Abstract
We used a surface trypsinolysis assay to probe accessibility of the membrane-proximal N-terminal tether peptides of Borrelia surface lipoproteins OspA and Vsp1. Our findings with both wild-type and mutant proteins are only compatible with the anchoring of these surface lipoproteins in the outer leaflet of the outer spirochetal membrane.
TEXT
Lipoproteins displayed on the surface of Borrelia spirochetes play crucial roles in the transmission of the bacteria by arthropod vectors as well as the ensuing development of Lyme borreliosis or relapsing fever in mammalian hosts (3, 6, 23). While it has been widely assumed that members of this important class of virulence factors are anchored peripherally via their triacyl moiety in the outer lipid bilayer leaflet of the outer membrane (OM), the supporting evidence has been quite circumstantial. For example, the phenotype of outer surface lipoprotein A (ospA)-deficient Borrelia burgdorferi cells “decorated” with recombinant lipidated OspA (L-OspA) was identical to that of OspA-expressing B. burgdorferi cells (10), and lipophilic fluorescent probes were shown to readily insert into the OM of B. burgdorferi (14). Also, the minimal lengths of N-terminal OspA “tether” peptides (Fig. 1) that were sufficient to target the red fluorescent reporter protein mRFP to the borrelial surface (28, 29) appeared to be too short to support anchoring in the periplasmic leaflet of the OM via a membrane-spanning peptide. However, Escherichia coli Lpp was recently shown to sequester into the OM and display its C terminus on the bacterial surface in its peptidoglycan-free form, i.e., to assume a variation of such an “inside anchor-outside protein,” or “in-out,” topology (13). As the precise definition of a borrelial surface lipoprotein's final topology is crucial for deciphering the steps required for lipoprotein surface display, we decided to revisit this issue by determining a surface lipoprotein's most-N-terminal amino acid residue accessible to surface proteolysis. One set of experiments built on the original observations by Barbour, Dunn, and colleagues that cell-associated as well as recombinant soluble OspA forms were resistant to trypsin (4, 15); the abundant Lys residues found throughout the protein are protected from trypsinolytic attack because of their confinement in a secondary structure or short loops (20, 33). These early studies as well as subsequent studies (9, 16) focused only on the remaining nontrypsinolyzed cell-associated OspA but did not investigate whether a trypsin-resistant OspA core was released into the reaction supernatant. Such a proteolysis-dependent protein core release had been observed for Borrelia lipoproteins belonging to the dimeric OspC/Vsp family (outer surface lipoprotein C/variable surface lipoprotein) (18, 33), which indicated that N-terminal tether peptide Lys residues were trypsin accessible in the context of an intact cell envelope. We therefore decided to first establish whether OspA was detectably released from cells upon trypsin treatment. As Borrelia turicatae Vsp1 is expressed and properly displayed on the surface of B. burgdorferi (34), we included it as a second model protein in the current study.
Fig. 1.
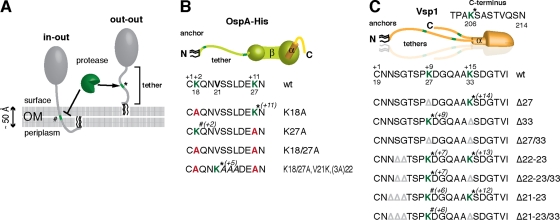
OspA and Vsp1 tether peptide mutant constructs. (A) Experimental approach of probing lipoprotein tether topology (“in-out” versus “out-out”; see text) by surface proteolysis. The position of a potential endopeptidase recognition site, e.g., a Lys residue recognized by trypsin, within the N-terminal tether peptide is shown in “in-out” and “out-out” topological scenarios. The accessible site is labeled with a *, while the protected site is labeled with a #. A dedicated OM pore likely required for an “in-out” topology has been omitted for clarity. (B and C) Cartoons of OspA tether peptide mutants (B) and Vsp1 tether peptide mutants (C) generated and used in this study. Triacyl anchor lipids are shown in black, OspA peptides are in green (N-terminal tether and β-sheets) and orange (C-terminal α-helix), epitope tag peptides are in yellow, and Vsp1 peptides are shown in orange. For clarity, one subunit of the Vsp1 dimer is emphasized. Tether peptides are shown in one-letter amino acid codes. Potential trypsin sites are indicated in green. Residues targeted in this study are highlighted in bold and colors. Gray Δ signs indicate deleted amino acid codons. Asterisks indicate trypsin sites that were determined to be surface accessible, while pound signs indicate sites that appeared to be protected from proteolysis. The mutant nomenclature follows earlier publications (28, 29). Briefly, modified residues are numbered according to their prolipoprotein position. These positions are indicated below the proteins' wt peptide sequence. Amino acid positions relative to the acyl-modified Cys residue (defined as +1) are shown above the wt peptide sequence. Italicized numbers in brackets next to the * or # signs above the mutant tether peptide sequences indicate the positions of the respective trypsin sites, relative to the +1 Cys.
