Abstract
Campylobacter jejuni is a leading cause of diarrheal disease in humans and an intestinal commensal in poultry and other agriculturally important animals. These zoonotic infections result in significant amounts of C. jejuni present in the food supply to contribute to disease in humans. We previously found that a transposon insertion in Cjj81176_1038, encoding a homolog of the Escherichia coli LivJ periplasmic binding protein of the leucine, isoleucine, and valine (LIV) branched-chain amino acid transport system, reduced the commensal colonization capacity of C. jejuni 81-176 in chicks. Cjj81176_1038 is the first gene of a six-gene locus that encodes homologous components of the E. coli LIV system. By analyzing mutants with in-frame deletions of individual genes or pairs of genes, we found that this system constitutes a LIV transport system in C. jejuni responsible for a high level of leucine acquisition and, to a lesser extent, isoleucine and valine acquisition. Despite each LIV protein being required for branched-chain amino acid transport, only the LivJ and LivK periplasmic binding proteins were required for wild-type levels of commensal colonization of chicks. All LIV permease and ATPase components were dispensable for in vivo growth. These results suggest that the biological functions of LivJ and LivK for colonization are more complex than previously hypothesized and extend beyond a role for binding and acquiring branched-chain amino acids during commensalism. In contrast to other studies indicating a requirement and utilization of other specific amino acids for colonization, acquisition of branched-chain amino acids does not appear to be a determinant for C. jejuni during commensalism.
INTRODUCTION
Campylobacter jejuni is a leading cause of bacterial diarrheal disease in the United States and a major cause of enteritis worldwide (7, 33). The severity of diarrheal disease caused by C. jejuni can range from a mild, watery diarrhea to a profuse bloody, inflammatory enteritis. A postinfectious sequela occurring in approximately 1 in 1,000 cases of C. jejuni disease is Guillain-Barré syndrome, which manifests as a transient paralysis of the peripheral nervous system. In contrast to infection of humans, C. jejuni is a commensal organism in the intestinal tracts of many animals and birds. As such, a majority of sporadic cases of C. jejuni diarrhea are due to consumption or handling of contaminated meats, especially those of poultry (9).
Due to the ability of C. jejuni to promote commensalism upon infection of poultry, important insights into colonization factors of C. jejuni required for in vivo growth and persistence have been gained by analyzing a natural chick model of infection. Upon infection of 1-day-old chicks, C. jejuni primarily colonizes the lower gastrointestinal tract, including the ceca and large intestines, and can persist in this niche for several weeks to months (4, 14, 29, 37). We previously exploited this model to identify genes of C. jejuni required for optimal levels of commensal colonization of the chick ceca by using a negative-selection procedure involving signature-tagged transposon (Tn) mutants of C. jejuni strain 81-176 (14). This study identified 29 mutants with reduced colonization capacities and provided a foundation for more in-depth studies into potential colonization factors of C. jejuni. From this study and subsequent studies, a better understanding of C. jejuni cytochrome c peroxidases and glycosylated proteins that are involved in commensal colonization has been acquired (5, 20, 21).
One C. jejuni mutant identified in this selection procedure contained a transposon insertion in Cj1019c (as designated in the C. jejuni NCTC11168 genome sequence [34] and Cjj81176_1038 in the C. jejuni 81-176 genome sequence [8]). Preliminary analysis showed that this Tn mutant demonstrated a 100- to 106-fold reduction in commensal colonization of the ceca of 1-day-old chicks relative to wild-type C. jejuni 81-176. Bioinformatic analysis indicated that Cjj81176_1038 is homologous to Escherichia coli livJ. In E. coli, LivJ is part of the LIV system that is responsible for the transport of branched-chain amino acids, such as leucine, isoleucine, and valine (LIV), into the bacterial cell (1, 38). Thus, identification of this mutant suggests that acquisition of these types of amino acids in the avian host may be necessary for C. jejuni to promote in vivo growth.
