Abstract
Upon starvation, Bacillus subtilis cells switch from growth to sporulation. It is believed that the N-terminal sensor domain of the cytoplasmic histidine kinase KinA is responsible for detection of the sporulation-specific signal(s) that appears to be produced only under starvation conditions. Following the sensing of the signal, KinA triggers autophosphorylation of the catalytic histidine residue in the C-terminal domain to transmit the phosphate moiety, via phosphorelay, to the master regulator for sporulation, Spo0A. However, there is no direct evidence to support the function of the sensor domain, because the specific signal(s) has never been found. To investigate the role of the N-terminal sensor domain, we replaced the endogenous three-PAS repeat in the N-terminal domain of KinA with a two-PAS repeat derived from Escherichia coli and examined the function of the resulting chimeric protein. Despite the introduction of a foreign domain, we found that the resulting chimeric protein, in a concentration-dependent manner, triggered sporulation by activating Spo0A through phosphorelay, irrespective of nutrient availability. Further, by using chemical cross-linking, we showed that the chimeric protein exists predominantly as a tetramer, mediated by the N-terminal domain, as was found for KinA. These results suggest that tetramer formation mediated by the N-terminal domain, regardless of the origin of the protein, is important and sufficient for the kinase activity catalyzed by the C-terminal domain. Taken together with our previous observations, we propose that the primary role of the N-terminal domain of KinA is to form a functional tetramer, but not for sensing an unknown signal.
INTRODUCTION
Bacterial cells are directly exposed to the environment. Hence, they must sense and respond rapidly to changes in their local environment in order to survive. One prevailing strategy to overcome this problem is to utilize the two-component system composed of a sensor histidine kinase (HK) and a response regulator (RR) (35, 41). In the most typical case, the HK is a transmembrane protein with the N-terminal sensor domain often situated in the extracytoplasmic compartment, such as the periplasm, inner or outer membrane, or even extracellular space, and the C-terminal autokinase domain, containing a conserved phospho-accepting histidine residue, normally resides in the cytosol (8, 35, 41). In contrast, the RR comprises a regulatory domain that includes a phospho-accepting aspartate residue at the N terminus, followed by an associated effector domain typically containing a DNA-binding motif, so that it becomes active as a transcription factor upon phosphorylation. Thus, using these two components, various environmental signals can be detected by the HK and transmitted as phosphate groups to the RR for cellular adaptation.
The individual sensor domains of HKs are highly variable and thus lack sequence homology with other HKs (8, 44, 45). Therefore, it is believed that a variety of environmental signals can be detected, with a high degree of sensitivity and specificity, by the unique amino acid sequence motif localized in the sensor domain. In fact, two-component systems have been found in diverse sensory processes, including sporulation, chemotaxis, osmolarity, metabolism of oxygen, nitrogen, and phosphate, induction of transport systems, and light and temperature sensing (1, 5, 24–26, 30, 33, 34, 36, 42, 49).
Among these sensory systems in bacteria, sporulation in Bacillus subtilis is not controlled by a simple two-component system, but an expanded version using four components, termed a phosphorelay (25, 26, 36). The phosphorelay is comprised of the primary sensor histidine kinase, KinA, two intermediate phosphotransferases, Spo0F and Spo0B, and the ultimate response regulator, Spo0A, as the master transcription factor (3). Under nutrient starvation culture conditions, KinA autophosphorylates followed by a transfer of a phosphate moiety to Spo0A in a His-Asp-His-Asp sequence through a four-component phosphorelay. When the active phosphorylated form of Spo0A (Spo0A∼P) reaches a certain concentration, the cells start to divide asymmetrically to give rise to two distinct cell types, a smaller forespore and a larger mother cell (6, 15).
During that process, Spo0A∼P works by directly or indirectly activating or repressing the transcription of hundreds of genes (13, 31). Among them, genes that do not contribute to sporulation directly (e.g., competence, cannibalism, and biofilm) are activated at early times with a low dose of Spo0A∼P, while genes that play a direct role in sporulation, such as spoIIG (an operon for the synthesis of σE, the mother cell-specific sigma factor) and spoIIA (an operon for the synthesis of σF, the forespore-specific sigma factor), are activated at later times with a high dose of Spo0A∼P. Thus, the progressive increase in the activated Spo0A (Spo0A∼P) explains the temporal and spatial expression patterns of the low- and high-threshold Spo0A-regulated genes (15, 18).
Genes for phosphorelay components are also the subject of Spo0A control both directly and indirectly (19, 25, 43). When the phosphorelay starts to be activated in sporulating cells, the low concentration of Spo0A∼P results in repression of transcription of the abrB gene, which encodes a transcription regulator, AbrB. Transcription of the gene (sigH, also known as spo0H) for σH, an alternative σ subunit of RNA polymerase (RNAP), is negatively regulated by AbrB, and thus the decrease in the level of AbrB protein leads to derepression of the transcription of the sigH. The subsequent increase in the concentration of σH RNAP leads to transcription of genes for KinA, Spo0F, and Spo0A. In addition to σH RNAP, Spo0A∼P also acts as the positive regulator for the transcription of spo0F and spo0A. Therefore, Spo0A, per se, is subject to control at the level of its synthesis by a self-reinforcing closed cycle in the phosphorelay network (4, 19). The nature of the initial event of this regulatory circuit is not known.
Although the individual sensor domains of HKs are unique, recent studies have revealed the PAS domain as one of the representative structural motifs found in the sensor domains (21, 45). The PAS domain, which is highly variable at the sequence level but structurally conserved, was initially identified as a common motif among the Drosophila melanogaster clock protein PER, mammalian ARNT (a dimerization partner of the dioxin receptor), and SIM (the product of the single-minded gene) (7, 21, 32). The PAS domain is now known to be involved in the protein-protein interaction and, in some cases, also in the binding of small ligands (44, 45). However, how the PAS domains function to activate the HK in response to ligand binding has been largely unknown (41, 44). In this regard, it is believed that the autophosphorylation activity of KinA is induced when the N-terminal sensor domain, containing three PAS domains, receives a hypothetical signal(s) that originates only under starvation conditions, and thus this “signal-sensing” step is crucial to trigger the sporulation phosphorelay (38). However, KinA does not have a transmembrane domain and localizes in the cytosol, suggesting that the sensor domain recognizes the signal(s), which might be transported from the nutrient-limited environment to the cytosol or synthesized endogenously in response to starvation (25, 26, 36). More importantly, no starvation signal(s) of either an extracellular or intracellular nature acting directly on KinA has been identified. Thus, the molecular mechanisms of the signal sensing by the cytoplasmic KinA and the subsequent signal transduction by the phosphorelay system are not fully understood.
To address these problems, several studies using primarily biochemical approaches have been attempted, but the results are conflicting, perhaps due to the different methodologies employed. Stephenson and Hoch claimed that in an in vitro system, ATP bound to the most-N-terminal PAS-A domain, and it might not serve as a signal but rather as a phosphate source for the phosphorylation at the histidine residue in the C-terminal domain (39). Lee et al. reported that amino acid substitution mutations in the PAS-A domain affected the activity of KinA, suggesting that PAS-A is required for the kinase activity (28).