OspA and Vsp1 tether peptide mutants (Fig. 1B and C; Table 1) were generated by modifying existing OspA and Vsp1 expression plasmids through oligo-mediated site-directed mutagenesis (QuikChange; Stratagene) (Table 2) as described previously (18, 28, 29). Following established protocols (26, 30), the resulting plasmids were used to transform B. burgdorferi B313, an OspA-deficient clone of type strain B31 (ATCC 35210) (25, 34, 35). Transformants were grown in selective BSK-II medium in a 5% CO2 atmosphere (2, 32). For surface proteolysis assays we used standard final trypsin or proteinase K concentrations of 200 μg/ml; remaining cell-associated proteins were harvested, and cell pellets were washed as described previously (9, 29). Where indicated, trypsinolysis reaction supernatants were further purified by Triton X-114 fractionation (8, 9, 22, 29) to remove potential contamination by membrane vesicle-associated OspA; only the aqueous fraction was loaded for analysis. Protein preparations were separated by sodium dodecyl sulfate–12% polyacrylamide electrophoresis (SDS-PAGE) as described previously (29) and probed for OspA or Vsp1 by Western immunoblotting using mouse monoclonal antibody (MAb) H5332 or 1H12, respectively (5, 11). Alkaline phosphatase (AP)-conjugated goat anti-mouse IgG (H+L) secondary antibody (Sigma) and CDP-Star AP substrate (GE Healthcare Life Sciences) were used for chemiluminescence detection using a Fuji LAS-4000 fluorescence imager. Flagellar protein FlaB served as loading control and was detected with an anti-FlaB rat polyclonal antiserum (1) (a gift from M. Caimano) and AP-conjugated rabbit anti-rat IgG (H+L) (Sigma).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| B. burgdorferi strain B313 | Clone of B31 ATCC 35210; cp26, cp32-1, cp32-2/7, cp32-3, lp17; OspA− | 25 |
| Plasmids | ||
| pSC0999 | pBSV2::PflaBospA-His lacking pBSV2 MCS BamHI/XmaI | 29 |
| pSC2007 | pBSV2::PflaBospA-K18A | This study |
| pSC2008 | pBSV2::PflaBospA-His, K27A | This study |
| pSC2009 | pBSV2::PflaBospA-His, K18/27A | This study |
| pSC2010 | pBSV2::PflaBospA-His, K18/27A, V21K, (3A)22 | This study |
| pVsp1 | pBSV2::PflaBVsp1 | 34 |
| pOSK216 | pBSV2::PflaBVsp1Δ27 | This study |
| pOSK221 | pBSV2::PflaBVsp1Δ33 | This study |
| pSC5001 | pBSV2::PflaBVsp1Δ27/33 | This study |
| pOSK289 | pBSV2::PflaBVsp1Δ22-23 | 18 |
| pSC5002 | pBSV2::PflaBVsp1Δ22-23/33 | This study |
| pOSK291 | pBSV2::PflaBVsp1Δ21-23 | 18 |
| pSC5004 | pBSV2::PflaBVsp1Δ21-23/33 | This study |
MCS, multiple-cloning site.
