbiol. 62:537–551 [DOI] [PubMed] [Google Scholar]
- 6. Aury J. M., et al. 2008. High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies. BMC Genomics 9:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartetzko V., et al. 2009. The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol. Plant Microbe Interact. 22:655–664 [DOI] [PubMed] [Google Scholar]
- 8. Bentley D. R. 2006. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 16:545–552 [DOI] [PubMed] [Google Scholar]
- 9. Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Büttner D., Bonas U. 2003. Common infection strategies of plant and animal pathogenic bacteria. Curr. Opin. Plant Biol. 6:312–319 [DOI] [PubMed] [Google Scholar]
- 11. Büttner D., Bonas U. 2010. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34:107–133 [DOI] [PubMed] [Google Scholar]
- 12. Chan J. W., Goodwin P. H. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17:489–508 [DOI] [PubMed] [Google Scholar]
- 13. Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke C. R., Cai R., Studholme D. J., Guttman D. S., Vinatzer B. A. 2010. Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol. Plant Microbe Interact. 23:198–210 [DOI] [PubMed] [Google Scholar]
- 15. Cubero J., Graham J. 2004. The leucine-responsive regulatory protein (lrp) gene for characterization of the relationship among Xanthomonas species. Int. J. Syst. Evol. Microbiol. 54:429–437 [DOI] [PubMed] [Google Scholar]
- 16. Cubero J., Graham J. H. 2002. Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 68:1257–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darling A. E., Mau B., Perna N. T. 2010. Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daubin V., Ochman H. 2004. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 14:1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dohm J. C., Lottaz C., Borodina T., Himmelbauer H. 2008. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 36:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dow J. M., Clarke B. R., Milligan D. E., Tang J. L., Daniels M. J. 1990. Extracellular proteases from Xanthomonas campestris pv. campestris, the black rot pathogen. Appl. Environ. Microbiol. 56:2994–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Egel D. S., Graham J. H., Stall R. E. 1991. Genomic relatedness of Xanthomonas campestris strains causing diseases of citrus. Appl. Environ. Microbiol. 57:2724–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Etchegaray A., Silva-Stenico M. E., Moon D. H., Tsai S. M. 2004. In silico analysis of nonribosomal peptide synthetases of Xanthomonas axonopodis pv. citri: identification of putative siderophore and lipopeptide biosynthetic genes. Microbiol. Res. 159:425–437 [DOI] [PubMed] [Google Scholar]
- 23. Fenselau S., Bonas U. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc. Spa, and Fli secretion systems. Mol. Plant Microbe Interact. 8:845–854 [DOI] [PubMed] [Google Scholar]
- 24. Furutani A., et al. 2009. Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 22:96–106 [DOI] [PubMed] [Google Scholar]
- 25. Gabriel D. W., Kingsley M. T., Hunter J. E., Gottwald T. 1989. Reinstatement of Xanthomonas citri (ex Hasse) and X. phaseoli (ex Smith) to species and reclassification of all X. campestris pv. citri strains. Int. J. Syst. Bacteriol. 39:9 [Google Scholar]
- 26. Gao F., Zhang C. T. 2006. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 34:W686–W691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garavaglia B. S., et al. 2010. Shedding light on the role of photosynthesis in pathogen colonization and host defense. Commun. Integr. Biol. 3:382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garavaglia B. S., et al. 2010. A plant natriuretic peptide-like molecule of the pathogen Xanthomonas axonopodis pv. citri causes rapid changes in the proteome of its citrus host. BMC Plant Biol. 10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gent D. H., et al. 2005. Pathogenic and genetic relatedness among Xanthomonas axonopodis pv. allii and other pathovars of X. axonopodis. Phytopathology 95:918–925 [DOI] [PubMed] [Google Scholar]
- 30. Gottig N., Garavaglia B. S., Garofalo C. G., Orellano E. G., Ottado J. 2009. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One 4:e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gottwald T. R., Alvarez A. M., Hartung J. S., Benedict A. A. 1991. Diversity of Xanthomonas campestris pv. citrumelo strains associated with epidemics of citrus bacterial spot in Florida citrus nurseries: correlation of detached leaf, monoclonal antibody, and restriction fragment length polymorphism assays. Phytopathology 81:5 [Google Scholar]