More recently, our in vivo studies showed that (i) the most N-terminal PAS domain of KinA (PAS-A), which was originally believed to be essential for signal sensing (28, 39), is dispensable (11, 12); (ii) induction of the synthesis of KinA beyond a certain level results in the increase of Spo0A activity above the threshold, thereby allowing sporulation to proceed efficiently, irrespective of nutrient availability (10). These results suggest that the activity of KinA is not regulated in response to the unknown sporulation signal(s), but rather by a threshold level of the kinase, which is primarily important for entry into sporulation. Currently, the control mechanism(s) responsible for the increase in the KinA protein level during sporulation is unclear.
Toward conclusive determination of whether the N-terminal “sensor” domain of KinA indeed acts as the true sensor for an unidentified sporulation-specific signal(s), here we report the replacement of the N-terminal domain of KinA with an unrelated protein segment containing two PAS domains, the N-terminal domain of YdaM derived from Escherichia coli (48). We found that the resulting chimeric protein, YdaMN-KinAC, formed a tetrameric homocomplex, as was observed for KinA (12), and triggered a massive entry into sporulation, irrespective of nutrient availability and in a manner dependent on the concentration of the chimeric kinase, which is consistent with our threshold model (10).
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
All B. subtilis strains (see Table S1 of the supplemental material) were derived from the prototrophic strain PY79 (51). Details of the constructions are available upon request. All plasmid constructions were performed in Escherichia coli DH5α using standard methods. The E. coli BL21(DE3) pET vector system (Novagen) was used for protein overexpression. The plasmids used in this study are listed in Table S2 of the supplemental material. Oligonucleotides used for plasmid construction are listed in Table S3 of the supplemental material.
Media and culture conditions.
To induce the synthesis of KinA, the chimeric protein, or green fluorescent protein (GFP) under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible hyper-spank promoter (Phy-spank) (16) in the engineered B. subtilis cells, IPTG was added to Luria-Bertani (LB) cultures during the exponential growth phase (optical density at 600 nm [OD600], 0.5) as the rich medium conditions. To induce protein synthesis under normal sporulation conditions, the engineered cells were grown in casein hydrolysate (CH) medium at 37°C. At the mid-exponential phase of growth (OD600, 0.5) in CH medium, cells were suspended in Sterlini-Mandelstam (SM) medium (40) supplemented with IPTG.
Sporulation efficiency and β-galactosidase assays.
Sporulation efficiency was determined in 16-h cultures as CFU per milliliter after incubation at 80°C for 10 min compared with the CFU of the pre-heat treatment sample. Assays of β-galactosidase activity were performed as described previously (12).
Immunoblot analysis.
Whole-cell lysates for immunoblot analysis were prepared by sonication. Immunoblot analysis was performed as described previously (12). Polyclonal rabbit antibodies against GFP (17) were used to detect GFP-tagged proteins. σA was detected by a polyclonal rabbit anti-σA antibdoy and served as an internal standard control (14).
Protein cross-linking.
Protein cross-linking with bis-maleimidohexane (BMH; Pierce) was performed as described previously (12).
Protein purification.
All His-tagged proteins were expressed in E. coli BL21(DE3). All proteins except YdaMN-KinAC were soluble, and thus purification steps were carried out at 4°C as described previously (18, 20). Renaturation of YdaMN-KinAC from the inclusion body and the following purification steps were performed as described previously (20).
In vitro phosphorylation.
Phosphorylation reactions were performed as described previously (18).
Microscopy.
Cells expressing GFP were examined by fluorescence microscopy as described previously (12). The microscope system control and image processing were performed using SlideBook image analysis software (Intelligent Imaging Innovations, Inc.).
RESULTS AND DISCUSSION
Induction of the synthesis of the chimeric protein triggers sporulation.
Based on our prior results (10–12), we hypothesized that if the sensor domain of KinA were replaced with a domain that is unrelated to sporulation but involved in tetramer formation, the resulting chimeric protein would be active as a kinase to trigger sporulation in a concentration-dependent manner, irrespective of nutrient availability.
To test this hypothesis, we constructed a chimeric protein in which the three N-terminal PAS domains of KinA were replaced with an amino acid sequence possibly involved in protein-protein interactions but derived from an unrelated bacterial species. By using the help of an E. coli genome database (E. coli wiki) in combination with the SMART (simple modular architecture research tool) online tool (37), we identified YdaM, which carries an N-terminal domain containing two PAS domains. YdaM exhibits diguanylate cyclase (DGC) activity in vitro that produces bis-(3′-5′)-cyclic-diguanosine monophosphate (c-di-GMP) from GTP and plays an antagonistic role in the expression of the biofilm-associated curli fimbriae (48). The catalytic activity resides in the C-terminal GGDEF domain of the protein. Thus, as a first attempt, we constructed a chimeric protein in which the N-terminal domain of KinA was replaced with an N-terminal domain of YdaM, which does not contain the catalytic active site of DGC. Then, we examined the functionality of the YdaMN-KinAC chimeric protein in vivo.
To reduce the possible variation in the level of gene expression among the constructs (e.g., regulation at the transcriptional or translational level) and to analyze the structure-function relationship of the chimeric protein in comparison with KinA, the genes for YdaMN-KinAC and its derivatives were placed under the control of an IPTG-inducible hyper-spank promoter (Phy-spank), followed by a sequence including the optimal ribosome-binding site in B. subtilis, as was reported for KinA (12, 17). Then, each of the resulting genes was introduced into the nonessential amyE locus of the chromosomal DNA as a single copy. To avoid the phosphate transfer from the endogenous KinA and another sporulation kinase, KinB (46), these two genes were knocked out in all the strains used for this assay. Thus, in this genetic background (ΔkinA and ΔkinB), the true activity of the chimeric protein as the sporulation kinase could be examined in comparison with the wild-type (KinA) control.
First, we examined the influence of the concentration of IPTG on the efficiency of sporulation under nutrient-rich conditions in LB medium, under which the starvation signal is presumably absent (10). Cells expressing the YdaMN-KinAC protein (here referred to as the YdaMN-KinAC strain) sporulated with high efficiency (∼10−1 [spores per viable cells]) when the protein synthesis was induced by increasing the concentration of IPTG (Table 1). By inducing with at least 200 μM IPTG, the sporulation efficiency was comparable (in a range of 10−1) to that of the sporulating wild-type cells under normal sporulation conditions in SM medium. We note that the absolute number of spores in the YdaMN-KinAC strain was approximately 10% (∼107/ml) of that in the wild-type strain (which lacked the IPTG-inducible construct), and the number of viable cells of the YdaMN-KinAC strain decreased in an IPTG concentration-dependent manner, although the reason for the reduced viable number in this strain is not clear. The sporulation efficiency of YdaMN-KinAC became similar to that of the wild-type strain, as described above (Table 1). In contrast, without IPTG, the sporulation efficiency was significantly low (∼10−6). Under the same culture conditions, a strain expressing KinA under the same expression system as that of the YdaMN-KinAC strain (here referred to as the KinA strain) was examined as a control (12). The results indicated that, at 10 μM IPTG, sporulation was induced to the same levels as in the sporulating wild-type strain (∼10−1) (wt; that is, lacking the IPTG-inducible construct), while sporulation was not efficiently induced without IPTG (∼10−4) as reported previously (Table 1) (11, 12). The wild-type strain, which lacks the IPTG-inducible construct, showed a significantly low level of sporulation efficiency under nutrient-rich conditions in LB medium (∼10−5) as observed previously (data not shown) (10). Next, we confirmed that inducing the synthesis of the N terminus of YdaM (YdaMN) showed no significant effect on triggering sporulation under the same culture conditions (∼10−6 sporulation efficiency) (data not shown). As we already reported (11, 12), a strain expressing the C-terminal autokinase domain of KinA preferentially accepts phosphate from Spo0F∼P in a reverse phosphotransfer reaction, resulting in an inability to trigger massive entry into sporulation. These results indicate that the individual N- and C-terminal domains (YdaMN and KinAC, respectively) are not functional when expressed alone, but sporulation is triggered efficiently only when the fused chimeric protein is produced.