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′–3′) | Description |
|---|---|---|
| OspA-K18A-fwd | AGCCTTAATAGCATGTGCGCAAAATGTTAGCAGCCTTGAC | K18A forward mutagenic primer |
| OspA-K18A-rev | TGCTAACATTTTGCGCACATGCTATTAAGGCTAATATTAG | K18A reverse mutagenic primer |
| OspA-K27A-fwd | AGCCTTGACGAGGCGAACAGCGTTTCAGTAGATTTGCC | K27A forward mutagenic primer |
| OspA-K27A-rev | GAAACGCTGTTCGCCTCGTCAAGGCTGCTAACATTTTG | K27A reverse mutagenic primer |
| OspA-K1827A-KAAA-fwd | AGCATGTGCGCAAAATGCTGCAGCTAAAGACGAGGCGAACAGCGTTTCAGTAG | K21AAA forward mutagenic primer |
| OspA-K1827A-KAAA-rev | GTTCGCCTCGTCTTTAGCTGCAGCATTTTGCGCACATGCTATTAAGC | K21AAA reverse mutagenic primer |
| 216-K27-fwd | CAGGAACTTCTCCTGATGGGCAAGCAGCTAAATCTGATGG | K27 deletion forward mutagenic primer |
| 216-K27-rev | CCATCAGATTTAGCTGCTTGCCCATCAGGAGAAGTTCCTG | K27 deletion reverse mutagenic primer |
| 221-K33-fwd | GATGGGCAAGCAGCTTCTGATGGCACTGTTATTGAC | K33 deletion forward mutagenic primer |
| 221-K33-rev | GTCAATAACAGTGCCATCAGAAGCTGCTTGCCCATC | K33 deletion reverse mutagenic primer |
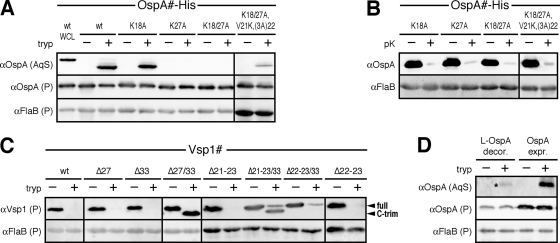
The Western immunoblotting results shown in Fig. 2 A indicate that trypsinolysis indeed released a smaller OspA core protein from the cell. Densitometric analysis of the cell-associated OspA protein bands was performed with the ImageJ software (NIH; http://rsbweb.nih.gov/ij), using the FlaB signal for normalization (19). This indicated that approximately 5% of total OspA protein was released (Fig. 2A). Raising the final trypsin concentration to 800 μg/ml in the assays did not significantly increase the efficiency of OspA cleavage (data not shown). The OspA core protein's water solubility and molecular mass difference of 1 to 2 kDa compared to the full-length OspAwt protein present in a whole-cell lysate (WCL) suggested cleavage close to OspA's N terminus, most likely between Lys27 and Asn28 (Fig. 1B). Indeed, replacement of Lys27 with an Ala residue in the OspAK27A mutant completely blocked trypsin-mediated release from the cells. Conversely, elimination of the alternate trypsin site at Lys18 in the single OspAK18A or double OspAK18/27A mutants did not alter the respective release phenotypes (Fig. 2A). A limited lysine-scanning mutagenesis of residues 21 to 24 showed that an introduced Lys at OspA position 21 [OspAK18/27A,V21K,(3A)22] was also cleaved by trypsin (Fig. 2A), albeit less efficiently (about 1% of total protein). As a control, an in situ proteinase K assay confirmed that all OspA tether mutants were surface exposed (Fig. 2B).
Fig. 2.