Six genes encode components of the E. coli LIV system, and their expression is largely regulated by the leucine-responsive protein (Lrp), which represses transcription of these genes in the presence of leucine (1, 3, 12). The liv genes are organized as two transcribed units, with livKHMGF separated from livJ by the gene yhhK, which encodes a hypothetical protein with an acetyltransferase domain (1, 23). The liv genes can be grouped into genes that encode proteins with similar functions, including livJ and livK, encoding periplasmic binding proteins; livH and livM, encoding the inner membrane permeases; and livG and livF, encoding cytoplasmic ATPases.
LivJ has been shown to bind leucine, isoleucine, and valine for subsequent transport into E. coli (2, 36, 38). LivK demonstrates a more limited specificity in E. coli and primarily facilitates the binding of leucine for transport (2, 10, 42). To mediate transport of these branched-chain amino acids into the cytoplasm, a single LIV permease and LIV ATPase are not sufficient. Both the LivH and LivM permeases and the LivG and LivF ATPases are required (1). Thus, the individual permeases and ATPases do not appear to have redundant functions in this system. As historically discovered in E. coli, LivJ- and LivK-facilitated transport of amino acids were classified into two high-affinity systems for branched-chain amino acid transport: the LIV-I system, which is dependent on LivJ-mediated binding to leucine, isoleucine, and valine, and the leucine-specific (LS) system, which is dependent on LivK-mediated binding to leucine (1, 10, 24, 38). The LIV-I system has also been shown to facilitate to a lesser extent the transport of serine, threonine, and alanine, which broadens the number of amino acids that can be acquired via LivJ and the other components of the LIV system (10, 38).
In C. jejuni 81-176, livJ (Cjj81176_1038) is the first gene of a predicted operon with five genes (Cjj81176_1037 through Cjj81176_1033) immediately downstream that are proposed to encode proteins homologous to the E. coli LIV transport components. The presence of these genes and the identification of a reduced colonization capacity of a livJ::Tn mutant suggest that C. jejuni may produce a branched-chain amino acid transport system similar to the E. coli LIV system that is required for acquisition of specific amino acids for in vivo growth and commensalism in poultry. Therefore, we constructed multiple mutants in C. jejuni 81-176 that lack each component of the putative LIV system. We also created double mutants (i.e., ΔlivJK, ΔlivHM, and ΔlivGF mutants) to eliminate both putative binding proteins, permeases, and ATPases of this system. These mutants were then tested for biochemical functionality in amino acid acquisition assays and commensal colonization of chicks to ascertain biological roles for in vivo growth. Our results uncovered surprisingly complex roles of specific LIV proteins for in vivo growth and commensalism. Whereas all LIV proteins were required for transport of specific amino acids in vitro, only the LivJ and LivK binding proteins were necessary for wild-type levels of commensal colonization of chicks. These results suggest that acquisition of branched-chain amino acids is not a limiting factor for the ability of C. jejuni to promote infection and commensalism of poultry. Second, LivJ and LivK likely possess additional roles outside branched-chain amino acid transport, which possibly include binding to and acquiring other factors present in the microenvironments during colonization of chicks to promote commensalism.
MATERIALS AND METHODS
Bacterial strains and general growth.
C. jejuni strain 81-176 is a clinical isolate capable of causing disease in human volunteers and promoting commensal colonization of the intestinal tract of chickens (6, 14, 22). DRH212 is a streptomycin-resistant (Smr) derivative of 81-176 with an rpsLSm allele (13). For many of the experiments in this work, C. jejuni strains were initially grown from frozen stocks on Mueller-Hinton (MH) agar containing trimethoprim (TMP) under microaerobic conditions (10% CO2, 5% O2, and 85% N2) at 37°C for 48 h followed by reinoculation onto MH agar containing TMP and growth for 16 h. Campylobacter defined medium (CDM+), which was made by the method of Leach et al. (25), contains defined concentrations of all 20 common amino acids. CDM-LIV was made similarly but lacked the amino acids leucine, isoleucine, and valine. CDM-L was made by omitting leucine from CDM+. When necessary, agar was added to CDM+, CDM-LIV, and CDM-L at a final concentration of 1.7%. Antibiotics for C. jejuni strains were used at the concentrations indicated: TMP, 10 μg ml−1; kanamycin, 50 μg ml−1; streptomycin, 0.1, 0.5, 1, 2, or 5 mg ml−1; and cefoperazone, 30 μg ml−1. All C. jejuni strains were stored in 85% MH broth, 15% glycerol at −80°C. E. coli DH5α was grown in Luria-Bertani (LB) agar or broth with antibiotics when needed at the concentrations indicated: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1. All E. coli strains were stored in 80% LB broth, 20% glycerol at −80°C.