Table 1.
Induction of synthesis of YdaMN-KinAC triggers sporulation in LB medium
| IPTG concn (μM) | YdaMN-KinACa |
KinAa |
||||
|---|---|---|---|---|---|---|
| CFU/ml |
Efficiencyb | CFU/ml |
Efficiencyb | |||
| Viable cells | Spores | Viable cells | Spores | |||
| 0 | 3.3 × 108 | 2.1 × 103 | 6.4 × 10−6 | 4.5 × 108 | 2.6 × 105 | 5.8 × 10−4 |
| 10 | 4.5 × 108 | 4.5 × 104 | 1.0 × 10−4 | 3.2 × 108 | 1.5 × 108 | 4.7 × 10−1 |
| 50 | 4.5 × 108 | 2.7 × 105 | 6.0 × 10−4 | 1.1 × 108 | 6.7 × 107 | 6.1 × 10−1 |
| 100 | 1.8 × 108 | 3.4 × 106 | 1.9 × 10−2 | 5.0 × 108 | 3.2 × 107 | 6.4 × 10−2 |
| 200 | 1.7 × 108 | 2.9 × 107 | 1.7 × 10−1 | 5.0 × 108 | 3.0 × 107 | 6.0 × 10−2 |
| 500 | 1.1 × 108 | 5.5 × 107 | 5.0 × 10−1 | 4.9 × 108 | 3.1 × 107 | 6.3 × 10−2 |
Strains used were MF4254 (YdaMN-KinAC) and MF4253 (KinA). Both strains harbor the PspoIIA-lacZ reporter and were also used for the experiments shown in Fig. 1.
Sporulation efficiency was determined in 16-h cultures, as CFU/ml after heat treatment by incubation at 80°C for 10 min, compared with CFU/ml in the pre-heat treatment sample.
Second, we repeated the above experiments, but this time under starvation conditions in SM medium. As shown in Table 2, we obtained essentially the same results as those under nutrient-rich conditions in LB medium. These results indicated that when the YdaMN-KinAC chimeric protein is expressed to a certain level, it is functional, if not fully but at least partially, to trigger sporulation, irrespective of starvation signal.
Table 2.
Induction of synthesis of YdaMN-KinAC and effect on sporulation in SM medium
| IPTG (μM) | YdaMN-KinACa |
KinAa |
Wild type |
||||||
|---|---|---|---|---|---|---|---|---|---|
| CFU/ml |
Efficiencyb | CFU/ml |
Efficiencyb | CFU/ml |
Efficiencyb | ||||
| Viable cells | Spores | Viable cells | Spores | Viable cells | Spores | ||||
| 0 | 3.8 × 108 | 2.7 × 108 | 7.1 × 10−1 | ||||||
| 10 | 1.2 × 108 | 5.0 × 101 | 4.2 × 10−7 | 3.0 × 108 | 1.7 × 108 | 5.7 × 10−1 | — | — | — |
| 20 | 1.3 × 108 | 3.1 × 102 | 2.4 × 10−6 | — | — | — | — | — | — |
| 50 | 2.9 × 108 | 6.6 × 104 | 2.3 × 10−4 | — | — | — | — | — | — |
| 100 | 4.2 × 108 | 4.8 × 106 | 1.1 × 10−2 | — | — | — | — | — | — |
| 200 | 3.1 × 108 | 5.3 × 107 | 1.7 × 10−1 | — | — | — | — | — | — |
Strains used were MF4254 (YdaMN-KinAC), MF4253 (KinA), and MF4276 (wt). All strains harbor the PspoIIA-lacZ reporter and were also used for the experiments shown in Fig. 1.
Sporulation efficiency was determined in 16-h cultures, as CFU per ml after heat treatment by incubation at 80°C for 10 min, compared with CFU of the pre-heat treatment sample. —, not determined.
Spo0A, the master regulator for entry into sporulation, is activated by the YdaMN-KinAC chimeric protein through phosphorelay.
We predicted that YdaMN-KinAC would function as a kinase. Due to technical limitations for direct measurement of the histidine autokinase activity in vivo, a reporter system consisting of the β-galactosidase gene (lacZ) fused to the Spo0A-directed promoter is widely used as an indirect measurement of the activity (12, 16). For this, we constructed a reporter system in which lacZ gene expression is driven by the Spo0A-dependent spoIIA promoter (23). We then introduced the reporter system into a strain that carried the IPTG-inducible YdaMN-KinAC to examine β-galactosidase activity in the presence of various concentration of IPTG. As a control, the KinA strain was examined under the same conditions as those for the YdaMN-KinAC strain. Using these systems, we examined the kinetics of Spo0A activation as an indirect assessment of the kinase activity when the synthesis of YdaMN-KinAC was induced with IPTG. The wild-type strain, which lacks the IPTG-inducible construct, served as another control.