Trypsin accessibility of wild-type and mutant OspA and Vsp1 tether peptides. (A) Western immunoblot analysis of recombinant B. burgdorferi cells expressing OspA tether trypsin-site mutants (see Fig. 1B). Cells were either incubated with trypsin (tryp +) or reaction buffer alone (tryp −). Cell-associated proteins were collected in the postreaction cell pellet (P), while proteins released from the cells were collected in the aqueous Triton X-114 fractionation phase of the reaction supernatant (AqS). Compared to the P samples, the AqS samples were overloaded approximately 20-fold to clearly visualize the protein. For protein size comparisons, the first lane was loaded with a WCL of B. burgdorferi expressing OspAwt-His (wt WCL). FlaB was used as a periplasmic, protease-inaccessible loading control. (B) Western immunoblot analysis of proteinase K (pK) accessibility assay products of all OspA tether trypsin site mutants (see Fig. 1B). Cells were either incubated with proteinase K (pK +) or reaction buffer alone. FlaB was used as a periplasmic, protease-inaccessible control. (C) Western immunoblot analysis of trypsin (tryp) accessibility assay products of Vsp1 tether mutants (see Fig. 1C). Arrowheads point to either full-length Vsp1 (full) or C-terminally trimmed Vsp1 (C-trim) (see Fig. 1 and references 33 and 34). Other labeling and controls are as for panel A. (D) Western immunoblots of trypsin accessibility assay products of B. burgdorferi cells decorated with purified recombinant lipidated OspA (L-OspA decor.). OspA-expressing cells (OspA expr.) were used as a control. An asterisk indicates the OspA core protein released from OspA-decorated cells. Other labeling is as for panel A.
In parallel experiments, surface trypsinolysis of B. burgdorferi expressing wild-type Vsp1 (Vsp1wt) confirmed the previously observed complete removal of the protein from the cellular envelope (Fig. 2C) (33, 34). Vsp1 tether mutants individually deleted for the Lys residues at positions 27 (Vsp1Δ27) and 33 (Vsp1Δ33) (Fig. 1C) were efficiently detached as well. In contrast, the Vsp1Δ27/33 double mutant remained associated with the cell envelope (Fig. 2C). As previously demonstrated, the slightly lower molecular mass of the observed cell-associated Vsp1 was due to the removal of the protein's C-terminal tail (Fig. 1C) (33, 34). This indicated that both the Vsp1 Lys27 and Lys33 residues localized to the surface. To determine if Lys residues more proximal to the N-terminal Cys remained accessible, we modified two described recombinant plasmids encoding surface-displayed Vsp1 tether mutants, Vsp1Δ21–23 and Vsp1Δ22–23 (18). Deletion of the Lys33 codon in both recombinant plasmids left only Lys27 as a potential target for trypsin, albeit at different positions relative to the N-terminal Cys at position +1 (Fig. 1). As expected, Vsp1Δ21–23 and Vsp1Δ22–23 were completely removed by proteolysis (Fig. 2C). The cleavage and release of Vsp1Δ22–23/33 were only minimally affected, showing about 15% residual cell-associated full-length OspA after proteolysis. Vsp1Δ21–23/33 remained associated with the cell, with about 70% of the total protein being C-terminally cleaved (Fig. 2C).
One conceivable interpretation of the divergent protease accessibilities of OspA and Vsp1 tether residues is that the two proteins ' prevalent tether topologies are dramatically different, the majority of OspA's tether peptides remaining sequestered from the bacterial surface, potentially in an “in-out” anchor topology (Fig. 1A). To further assess this possibility, we repeated the trypsin accessibility assays with B. burgdorferi B313 cells that were “decorated,” as described previously (10), with purified lipidated wt OspA (L-OspA; a gift from R. Huebner and A. G. Barbour). In accordance with the previously published data (10), densitometry showed that the overall OspA level in OspA-decorated cells was about 4 times lower than in OspA-expressing cells. Trypsin-mediated release of OspA core protein from OspA-decorated cells, however, was equally detectable and efficient as with OspA-expressing cells (Fig. 2D), showing a reduction of about 5% in cell-associated OspA after proteolysis. This indicated that endogenously expressed OspA and exogenously added OspA assumed identical steady-state topologies in the B. burgdorferi outer membrane.