- 32. Gottwald T. R., et al. 1988. Dynamics and spatial distribution of Xanthomonas campestris pv. citri group E strains in simulated nursery and new grove situations. Plant Dis. 72:7 [Google Scholar]
- 33. Gottwald T. R., Graham J. H. 1990. Spatial pattern analysis of epidemics of citrus bacterial spot in Florida nurseries. Phytopathology 80:10 [Google Scholar]
- 34. Gottwald T. R., Graham J. H., Civerolo E. L., Barrett H. C., Hearn C. J. 1993. Differential host range reaction of citrus and citrus relatives to citrus canker and citrus bacterial spot determined by leaf mesophyll susceptibility. Plant Dis. 77:6 [Google Scholar]
- 35. Graham J. H., Gottwald T. R. 1991. Research perspectives on eradication of citrus bacterial diseases in Florida. Plant Dis. 75:1193–1200 [Google Scholar]
- 36. Graham J. H., Gottwald T. R. 1990. Variation in aggressiveness of Xanthomonas campestris pv. citrumelo associated with citrus bacterial spot in Florida citrus nurseries. Phytopathology 80:7 [Google Scholar]
- 37. Graham J. H., Gottwald T. R., Fardelmann D. 1990. Cultivar-specific interactions for strains of Xanthomonas campestris from Florida that cause citrus canker and citrus bacterial spot. Plant Dis. 74:753–756 [Google Scholar]
- 38. Grant J. R., Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36:W181–W184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grant S. R., Fisher E. J., Chang J. H., Mole B. M., Dangl J. L. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60:425–449 [DOI] [PubMed] [Google Scholar]
- 40. Guidot A., et al. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo Y., Figueiredo F., Jones J., Wang N. 2011. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri. Mol. Plant Microbe Interact. 24:649–661 [DOI] [PubMed] [Google Scholar]
- 42. Gürlebeck D., Thieme F., Bonas U. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163:233–255 [DOI] [PubMed] [Google Scholar]
- 43. Hajri A., et al. 2009. A “repertoire for repertoire” hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One 4:e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han S. W., Lee S. W., Ronald P. C. 2011. Secretion, modification, and regulation of Ax21. Curr. Opin. Microbiol. 14:62–67 [DOI] [PubMed] [Google Scholar]
- 45. Hartung J. S., Civerolo E. S. 1989. Restriction fragment length polymorphisms distinguish Xanthomonas campestris strains isolated from Florida citrus nurseries from X. c. pv. citri. Mol. Plant Pathol. 79:7 [Google Scholar]
- 46. Hauben L., Vauterin L., Moore E. R., Hoste B., Swings J. 1999. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 49 (Pt. 4):1749–1760 [DOI] [PubMed] [Google Scholar]
- 47. Hildebrand D. C. 1971. Pectate and pectin gels for differentiation of Pseudomonas sp. and other bacterial plant pathogens. Phytopathology 61:7 [Google Scholar]
- 48. Huse S. M., Huber J. A., Morrison H. G., Sogin M. L., Welch D. M. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang W., et al. 2009. Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant Microbe Interact. 22:1401–1411 [DOI] [PubMed] [Google Scholar]
- 50. Jiang Y. 2007. The rice XA21-binding protein 25 is an ankyrin repeat-containing protein and required for XA21-mediated disease resistance Ph.D. dissertation. University of Florida, Gainesville, FL [Google Scholar]
- 51. Jones J. B., Stall R. E., Bouzar H. 1998. Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36:41–58 [DOI] [PubMed] [Google Scholar]
- 52. Kaewnum S., Prathuangwong S., Burr T. J. 2006. A pectate lyase homolog, xagP, in Xanthomonas axonopodis pv. glycines is associated with hypersensitive response induction on tobacco. Phytopathology 96:1230–1236 [DOI] [PubMed] [Google Scholar]
- 53. Kearney B., Staskawicz B. J. 1990. Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature 346:385–386 [DOI] [PubMed] [Google Scholar]
- 54. Kim J. F., Beer S. V. 1998. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180:5203–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim J. G., et al. 2009. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21:1305–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim J. G., et al. 2008. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell 20:1915–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kiraly Z., El-Zahaby H. M., Klement Z. 1997. Role of extracellular polysaccharide (eps) slime of plant pathogenic bacteria in protecting cells to reactive oxygen species. J. Phytopathol. 145:10 [Google Scholar]