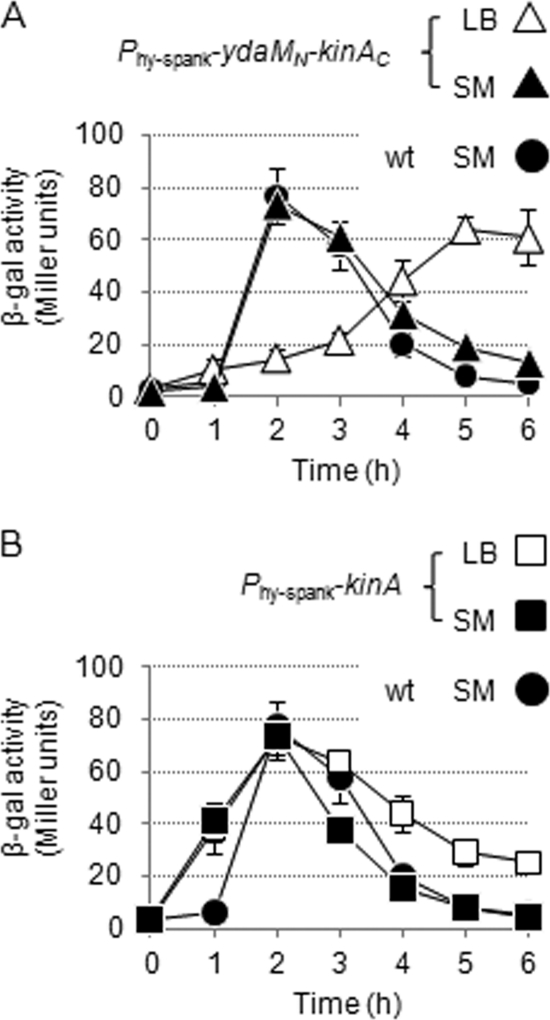
During the course of experiments, we detected the reporter activity in the YdaMN-KinAC strain at relatively low levels at early times (∼2 h), but the activity became significant at later times (∼5 h) after IPTG addition (200 μM) under nutrient-rich conditions in LB medium (Fig. 1A). In contrast, under normal sporulation conditions in SM medium, we found that the peak of the activity was shifted to around 2 h after IPTG addition (200 μM) (Fig. 1A). In the KinA strain, the reporter activities reached maximum levels at around 2 h after IPTG addition (10 μM) under either nutrient-rich conditions in LB medium or normal sporulation conditions in SM medium (Fig. 1B). In the absence of IPTG, little or no reporter activity was detected in either the YdaMN-KinAC or the KinA strains (data not shown). In the wild-type strain (which lacks the IPTG-inducible construct), the reporter activities reached maximum levels at around 2 h in SM medium (Fig. 1A and B), while little or no reporter activity was detected in LB medium (data not shown), as reported previously (10). We note that, in general, many genes involved in sporulation are controlled to express in a just-in-time manner. Thus, as observed in the experiements shown in Fig. 1 and 3 (see below), the reporter activity declined beyond the peak, even when the regulatory proteins were sufficiently present, perhaps due to the transcriptional repression at that point. These results indicate that, in the YdaMN-KinAC strain, activation of Spo0A is mediated by the chimeric protein but is delayed in LB medium compared with SM medium, suggesting that the chimeric protein is negatively regulated, perhaps through the N-terminal YdaMN domain, when the cells are grown under rich medium conditions. Alternatively, it remains possible that the PAS domain of YdaMN may respond to some stimuli, such as redox states (45) generated in SM medium, resulting in an early activation of the reporter gene. Nevertheless, these results are consistent with our prediction that sporulation is induced as a result of Spo0A activation mediated by induction of synthesis of the YdaMN-KinAC protein, irrespective of nutrient availability.
Fig. 1.
Activity of YdaMN-KinAC in cells cultured in LB and SM media. Cells of the IPTG-inducible YdaMN-KinAC (A) and KinA (B) strains harboring the PspoIIA-lacZ reporter construct were cultured in LB (open symbols) and SM (filled symbols) media and collected at the indicated times after the addition of IPTG (200 μM for YdaMN-KinAC [MF4254; triangle] and 10 μM for KinA [MF4253; square]). The wild-type strain (wt; MF4238) cultured in SM medium served as a control, and the identical data sets are displayed as filled circles in both panels A and B. The time courses of YdaMN-KinAC and KinA activities were shown as a measure of β-galactosidase activity of a Spo0A-dependent PspoIIA-lacZ reporter. All experiments were performed at least three times independently, and average values and standard deviations are plotted.
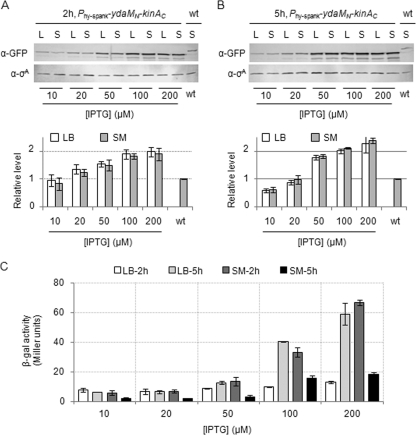
Fig. 3.
Protein levels and activities of YdaMN-KinAC in cells cultured in LB and SM media. Cells of the strain harboring the gene for YdaMN-KinAC-GFP under the control of the Phy-spank promoter (MF4201) were cultured in LB and SM media in the presence of the indicated concentrations of IPTG. Cell extracts were prepared from cells harvested at 2 h (A) and 5 h (B) after the addition of IPTG. Cell extracts were subjected to SDS-PAGE followed by immunoblot analysis with anti-GFP antibodies. The constitutively expressed σA protein was detected with anti-σA antibodies and used as a loading control as reported previously (14). The intensities of each band were quantified with an image analyzer (FluorChem; Alpha Innotech). Each of the protein levels was normalized to σA levels and then to the endogenous KinA-GFP level produced in the wild-type strain (wt; MF3593) cultured for 2 h in SM medium. Relative levels shown in the graph are after the normalization of the intensities. (C) Cells of the YdaMN-KinAC strain harboring the PspoIIA-lacZ reporter construct (MF4254) were cultured in LB and SM media and collected at 2 and 5 h after the addition of the indicated concentrations of IPTG. YdaMN-KinAC activity was measured as Spo0A activity based on the PspoIIA-lacZ reporter as described for Fig. 1. All experiments were performed at least three times independently, and average values and standard deviations are plotted.
To confirm the compartment-specific transcription of the sporulation genes in the individual cells expressing the YdaMN-KinAC protein, we performed fluorescence microscopy experiments. First, the gene for yellow fluorescent protein (YFP) was placed under the control of the Spo0A-dependent spoIIA promoter to express YFP in the sporulating cells before polar septation. Second, the gene for mCherry was fused to the σF-RNAP-controlled spoIIQ promoter to express mCherry in the forespore. Then, each of the two reporter gene constructs was integrated into the chromosome of the same strain as a single copy of each. These two reporter genes were introduced into the KinA strain for the control experiments. As shown in Fig. S1 of the supplemental material, each of the fluorescent reporters was expressed in a compartment-specific manner, but essentially displayed a similar time course pattern to that of the β-galactosidase reporter (Fig. 1A and B); timing of the expression of each sporulation gene in the YdaMN-KinAC strain was delayed in LB medium compared to SM medium.
Next, we investigated whether Spo0A is activated by the YdaMN-KinAC protein via the phosphotransferases Spo0F and Spo0B, as in the case of KinA (3). For this, the deletion mutation of the gene for Spo0F or Spo0B was introduced into the YdaMN-KinAC or KinA strain, each harboring the spoIIA promoter fusion to lacZ as described above. Exponential-phase cells of each strain grown in liquid LB were spotted on solid LB agar with no supplements, 5-bromo-4-chloro-3-indolyl-β-d-galacto-pyranoside (X-Gal), or both X-Gal and IPTG. In the wild-type background, the reporter activity was detected as blue colonies only in the presence of both X-Gal and IPTG, indicating that Spo0A is activated by either YdaMN-KinAC or KinA (see Fig. S2 in the supplemental material). When the deletion mutation of the gene for Spo0F or Spo0B was introduced into the YdaMN-KinAC strain, the capacity of the cells to activate Spo0A in response to the inducer was reduced significantly, similar to changes observed with the spo0A mutant in the corresponding KinA strain (see Fig. S2). These results indicated that YdaMN-KinAC exerts its effect through the phosphorelay, as similarly observed with the KinA strain (16). We note that a relatively small colony was obtained in the KinA strain with the deletion of spo0A, suggesting that cross talk between Spo0B and some unknown downstream factors may mediate this effect (see Fig. S2).