Together, these experiments show that N-terminal-proximal tether residues of OspA and Vsp1, two structurally and functionally different Borrelia surface lipoproteins, are accessible to in situ trypsinolysis, i.e., are displayed on the spirochetal surface. By proxy, this serves as corroborating evidence that borrelial surface lipoprotein anchors are ultimately inserted in the surface leaflet of the borrelial OM. The surface accessibility of OspA and Vsp1 proteolytic sites in the +5 or +7 positions, i.e., only 4 or 6 amino acids beyond the triacylated cysteines, is irreconcilable with the surface lipoproteins being anchored via a periplasmic anchor and an OM-spanning peptide; even in a completely disordered peptide conformation extending 3.8 Å per residue (21), a minimum of 13 amino acids would be required to span the 50-Å-thick OM lipid bilayer (17) (Fig. 1). Based on our current understanding of bacterial protein transport, retrograde absorption of lipidated OspA from the extracellular milieu to the spirochetal periplasm is unlikely. Therefore, the identical accessibility of OspA tether peptides in OspA-decorated and OspA-expressing B. burgdorferi cells all but excludes that this lipoprotein—in contrast to Vsp—assumes an “in-out” anchor topology. One plausible reason for the relative inaccessibility of OspA tether lysines in the context of an intact B. burgdorferi envelope could be the protection of OspA tether residues by the protein's documented interaction with other surface-exposed proteins, such as P66 (9). Yet, the simplest explanation for our findings is steric hindrance. Trypsin's diameter of 38 Å (31) corresponds to the length of a 10-amino-acid peptide in extended conformation. The peptidase's active site therefore may have easier access to lysine residues in the 21-amino-acid Vsp1 tether than in the only-12-amino-acid OspA tether (Fig. 1). OspA Lys18 is trypsin accessible in the purified lipoprotein (7), so steric effects may also explain the inaccessibility of membrane-proximal tether residues. Dimerization of Vsp1 might also hinder trypsin from acting on residues proximal to the membrane anchor. It is of note that Lp6.6, a lipoprotein localizing to the periplasmic leaflet of the B. burgdorferi OM (24), remains inaccessible to trypsin unless the envelope is detergent permeabilized (18).
The current data further define the topology of spirochetal surface lipoproteins, based on an approach that might be applicable to other systems. The precise molecular mechanism of OM crossing by spirochetal surface lipoproteins remains to be elucidated. Recent findings, including one study using lipoprotein fusions to a conditionally folding calmodulin moiety (12), support the transient anchoring of B. burgdorferi surface lipoproteins in the periplasmic leaflet of the OM (28). OM crossing by lipoproteins may therefore involve “flipping” of the lipid moiety within the hydrophobic environment of the lipid bilayer, while hydrophilic peptides are translocated through a protein channel or at least through interaction with a transmembrane protein (27) to be ultimately displayed on the bacterial surface.
Acknowledgments
This work was supported by the National Institutes of Health (grant R01 AI063261 to W.R.Z.).
We thank Eszter Adany for experimental support and Alan Barbour, Bob Huebner, and Melissa Caimano for reagents, as well as Joe Lutkenhaus and Ryan Schulze for comments on the manuscript.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Akins D. R., et al. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbour A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 3. Barbour A. G., Guo B. P. 2010. Pathogenesis of relapsing fever, p. 333–357 In Samuels D. S., Radolf J. D. (ed.), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 4. Barbour A. G., Tessier S. L., Hayes S. F. 1984. Variation in a major surface protein of Lyme disease spirochetes. Infect. Immun. 45:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barbour A. G., Tessier S. L., Todd W. J. 1983. Lyme disease spirochetes and Ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect. Immun. 41:795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bergström S., Zückert W. R. 2010. Structure, function and biogenesis of the Borrelia cell envelope, p. 139–166 In Samuels D. S., Radolf J. D. (ed.), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 7. Bouchon B., Klein M., Bischoff R., Van Dorsselaer A., Roitsch C. 1997. Analysis of the lipidated recombinant outer surface protein A from Borrelia burgdorferi by mass spectrometry. Anal. Biochem. 246:52–61 [DOI] [PubMed] [Google Scholar]
- 8. Brandt M. E., Riley B. S., Radolf J. D., Norgard M. V. 1990. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect. Immun. 58:983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bunikis J., Barbour A. G. 1999. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect. Immun. 67:2874–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bunikis J., Mirian H., Bunikiene E., Barbour A. G. 2001. Non-heritable change of a spirochaete's phenotype by decoration of the cell surface with exogenous lipoproteins. Mol. Microbiol. 40:387–396 [DOI] [PubMed] [Google Scholar]