In vitro growth assays.
After 16 h of growth on MH agar containing TMP, strains were resuspended to an optical density at 600 nm (OD600) of 1.0. Three milliliters of this culture was used to inoculate 50 ml of MH broth or CDM containing TMP. The cultures were incubated at 37°C under microaerobic conditions without shaking, and OD600 measurements were taken at 24 h postinoculation. Assays from three experiments were averaged to report the final OD600 readings, and standard errors were calculated.
Construction of C. jejuni 81-176 mutants.
All strains and plasmids used in this study are listed in Tables 1 and 2. Insertional inactivation of liv genes and ilvE was accomplished by first amplifying DNA fragments by PCR from C. jejuni 81-176 chromosomal DNA containing livJK, livHM, livGF, and ilvE with approximately 500 nucleotides of flanking DNA sequence with primers containing 5′ BamHI sites. After cloning into BamHI-digested pUC19 to create pDRH570 (containing livJK), pDRH569 (containing livHM), pDRH568 (containing livGF), and pSMS167 (containing ilvE), PCR-mediated mutagenesis was used to create an MscI restriction site within livJ, livG, and livF in plasmid pDRH570 or pDRH568 (Table 2) (30). Naturally occurring SpeI, EcoRV, and StyI sites were used to interrupt livK, livH, livM, and ilvE. A SmaI-digested kan-rpsL cassette from pDRH437 was cloned into the appropriate restriction sites, resulting in plasmids that were electroporated into C. jejuni 81-176 Smr (DRH212) to insertionally inactivate each liv gene or ilvE (Table 1) (13, 15). Transformants were recovered on MH agar containing 50 μg ml−1 kanamycin.
Table 1.
Bacterial strains used in this study
| Strain | Genotype/characteristic | Reference or source |
|---|---|---|
| C. jejuni 81-176 | Clinical isolate, wild-type C. jejuni | 6 |
| DRH212 | Smr derivative of C. jejuni 81-176 | 13 |
| SMS163 | DRH212 livJ::kan-rpsL | This study |
| SMS103 | DRH212 livK::kan-rpsL | This study |
| SMS101 | DRH212 livM::kan-rpsL | This study |
| SMS129 | DRH212 livH::kan-rpsL | This study |
| SMS227 | DRH212 livF::kan-rpsL | This study |
| SMS154 | DRH212 livG::kan-rpsL | This study |
| SMS223 | DRH212 ilvE::kan-rpsL | This study |
| SMS301 | DRH212 ΔlivJ | This study |
| SMS241 | DRH212 ΔlivK | This study |
| SMS130 | DRH212 ΔlivM | This study |
| SMS262 | DRH212 ΔlivH | This study |
| SMS257 | DRH212 ΔlivF | This study |
| SMS158 | DRH212 ΔlivG | This study |
| SMS270 | DRH212 ΔilvE | This study |
| SMS263 | DRH212 ΔlivJK | This study |
| SMS540 | DRH212 ΔlivHM | This study |
| SMS539 | DRH212 ΔlivGF | This study |
| E. coli DH5α | Strain used for routine cloning | Invitrogen |
Table 2.