All the above results are consistent with the idea that YdaMN-KinAC functions as a kinase. Therefore, to verify directly whether YdaMN-KinAC possesses autokinase activity and transfers phosphate to Spo0A through phosphorelay, we performed in vitro phosphorylation experiments with the purified protein components (see Fig. S3 in the supplemental material).
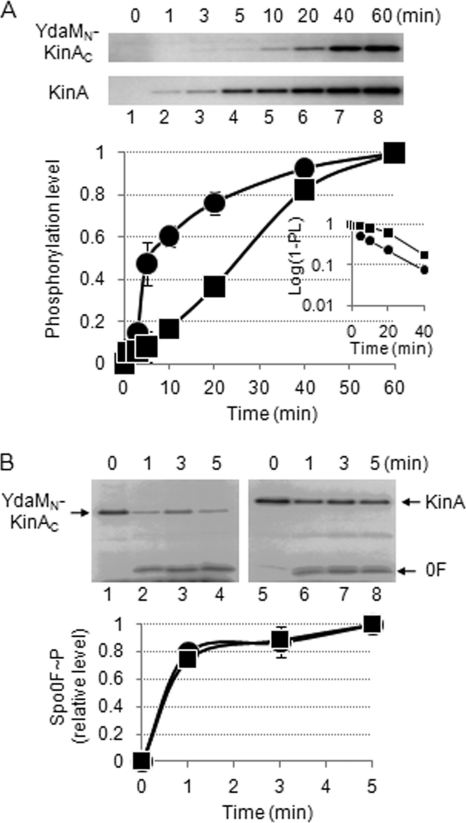
First, we determined the autophosphorylation activity of purified YdaMN-KinAC in the presence of radiolabeled ATP. Purified KinA was used as a control. We found that YdaMN-KinAC exhibited autophosphorylation activity, but the time course was delayed compared with that of KinA (Fig. 2A). Under our standard reaction conditions, the estimated values of kobs for the autophosphorylation of YdaMN-KinAC and KinA were calculated from the slopes shown in the inset of Fig. 2A; the values of (kobs) were approximately 0.046 min−1 and 0.066 min−1, respectively. The source of the different kinetic behaviors of these two proteins and how these differences are related to the process of sporulation are unknown. In the following sections, we examine several possibilities regarding the relationship between enzyme kinetics and cellular response.
Fig. 2.
Autophosphorylation and phosphotransfer activities of YdaMN-KinAC. (A) Autophosphorylation activities of YdaMN-KinAC and KinA were measured in an in vitro reaction. The purified YdaMN-KinAC and KinA were incubated with [γ-32P]ATP as described in Materials and Methods. At the indicated times, aliquots were removed from the reaction mixture, mixed with SDS-PAGE sample buffer, and analyzed by SDS-PAGE (top panels). The fractions of phosphorylation levels plotted on the y axis in the graph were defined as the ratio of each of the labeled proteins at the indicated time point to the maximum level of that present at the end point of the reaction and are expressed as the relative level (RL). YdaMN-KinAC∼P, filled squares; KinA∼P, filled circles. The inset graph indicates the semilogarithmic plot of the value of (1 − RL) as a function of time. The estimated values of kobs (pseudo-first-order rate constant) for the autophosphorylation of YdaMN-KinAC and KinA were calculated from the slopes. (B) The phosphotransfer activities from phosphorylated YdaMN-KinAC or KinA to Spo0F were measured in an in vitro reaction. The purified YdaMN-KinAC or KinA incubated with [γ-32P]ATP for 1 h was incubated with aliquots of Spo0F as described in Materials and Methods. At the indicated times, aliquots were removed from the reaction mixture, mixed with SDS-PAGE sample buffer, and analyzed by SDS-PAGE (top panels). The fraction of phosphorylation level (Spo0F∼P) plotted on the y axis in the graph was defined as the ratio of the labeled Spo0F at the indicated time point to the maximum level of that present at the end point of the reaction. Spo0F∼P with YdaMN-KinAC, filled squares; Spo0F∼P with KinA, filled circles.
Second, by mixing each of the phosphorelay components with YdaMN-KinAC for the in vitro reaction, we found that Spo0F was preferentially phosphorylated by YdaMN-KinAC, indicating that YdaMN-KinAC possesses the same substrate specificity as KinA and phosphorylates Spo0A via phosphorelay (see Fig. S3B and C in the supplemental material). Thus, these in vitro results confirmed the in vivo results shown in Fig. S2.
Third, we determined the rate of phosphotransfer from the phosphorylated YdaMN-KinAC or KinA to Spo0F. As expected from the published results for KinA (22), within 1 min, the labeled phosphoryl group of KinA was efficiently transferred to Spo0F. Similarly, phosphorylated YdaMN-KinAC donated the labeled phosphate to Spo0F efficiently, indicating that there was no significant difference between YdaMN-KinAC and KinA in their phosphotransfer abilities (Fig. 2B). All the above in vivo and in vitro results indicated that YdaMN-KinAC functions as a kinase to activate Spo0A in a phosphorelay-dependent manner. Thus, we refer to the chimeric protein as YdaMN-KinAC chimeric kinase.
As indicated above, our results so far suggest that sensing the redox changes by the PAS domain of YdaMN, or the different kinetic parameters between KinA and YdaMN-KinAC, may explain, in part, the reason why the YdaMN-KinAC strain requires a longer time period (approximately 3 h more) to achieve the threshold of Spo0A activity in LB medium than in SM medium. However, due to the difficulties in investigating these problems, the exact mechanism(s) is still unclear. Alternatively, we hypothesized that (i) the protein level of YdaMN-KinAC, (ii) the local subcellular concentration of YdaMN-KinAC, (iii) the tetramer formation of YdaMN-KinAC, or (iv) some combination or all of the above three factors is preferentially reduced in LB medium, leading to the delay in the activation of phosphorelay. To test these hypotheses, the following experiments were performed.
Similar levels of the chimeric kinase protein are detected in cells cultured in either LB or SM.
First, to determine whether YdaMN-KinAC is produced at different levels in LB and SM media, we performed quantitative immunoblot analyses using a C-terminal GFP-tagged YdaMN-KinAC under the control of Phy-spank. We confirmed that the YdaMN-KinAC-GFP construct showed essentially similar levels of sporulation efficiency as the strain expressing the untagged protein, indicating that YdaMN-KinAC-GFP is functional. As controls, we used strains harboring the gene for KinA-GFP under the control either of its own promoter at the endogenous locus or Phy-spank, as reported previously (16).