- 11. Cadavid D., Pennington P. M., Kerentseva T. A., Bergström S., Barbour A. G. 1997. Immunologic and genetic analyses of VmpA of a neurotropic strain of Borrelia turicatae. Infect. Immun. 65:3352–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen S., Zückert W. R. Probing the Borrelia burgdorferi surface lipoprotein secretion pathway using a conditionally folding protein domain. J. Bacteriol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cowles C. E., Li Y., Semmelhack M. F., Cristea I. M., Silhavy T. J. 2011. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79:1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox D. L., Radolf J. D. 2001. Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology 147:1161–1169 [DOI] [PubMed] [Google Scholar]
- 15. Dunn J. J., Lade B. N., Barbour A. G. 1990. Outer surface protein A (OspA) from the Lyme disease spirochete, Borrelia burgdorferi: high level expression and purification of a soluble recombinant form of OspA. Protein Expr. Purif. 1:159–168 [DOI] [PubMed] [Google Scholar]
- 16. Jiang W., Gorevic P. D., Dattwyler R. J., Dunn J. J., Luft B. J. 1994. Purification of Borrelia burgdorferi outer surface protein A (OspA) and analysis of antibody binding domains. Clin. Diagn. Lab. Immunol. 1:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kudryashev M., et al. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol. Microbiol. 71:1415–1434 [DOI] [PubMed] [Google Scholar]
- 18. Kumru O. S., Schulze R. J., Rodnin M. V., Ladokhin A. S., Zückert W. R. 2011. Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J. Bacteriol. 193:2814–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumru O. S., Schulze R. J., Slusser J. G., Zückert W. R. 2010. Development and validation of a FACS-based lipoprotein localization screen in the Lyme disease spirochete Borrelia burgdorferi. BMC Microbiol. 10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H., Dunn J. J., Luft B. J., Lawson C. L. 1997. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc. Natl. Acad. Sci. U. S. A. 94:3584–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller W. G., Goebel C. V. 1968. Dimensions of protein random coils. Biochemistry 7:3925–3935 [DOI] [PubMed] [Google Scholar]
- 22. Nally J. E., Timoney J. F., Stevenson B. 2001. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 69:400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Norris S. J., Coburn J., Leong J. M., Hu L. T., Höök M. 2010. Pathobiology of Lyme disease Borrelia, p. 299–332 In Samuels D. S., Radolf J. D. (ed.), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 24. Promnares K., et al. 2009. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol. Microbiol. 74:112–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sadziene A., Thomas D. D., Barbour A. G. 1995. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect. Immun. 63:1573–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Samuels D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanyal S., Menon A. K. 2009. Flipping lipids: why an' what's the reason for? ACS Chem. Biol. 4:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schulze R. J., Chen S., Kumru O. S., Zückert W. R. 2010. Translocation of Borrelia burgdorferi surface lipoprotein OspA through the outer membrane requires an unfolded conformation and can initiate at the C-terminus. Mol. Microbiol. 76:1266–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schulze R. J., Zückert W. R. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 59:1473–1484 [DOI] [PubMed] [Google Scholar]
- 30. Stewart P. E., Thalken R., Bono J. L., Rosa P. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714–721 [DOI] [PubMed] [Google Scholar]
- 31. Stroud R. M., Kay L. M., Dickerson R. E. 1974. The structure of bovine trypsin: electron density maps of the inhibited enzyme at 5 angström and at 2-7 angström resolution. J. Mol. Biol. 83:185–208 [DOI] [PubMed] [Google Scholar]
- 32. Zückert W. R. 2007. Laboratory maintenance of Borrelia burgdorferi. Curr. Protoc. Microbiol. 12:Unit 12C.1 [DOI] [PubMed] [Google Scholar]
- 33. Zückert W. R., Kerentseva T. A., Lawson C. L., Barbour A. G. 2001. Structural conservation of neurotropism-associated VspA within the variable Borrelia Vsp-OspC lipoprotein family. J. Biol. Chem. 276:457–463 [DOI] [PubMed] [Google Scholar]
- 34. Zückert W. R., Lloyd J. E., Stewart P. E., Rosa P. A., Barbour A. G. 2004. Cross-species surface display of functional spirochetal lipoproteins by recombinant Borrelia burgdorferi. Infect. Immun. 72:1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zückert W. R., Meyer J., Barbour A. G. 1999. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect. Immun. 67:3257–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]