Relevant plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pDRH437 | Source of kan-rpsL cassette | 15 |
| pDRH568 | pUC19 containing livGF locus on 2.4-kb DNA fragment amplified from C. jejuni 81-176 chromosomal DNA | This study |
| pDRH569 | pUC19 containing livHM locus on 3-kb DNA fragment amplified from C. jejuni 81-176 chromosomal DNA | This study |
| pDRH570 | pUC19 containing livJK locus on 3.4-kb DNA fragment amplified from C. jejuni 81-176 chromosomal DNA | This study |
| pDRH611 | kan-rpsL cassette inserted into EcoRV site of livM in pDRH569 | This study |
| pDRH616 | kan-rpsL cassette inserted into SpeI site of livK in pDRH570 | This study |
| pSMS119 | kan-rpsL cassette inserted into StyI site of livH in pDRH569 | This study |
| pSMS121 | pDRH570 containing livK with in-frame deletion from start codon to codon 368 | This study |
| pSMS123 | pDRH569 containing livM with in-frame deletion from codon 2 to codon 358 | This study |
| pSMS125 | pDRH569 containing livH with in-frame deletion from start codon to codon 291 | This study |
| pSMS127 | pDRH568 with an MscI site in livG created by site-directed mutagenesis | This study |
| pSMS131 | kan-rpsL cassette inserted into StyI site of ilvE in pSMS167 | This study |
| pSMS136 | kan-rpsL cassette inserted into MscI site of livG in pSMS127 | This study |
| pSMS167 | pUC19 containing ilvE locus on 2.4-kb DNA fragment amplified from C. jejuni 81-176 chromosomal DNA | This study |
| pSMS142 | pSMS167 containing ilvE with in-frame deletion from start codon to codon 302 | This study |
| pSMS146 | kan-rpsL cassette inserted into MscI site of livJ in pSMS149 | This study |
| pSMS147 | pDRH570 containing livJ with in-frame deletion including start and stop codons | This study |
| pSMS149 | pDRH570 with an MscI site in livJ created by site-directed mutagenesis | This study |
| pSMS151 | pDRH568 containing livF with in-frame deletion from codon 39 to stop codon | This study |
| pSMS153 | pDRH568 containing livG with in-frame deletion from codon to codon 245 | This study |
| pSMS171 | pDRH568 with an MscI site in livF created by site-directed mutagenesis | This study |
| pSMS206 | kan-rpsL cassette inserted into MscI site of livF in pSMS171 | This study |
| pSMS214 | pDRH570 with in-frame fusion of livJ and livK genes by linking the start codon of livJ to codon 367 of livK | This study |
| pSMS514 | pDRH569 with in-frame fusion of livM and livH genes by linking the start codon of livH to codon 349 of livM | This study |
| pSMS517 | pDRH568 with in-frame fusion of livF and livG genes by linking codon 2 of livG to codon 229 of livF | This study |
PCR-mediated mutagenesis was used to create an in-frame deletion of each gene including livJ (pSMS147), livK (pSMS121), livH (pSMS125), livM (pSMS123), livG (pSMS153), livF (pSMS151), and ilvE (pSMS142) (Table 2) (30). These plasmids were electroporated into the respective kan::rpsL insertionally inactivated mutants to replace the interrupted gene with the in-frame deletion (13, 15). Transformants were recovered on MH agar containing 0.5, 1, 2, and 5 mg ml−1 streptomycin. In-frame deletion mutants were confirmed by colony PCR, resulting in the recovery of C. jejuni 81-176 Smr ΔlivJ (SMS301), ΔlivK (SMS241), ΔlivH (SMS262), ΔlivM (SMS130), ΔlivG (SMS158), ΔlivF (SMS257), and ΔilvE (SMS270) (Table 1) mutants. To make paired double liv mutants, PCR-mediated mutagenesis was used to construct in-frame deletions of adjacent genes, livJK (pSMS214), livHM (pSMS514), and livGF (pSMS517) (Table 2). These plasmids were then electroporated into appropriate kan::rpsL insertionally inactivated mutant strains (13, 15). Transformants were recovered on MH agar containing 0.5, 1, 2, and 5 mg ml−1 streptomycin. Colony PCR was used to confirm construction of C. jejuni 81-176 Smr ΔlivJK (SMS263), ΔlivHM (SMS540), and ΔlivGF (SMS539) (Table 1) mutants.
Motility assays.
Motility phenotypes of C. jejuni strains were assessed as previously described (19). Briefly, after growth from freezer stocks, strains were restreaked and grown at 37°C under microaerobic conditions for 16 h on MH agar containing TMP. Strains were then suspended in MH broth to an OD600 of 0.8 and stabbed into semisolid MH motility agar using a sterilized inoculating needle. The plates were incubated for 24 h at 37°C under microaerobic conditions and then visualized for motility.