Cells harboring the gene for YdaMN-KinAC-GFP under the control of Phy-spank were grown in the presence of various IPTG concentrations in LB or SM medium. Cell extracts from these two strains were prepared at 2 h and 5 h after IPTG addition. Then, we measured the levels of the GFP-fusion protein with immunoblot analysis using anti-GFP antibodies. For the control, cell extracts were prepared from the wild-type strain (harboring KinA-GFP at the endogenous locus) cultured for 2 h under normal sporulation conditions in SM medium and used for the standard sample. A constitutively expressed sigma factor, σA, was used as a loading and normalization control (14). In response to the concentration of added IPTG, the protein levels of YdaMN-KinAC-GFP increased similarly in both LB and SM media (Fig. 3A and B). At the effective concentration of IPTG (200 μM) to trigger sporulation, levels of YdaMN-KinAC-GFP protein in both LB and SM media reached approximately 2-fold above the endogenous KinA-GFP levels in the wild-type sporulating cells (Fig. 3A and B). These protein levels were approximately constant from 2 to 5 h during the culture period, although the protein levels in the presence of low concentrations of IPTG (10 and 20 μM) were slightly decreased at 5 h. For comparison of the activities of the chimeric kinase with the protein levels, the activity data were plotted as a function of IPTG concentration (Fig. 3C). We predicted that if the signal to be sensed by YdaMN was produced only under starvation conditions in SM medium, the YdaMN-KinAC kinase, even at relatively lower protein levels, would be more active in SM medium than in LB medium. However, the results indicated that there was no significant medium-dependent effect on the activity itself (Fig. 3C), essentially as observed for KinA (data not shown) (10), except the delay of the peak activity was observed only in the YdaMN-KinAC strain in LB medium (Fig. 1A). As reported previously (10), we noted that little or no Spo0A activity could be detected in the wild-type strain (which lacks the IPTG-inducible construct) under the LB conditions, even at a later growth stage at which Spo0A activity was detectable in the YdaMN-KinAC strain. These results suggest that the YdaMN-KinAC kinase triggers sporulation efficiently in a concentration-dependent manner, irrespective of nutrient availability. As discussed in the above section, the chimeric protein might be positively or negatively regulated, depending on nutrient availability, and resulted in the delay of peak activity in LB medium compared with SM medium.
The chimeric kinase distributes uniformly throughout the cytoplasm, similar to wild-type KinA.
Second, to investigate whether the subcellular localization of YdaMN-KinAC is related to the shift in the peak activity, possibly by increasing the local concentration of the kinase in SM medium at earlier times than in LB medium, we examined the cellular distribution pattern of the GFP-fusion protein with fluorescence microscopy, under the same culture conditions as above. As shown in Fig. S4 of the supplemental material, we found that YdaMN-KinAC-GFP localized uniformly in the cytosolic compartment at the effective IPTG concentration (200 μM) in both LB and SM media, and no significant differences in localization patterns were detected between these two conditions, as was observed in the KinA-GFP strain (9, 16). These results suggest that the shift in the peak activity was not due to the decreased local concentration of the chimeric kinase in LB medium.
The chimeric kinase forms a homotetramer, similar to wild-type KinA.
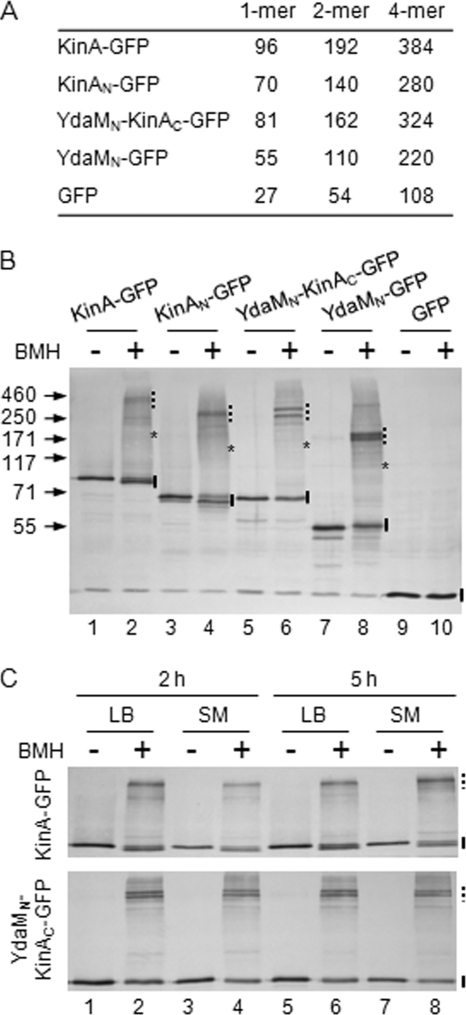
Recently, using biochemical means, it has become evident that KinA forms a homotetramer as a stable and functional kinase (12). In the light of these results, we wonder whether the YdaMN-KinAC chimeric kinase forms a tetramer as a functional unit and the tetramer formation is preferentially reduced in the cells cultured in LB medium, resulting in the delay of the peak activity. To address these issues, we performed chemical cross-linking experiments. For this, crude cell extracts were prepared from cells expressing YdaMN-KinAC-GFP in LB and SM media and incubated in the presence or absence of BMH, a thiol-specific cross-linker (12). The N terminal of YdaM (YdaMN) has three cysteines (C88, C172, and C234) that are potentially cross-linkable by BMH, if they are located near each other in the complex. We note that the C terminus of KinA (KinAC) has no cysteine residues, so that it cannot be used for the cross-linking reaction with BMH and thus, the complex formation, if detected, is absolutely dependent on the N terminus. The calculated molecular masses of each of the protein samples are listed in Fig. 4A. The crude extracts containing YdaMN-KinAC-GFP (YdaMN-KinAC) were prepared from cells grown in LB medium. Then, the samples were treated with or without cross-linker. In the presence of cross-linker, larger bands at around 300 kDa were detected (Fig. 4B, lane 6). In contrast, only a discrete band at around monomer size (estimated size, 80 kDa) was detected in the absence of the cross-linker (Fig. 4B, lane 5). Furthermore, YdaMN-GFP migrated at around 200 kDa in the presence of the cross-linker (Fig. 4B, lane 8), while only the monomer (55 kDa) was detected in the absence of the cross-linker (Fig. 4B, lane 7). These shifted bands at high molecular mass positions corresponded well with the tetramer sizes of each of the tested samples (Fig. 4A). As positive controls, we used both KinA-GFP and KinAN-GFP to verify that the results were reproducible as reported previously (12), showing that KinA predominantly forms a tetramer (Fig. 4B, lanes 1 to 4). As a negative control, both in the presence and absence of the cross-linker, only a discrete single band corresponding to the monomer size of GFP was detected in the crude extracts from cells expressing GFP alone, suggesting that two cysteine residues in GFP are not involved in complex formation under our tested conditions (Fig. 4B, lanes 9 and 10) (50). Our previous results indicate that neither Spo0F nor Spo0A is detected in the immunoprecipitated fraction, suggesting that KinA does not form a stable complex with other phosphorelay components (12). Therefore, some minor bands detected in the cross-linked samples might not be the complex with the other phosphorelay component. Rather, they might be due to the formation of nonspecific protein complexes, such as inter- or intramolecular cross-linking formations in different ways, depending on the sulfhydryl involved.
Fig. 4.