- 58. Laia M. L., et al. 2009. New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC Microbiol. 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 60. Latreille P., et al. 2007. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics 8:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li J., Wang N. 2011. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol. Plant Pathol. 12:381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin R. H., Peng C. W., Lin Y. C., Peng H. L., Huang H. C. 2011. The xopE2 effector protein of Xanthomonas campestris pv. vesicatoria is involved in virulence and in the suppression of the hypersensitive response. Bot. Stud. 52:18 [Google Scholar]
- 63. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lu H., et al. 2008. Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS One 3:e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Margulies M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Metz M., et al. 2005. The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 41:801–814 [DOI] [PubMed] [Google Scholar]
- 67. Minsavage G. V., et al. 1990. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol. Plant Microbe Interact. 3:7 [Google Scholar]
- 68. Moreira L. M., et al. 2010. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Napoli C., Staskawicz B. 1987. Molecular characterization and nucleic acid sequence of an avirulence gene from race 6 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Newman M. A., von Roepenack E., Daniels M., Dow M. 2000. Lipopolysaccharides and plant responses to phytopathogenic bacteria. Mol. Plant Pathol. 1:25–31 [DOI] [PubMed] [Google Scholar]
- 71. Nimchuk Z. L., Fisher E. J., Desveaux D., Chang J. H., Dangl J. L. 2007. The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol. Plant Microbe Interact. 20:346–357 [DOI] [PubMed] [Google Scholar]
- 72. Noël L., Thieme F., Nennstiel D., Bonas U. 2001. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 41:1271–1281 [DOI] [PubMed] [Google Scholar]
- 73. Noël L., Thieme F., Nennstiel D., Bonas U. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ochman H., Lawrence J. G., Groisman E. A. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304 [DOI] [PubMed] [Google Scholar]
- 75. Palmer L. E., McCombie W. R. 2002. On the importance of being finished. Genome Biol. 3:COMMENT201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Park D. S., et al. 2006. Sensitive and specific detection of Xanthomonas axonopodis pv. citri by PCR using pathovar specific primers based on hrpW gene sequences. Microbiol. Res. 161:145–149 [DOI] [PubMed] [Google Scholar]
- 77. Pati A., et al. 2010. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat. Methods 7:455–457 [DOI] [PubMed] [Google Scholar]
- 78. Patil P., Bogdanove A., Sonti R. 2007. The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Petnicki-Ocwieja T., et al. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 99:7652–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Potnis N., et al. 2011. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rajeshwari R., Jha G., Sonti R. V. 2005. Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant Microbe Interact. 18:830–837 [DOI] [PubMed] [Google Scholar]
- 82. Ravin N. V., Kuprianov V. V., Gilcrease E. B., Casjens S. R. 2003. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 31:6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ray S. K., Rajeshwari R., Sonti R. V. 2000. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant Microbe Interact. 13:394–401 [DOI] [PubMed] [Google Scholar]
- 84. Records A. R. 2011. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24:751–757 [DOI] [PubMed] [Google Scholar]
- 85. Rigano L., et al. 2007. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol. Plant Microbe Interact. 20:1222–1230 [DOI] [PubMed] [Google Scholar]
- 86. Roden J. A., et al. 2004. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. U. S. A. 101:16624–16629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ryan R. P., et al. 2011. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat. Rev. Microbiol. 9:344–355 [DOI] [PubMed] [Google Scholar]
- 88. Saddler G. S., Bradbury J. F. 2005. Family I. Xanthomonadaceae fam. nov., p. 63 In Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria), 2nd ed. Springer, New York, NY [Google Scholar]