Analysis of the commensal colonization capacity of C. jejuni mutants.
One-day-old White Leghorn strain Δ chicks were orally infected with wild-type or mutant C. jejuni 81-176 strains as previously described (5, 14). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 days at 37.8°C with appropriate humidity and rotation in a Sportsman II Model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12 to 36 h after hatching, chicks were orally gavaged with 100 μl of phosphate-buffered saline (PBS) containing approximately 100 or 104 CFU of wild-type or mutant C. jejuni 81-176 Smr strains. To prepare strains for infection, after growth from frozen stocks, strains were restreaked and grown at 37°C under microaerobic conditions for 16 h on MH agar containing TMP. Bacteria were resuspended from plates and diluted to the appropriate inoculum in PBS. Dilutions of the inocula were plated on MH agar containing TMP to determine the number of bacteria used to inoculate chicks. At day 7 postinfection, chicks were sacrificed and the cecal contents were recovered. Cecal contents were suspended to 0.1 mg ml−1 in PBS, and serial dilutions were spread on MH agar containing TMP and cefoperazone to determine the number of C. jejuni bacteria per gram of cecal content. Statistical analysis of results from colonization experiments was performed by the Mann-Whitney U test.
Amino acid transport assays.
After growth from frozen stocks, strains were grown at 37°C for 16 h under microaerobic conditions on MH agar or CDM-LIV agar. Strains were harvested from plates in 1× M9 medium (90.2 mM Na2HPO4, 22 mM KH2PO4, 8.56 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4, 22.2 mM glucose, 10 μM CaCl2, pH 7.4) and resuspended to an OD600 of 1.0. For leucine transport, assays were also performed in 1× M9 with 0.5% lactic acid. An aliquot of 150 μl was combined with 1.5 ml 1× M9 medium and warmed at 37°C for 5 min before addition of 1 μl of 3H-labeled amino acid. The specific activity of each radiolabeled amino acid was as indicated: leucine, 144 Ci/mmol; isoleucine, 60 Ci/mmol; valine, 60 Ci/mmol; proline, 75.2 Ci/mmol; glutamic acid, 49.9 Ci/mmol; serine, 24.9 Ci/mmol; aspartic acid, 13 Ci/mmol; threonine, 20 Ci/mmol; and asparagine, 400 Ci/mmol. At 0, 1, 2, and 5 min after addition of the radiolabeled amino acid, a 100-μl sample was withdrawn and filtered rapidly through a Millipore filter apparatus containing 0.22-μm microbiological membranes (Fisher Scientific) that had been prewashed with 5 ml of 1× M9 medium. Samples were washed twice with 5 ml of 1× M9 medium to remove excess 3H-labeled amino acid not associated with bacteria. Membranes were dissolved in 1 ml of methanol in scintillation vials, and 7 ml of scintillation fluid was then added. The amount of radioactivity associated with each strain was measured by a scintillation counter. Disintegrations per minute for transport of each amino acid by each strain were calculated as the counts per minute divided by the efficiency of the scintillation counter used for analyses, which was measured as 44.82%. The amount of transport is reported as pmol of amino acid transported per mg (dry weight) of cells. Strains were analyzed in triplicate for transport of leucine, valine, isoleucine, proline, glutamic acid, serine, aspartic acid, threonine, and asparagine.
Qualitative reverse transcription-PCR (RT-PCR) and quantitative real-time RT-PCR analysis.
C. jejuni strains were grown from freezer stocks as described above and restreaked onto MH agar, CDM+ agar, or CDM-LIV agar. After 16 h of growth at 37°C under microaerobic conditions, strains were resuspended from plates in PBS and pelleted in a Microfuge tube. RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For qualitative RT-PCR analysis, cDNA was generated using RNA (5 μg) from each strain, 250 nmol of random primers (Invitrogen), and Superscript II reverse transcriptase (Invitrogen). Equal amounts of cDNA products were used in PCR with appropriate primers to amplify a 250-bp fragment of each liv gene. Control reactions with mixtures lacking reverse transcriptase were performed as described above. Amplification of each liv gene from genomic DNA of wild-type C. jejuni 81-176 served as a positive control.