Cross-link analysis of YdaMN-KinAC complex formation in cells cultured in LB and SM media. (A) Molecular masses and possible multimeric combinations of each sample. (B) Cells harboring the IPTG-inducible GFP-tagged protein construct were cultured in LB in the presence of IPTG (10 μM for KinA-GFP and GFP, 200 μM for YdaMN-KinAC-GFP). Cell extracts were prepared from each strain 2 h after IPTG addition and processed for the BMH cross-linking reaction as described in Materials and Methods (BMH+, even lanes). Samples processed in the absence of BMH were used as controls (BMH-, odd lanes). Strains expressing KinA-GFP (MF3352; lanes 1 and 2), KinAN-GFP (MF3360; lanes 3 and 4), YdaMN-KinAC-GFP (MF4201; lanes 5 and 6), YdaMN-GFP (MF4080; lanes 7 and 8), and GFP alone (MF2732; lanes 9 and 10) were examined for cross-linking experiments. Samples were separated by SDS-PAGE, followed by immunoblot analysis with anti-GFP antibodies. Solid bars, asterisks, and dashed bars depict the expected sizes of monomer, dimer, and tetramer forms, respectively. (C) Cells expressing KinA-GFP (top panel) or YdaMN-KinAC-GFP (bottom panel) were cultured in LB (lanes 1, 2, 5, and 6) and SM (lanes 3, 4, 7, and 8) and collected at 2 h (lanes 1 to 4) or 5 h (lanes 5 and 6) after IPTG addition (10 μM for KinA-GFP, 200 μM for YdaMN-KinAC-GFP). Cell extracts were processed as described for panel B. Even and odd lanes contain samples with and without BMH, respectively. Solid and dashed bars depict the expected sizes of monomer and tetramer forms, respectively. Molecular masses (in kDa) of the protein size marker (Invitrogen) are indicated to the left.
Next, we examined whether the functional tetramer of YdaMN-KinAC-GFP is preferentially formed in SM medium (compared to LB medium), which may explain the shift in the peak activity shown in Fig. 1A. Cells expressing YdaMN-KinAC-GFP were grown in LB and SM media for 2 h and 5 h in the presence of IPTG (200 μM). Then, crude cell extracts were prepared and incubated in the presence or absence of cross-linker. As shown in Fig. 4C, in the presence of cross-linker, the tetramer form of YdaMN-KinAC-GFP was detected in each of the tested conditions and there were no significant differences of the complex formations in all tested samples prepared from cells grown in LB and SM media. These results suggest that the shift in the peak activity is not due to the destabilization of the complex formation in LB medium. In conclusion, although various attempts were made, the mechanism by which the timing of the chimeric kinase activity is regulated either positively or negatively, depending on the medium used, remains unknown.
Autophosphorylation activity of the chimeric kinase is independent of enzyme concentration, similar to wild-type KinA.
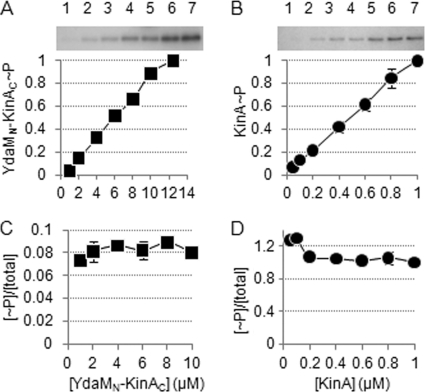
We determined whether the autophosphorylation of the chimeric kinase occurs in a protein concentration-dependent manner. Generally, an intramolecular reaction would exhibit saturation kinetics, so that the activity of the autophosphorylation reaction would not be enzyme concentration dependent (27). In other words, if the activity of the tetrameric kinase is dependent on the protein concentration, the functional subunit assembly might be dependent on the concentration of the individual protomers. In order to test this, the autophosphorylation activity was monitored as a function of enzyme concentration over a 10-fold range of concentrations (Fig. 5). A plot of the activity at various concentrations showed a constant level, indicating that the rate of YdaMN-KinAC autophosphorylation is independent of enzyme concentration (Fig. 5AC). As a control, we examined the purified KinA, as described above, and obtained essentially reproducible results as reported in the literature (Fig. 5B and D) (22). In our previous study with a dominant negative assay, we showed that autophosphorylation of KinA is in a transphosphorylation manner (12). Taking together all the data presented above and considering the similar properties of YdaMN-KinAC and KinA, we suggest that the autophosphorylation activity of each kinase is likely to result from an intramolecular transautophosphorylation in a stable tetramer as a functional unit.
Fig. 5.
YdaMN-KinAC is autophosphorylated in a protein concentration-independent manner in vitro. (A) The indicated concentrations (in μM) of purified YdaMN-KinAC were incubated with [γ-32P]ATP as described in Materials and Methods. Reaction mixtures were analyzed by SDS-PAGE and autoradiography (top panel). Phosphorylation levels of YdaMN-KinAC (YdaMN-KinAC∼P) plotted on the y axis were defined as the ratio of [32P]YdaMN-KinAC present at the indicated concentration of YdaMN-KinAC versus the level of [32P]YdaMN-KinAC at the highest concentration of YdaMN-KinAC (lane 7). (B) The indicated concentrations of purified KinA were examined as described for panel A. (C) Phosphorylation levels of YdaMN-KinAC shown in panel A at the indicated concentrations of YdaMN-KinAC were divided by the same range of concentrations. Thus, the ratio of phosphorylated YdaMN-KinAC over total YdaMN-KinAC is expressed as [∼P]/[total] on the y axis, and the value is plotted as a function of the protein concentration on the x axis. (D) The data for KinA autophosphorylation were processed as described for panel C.
Concluding remarks and perspectives.
Evidence is presented in this study that the replacement of the sensor domain of KinA with a foreign domain of YdaM from E. coli results in a massive induction of sporulation initiation by a concentration-dependent mechanism through phosphorelay. The resulting chimeric kinase forms a homotetramer, as was reported for KinA (10–12). Although the chimeric kinase shows delayed activation of Spo0A under nutrient-rich conditions compared with starvation conditions, with unknown mechanisms, the peak activity levels of Spo0A (as an indirect measure of the kinase activity) and the resulting sporulation efficiencies are essentially the same under these two conditions. These results and our published studies (10–12) suggest that, regardless of the origin of the protein, tetramer formation mediated by the N-terminal domain is sufficient for the kinase activity catalyzed by the C-terminal domain, irrespective of nutrient availability. The prior studies of the well-known sensor kinases, such as the EnvZ osmosensor and the nitrogen sensor NRII kinase/phosphatase, indicate that, even under conditions of overexpression from multicopy plasmids, the signal (ligand) is still required for the proper response of the kinases (2, 47). More recently, overexpression of KinC, an alternative histidine kinase involved in biofilm formation, does not induce massive pellicle formation (biofilm growth) (29). These prior results suggest that the overexpression of the kinase is not capable of overriding the sensor control in vivo, if a ligand is required for the activation process. Thus, our results suggest that massive entry into sporulation by inducing the synthesis of the chimeric kinase is not simply due to the overexpression of the chimeric kinase. Rather, our data support a model in which the threshold level of the kinase, which is constitutively active and signal independent, acts as a molecular switch regulating the entry into sporulation. The mechanism(s) that controls the cellular level of the kinase remains to be determined.