- 89. Salomon D., Dar D., Sreeramulu S., Sessa G. 2011. Expression of Xanthomonas campestris pv. vesicatoria type III effectors in yeast affects cell growth and viability. Mol. Plant Microbe Interact. 24:305–314 [DOI] [PubMed] [Google Scholar]
- 90. Schaad N. W., et al. 2006. Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29:690–695 [DOI] [PubMed] [Google Scholar]
- 91. Schubert T. S. 1991. Recent history of citrus canker eradication programs in Florida. Newsl. Fla. Phytopathol. Soc. 2:6 [Google Scholar]
- 92. Schubert T. S., et al. 2001. Meeting the challenge of eradicating citrus canker in Florida - again. Plant Dis. 85:17. [DOI] [PubMed] [Google Scholar]
- 93. Shiotani H., Fujikawa T., Ishihara H., Tsuyumu S., Ozaki K. 2007. A pthA homolog from Xanthomonas axonopodis pv. citri responsible for host-specific suppression of virulence. J. Bacteriol. 189:3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Siciliano F., et al. 2006. Analysis of the molecular basis of Xanthomonas axonopodis pv. citri pathogenesis in Citrus limon. Electron. J. Biotechnol. 9:5 [Google Scholar]
- 95. Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Simpson A. J., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature 406:151–159 [DOI] [PubMed] [Google Scholar]
- 97. Slater H., Alvarez-Morales A., Barber C. E., Daniels M. J., Dow J. M. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986–1003 [DOI] [PubMed] [Google Scholar]
- 98. Sun M. 1984. The mystery of Florida's citrus canker: scientists haven't seen this bacterial strain before, but it's already led to the destruction of millions of seedlings. Science 226:322–323 [DOI] [PubMed] [Google Scholar]
- 99. Swarup S., Yang Y., Kingsley M. T., Gabriel D. W. 1992. An Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant Microbe Interact. 5:204–213 [DOI] [PubMed] [Google Scholar]
- 100. Swofford D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). 4.0 ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- 101. Swords K. M., Dahlbeck D., Kearney B., Roy M., Staskawicz B. J. 1996. Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 178:4661–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Szczesny R., et al. 2010. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol. 187:983–1002 [DOI] [PubMed] [Google Scholar]
- 103. Szurek B., Rossier O., Hause G., Bonas U. 2002. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46:13–23 [DOI] [PubMed] [Google Scholar]
- 104. Thieme F. 2008. Genombasierte Identifizierung neuer potentieller Virulenzfaktoren von Xanthomonas campestris pv. vesicatoria. Ph.D. dissertation. Mathematisch-Naturwissenschaftlich-Technische Fakultät der Martin-Luther Universität, Halle-Wittenberg, Germany [Google Scholar]
- 105. Thieme F., et al. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Thieme F., et al. 2007. New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant Microbe Interact. 20:1250–1261 [DOI] [PubMed] [Google Scholar]
- 107. Timmer L. W., Gottwald T. R., Zitko S. E. 1991. Bacterial exudation from lesions of asiatic citrus canker and citrus bacterial spot. Plant Dis. 75:4 [Google Scholar]
- 108. Tyson G. W., et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 109. Vauterin L., Hoste B., Kersters K., Swings J. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:18 [Google Scholar]
- 110. Vauterin L., Rademaker J., Swings J. 2000. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 90:677–682 [DOI] [PubMed] [Google Scholar]
- 111. Wei C. F., et al. 2007. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51:32–46 [DOI] [PubMed] [Google Scholar]
- 112. Welch R. A. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521–528 [DOI] [PubMed] [Google Scholar]
- 113. White F. F., Potnis N., Jones J. B., Koebnik R. 2009. The type III effectors of Xanthomonas. Mol. Plant Pathol. 10:749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Williams A. W., et al. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yan Q., Wang W. 2011. High-throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol. Plant Microbe Interact. doi:10.1094/MPMI-05-11-0121 [DOI] [PubMed] [Google Scholar]
- 116. Yang B., White F. F. 2004. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant Microbe Interact. 17:1192–1200 [DOI] [PubMed] [Google Scholar]
- 117. Zhao B., et al. 2004. The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol. Plant Microbe Interact. 17:771–779 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.