Supplementary Material
ACKNOWLEDGMENTS
We thank Oleg Igoshin, Jatin Narula, Ryutaro Utsumi, Yoko Eguchi, Daniel Kearns, Kumaran Ramamurthi, and all members of the Fujita laboratory for valuable discussions. We also thank James Varughese, Jeannette Dukes, Michael Evangelopoulos, and Linh Tran for technical assistance.
This work was supported by the Norman Hackerman Advanced Research Program (003652-0072-2007), the Welch Foundation (E-1627), and the National Science Foundation (MCB-0920463).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 16 September 20111.
REFERENCES
- 1. Aguilar P. S., Hernandez-Arriaga A. M., Cybulski L. E., Erazo A. C., de Mendoza D. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atkinson M. R., Ninfa A. J. 1993. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRII or NtrB). J. Bacteriol. 175: 7016–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burbulys D., Trach K. A., Hoch J. A. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64: 545–552 [DOI] [PubMed] [Google Scholar]
- 4. Chibazakura T., Kawamura F., Takahashi H. 1991. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J. Bacteriol. 173: 2625–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christie J. M., Salomon M., Nozue K., Wada M., Briggs W. R. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. U. S. A. 96: 8779–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chung J. D., Stephanopoulos G., Ireton K., Grossman A. D. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 176: 1977–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crews S. T. 1998. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 12: 607–620 [DOI] [PubMed] [Google Scholar]
- 8. Dutta R., Qin L., Inouye M. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34: 633–640 [DOI] [PubMed] [Google Scholar]
- 9. Eswaramoorthy P., Dinh J., Duan D., Igoshin O. A., Fujita M. 2010. Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells. Microbiology 156: 2294–2304 [DOI] [PubMed] [Google Scholar]
- 10. Eswaramoorthy P., et al. 2010. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J. Bacteriol. 192: 3870–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eswaramoorthy P., Fujita M. 2010. Systematic domain deletion analysis of the major sporulation kinase in Bacillus subtilis. J. Bacteriol. 192: 1744–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eswaramoorthy P., Guo T., Fujita M. 2009. In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis. J. Bacteriol. 191: 5358–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fawcett P., Eichenberger P., Losick R., Youngman P. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97: 8063–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujita M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5: 79–88 [DOI] [PubMed] [Google Scholar]
- 15. Fujita M., Gonzalez-Pastor J. E., Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187: 1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujita M., Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19: 2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujita M., Losick R. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 43: 27–38 [DOI] [PubMed] [Google Scholar]
- 18. Fujita M., Losick R. 2003. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 17: 1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujita M., Sadaie Y. 1998. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J. Biochem. 124: 98–104 [DOI] [PubMed] [Google Scholar]
- 20. Fujita M., Sadaie Y. 1998. Promoter selectivity of the Bacillus subtilis RNA polymerase σA and σH holoenzymes. J. Biochem. 124: 89–97 [DOI] [PubMed] [Google Scholar]
- 21. Galperin M. Y., Nikolskaya A. N., Koonin E. V. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203: 11–21 [DOI] [PubMed] [Google Scholar]
- 22. Grimshaw C. E., et al. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 37: 1365–1375 [DOI] [PubMed] [Google Scholar]
- 23. Hatt J. K., Youngman P. 1998. Spo0A mutants of Bacillus subtilis with sigma factor-specific defects in transcription activation. J. Bacteriol. 180: 3584–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heermann R., Altendorf K., Jung K. 1998. The turgor sensor KdpD of Escherichia coli is a homodimer. Biochim. Biophys. Acta 1415: 114–124 [DOI] [PubMed] [Google Scholar]
- 25. Hoch J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47: 441–465 [DOI] [PubMed] [Google Scholar]
- 26. Hoch J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3: 165–170 [DOI] [PubMed] [Google Scholar]
- 27. Kuret J., Schulman H. 1985. Mechanism of autophosphorylation of the multifunctional Ca2+/calmodulin-dependent protein kinase. J. Biol. Chem. 260: 6427–6433 [PubMed] [Google Scholar]
- 28. Lee J., et al. 2008. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry 47: 4051–4064 [DOI] [PubMed] [Google Scholar]
- 29. Lopez D., Kolter R. 2010. Functional microdomains in bacterial membranes. Genes Dev. 24: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malpica R., Franco B., Rodriguez C., Kwon O., Georgellis D. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101: 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Molle V., et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50: 1683–1701 [DOI] [PubMed] [Google Scholar]
- 32. Nambu J. R., Lewis J. O., Wharton K. A., Jr., Crews S. T. 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67: 1157–1167 [DOI] [PubMed] [Google Scholar]
- 33. Neiditch M. B., et al. 2006. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ninfa E. G., Atkinson M. R., Kamberov E. S., Ninfa A. J. 1993. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J. Bacteriol. 175: 7024–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parkinson J. S. 1993. Signal transduction schemes of bacteria. Cell 73: 857–871 [DOI] [PubMed] [Google Scholar]
- 36. Perego M., Hoch J. A. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases. American Society for Microbiology, Washington, DC [Google Scholar]
- 37. Schultz J., Milpetz F., Bork P., Ponting C. P. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stephenson K., Hoch J. A. 2002. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 46: 297–304 [DOI] [PubMed] [Google Scholar]
- 39. Stephenson K., Hoch J. A. 2001. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98: 15251–15256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sterlini J. M., Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69: 183–215 [DOI] [PubMed] [Google Scholar]
- 42. Stock J. B., Levit M. N., Wolanin P. M. 2002. Information processing in bacterial chemotaxis. Sci. STKE 2002: pe25. [DOI] [PubMed] [Google Scholar]
- 43. Strauch M. A., Wu J. J., Jonas R. H., Hoch J. A. 1993. A positive feedback loop controls transcription of the spo0F gene, a component of the sporulation phosphorelay in Bacillus subtilis. Mol. Microbiol. 7: 967–974 [DOI] [PubMed] [Google Scholar]
- 44. Szurmant H., White R. A., Hoch J. A. 2007. Sensor complexes regulating two-component signal transduction. Curr. Opin. Struct. Biol. 17: 706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor B. L., Zhulin I. B. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trach K. A., Hoch J. A. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol. Microbiol. 8: 69–79 [DOI] [PubMed] [Google Scholar]
- 47. Waukau J., Forst S. 1999. Identification of a conserved N-terminal sequence involved in transmembrane signal transduction in EnvZ. J. Bacteriol. 181: 5534–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weber H., Pesavento C., Possling A., Tischendorf G., Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62: 1014–1034 [DOI] [PubMed] [Google Scholar]
- 49. Wolanin P. M., Webre D. J., Stock J. B. 2003. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry 42: 14075–14082 [DOI] [PubMed] [Google Scholar]
- 50. Yang F., Moss L. G., Phillips G. N., Jr 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14: 1246–1251 [DOI] [PubMed] [Google Scholar]
- 51. Youngman P. J., Perkins J. B., Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. U. S. A. 80: 2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.