For quantitative real-time RT-PCR analysis, RNA was isolated as described above, DNase treated, and then diluted to a final concentration of 50 ng μl−1. Real-time RT-PCR analysis was performed in a 25-μl volume with 1× Sybr green PCR master mix (Applied Biosystems), 0.2 μM forward and reverse primers, and 0.25 μg RNA. RT-positive samples contained 0.1 μl of Multiscribe reverse transcriptase (Applied Biosystems). Reactions were performed on a 7500 real-time PCR system (Applied Biosystems) under the following conditions: 42°C for 30 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Results were normalized using gyrA or 16S rRNA as a control, and analysis was performed using the threshold cycle (ΔΔCT) method. The level of gene expression from samples grown on MH agar was calibrated to 1.
RESULTS
Description of the liv operon.
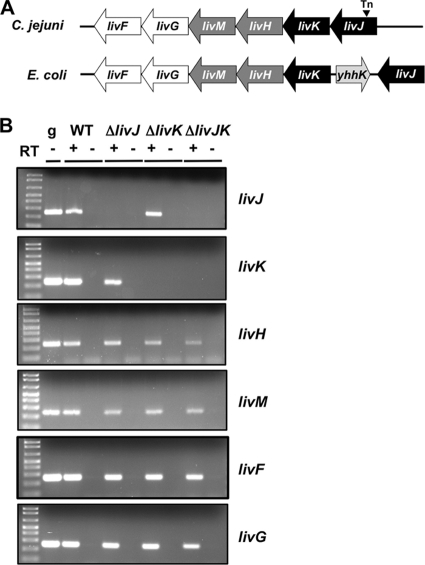
We previously identified a C. jejuni 81-176 mutant with a Tn insertion in Cjj81176_1038 that was attenuated for commensal colonization of the chick ceca (14). Cjj81176_1038 is homologous to livJ of E. coli and appears to be the first gene of a locus containing genes encoding additional proteins similar to other components of the E. coli LIV system (Fig. 1). Due to these homologies and functional assays presented below, we confirmed the annotation of these genes as livJKHMGF in the genome sequence of C. jejuni 81-176 (8). The composition of the putative C. jejuni LIV system appears to be similar to that for E. coli, with two periplasmic binding proteins (LivJ and LivK), two inner membrane permeases (LivH and LivM), and two cytoplasmic ATPases (LivG and LivF) (Fig. 1). The C. jejuni LIV proteins are homologous to the equivalent LIV proteins of E. coli with 27 to 46% identity and 45 to 66% similarity between respective proteins.
Fig. 1.
Organization of the liv loci of C. jejuni and E. coli and analysis of constructed C. jejuni liv mutants. The LIV locus of C. jejuni 81-176 contains six consecutive genes. In E. coli, livJ is separated from the other liv genes by the gene yhhK (light gray arrow) (1). The genes of the LIV system in these bacteria include livJ and livK (encoding the LIV binding proteins; black arrows), livH and livM (encoding the inner membrane permeases; dark gray arrows), and livG and livF (encoding the cytoplasmic ATPases; white arrows). The triangle indicates the site of the insertion of the signature-tagged Tn in the 81-176 mutant that was previously identified to have a reduced ability to colonize the chick ceca (14). (B) Qualitative reverse transcriptase PCR analysis of expression of liv genes in wild-type (WT) C. jejuni and ΔlivJ, ΔlivK, and ΔlivJK mutants. RNAs from C. jejuni strains were used in reactions with or without reverse transcriptase (RT) to generate cDNA. Each gene was amplified from cDNA using gene-specific primers. A positive control for amplification of each gene was performed by PCR with wild-type C. jejuni 81-176 genomic DNA (g).
To analyze the requirement of the system for a role in amino acid acquisition and in vivo growth, we created single, in-frame deletions of each liv gene. In addition, we deleted pairs of genes encoding proteins likely involved at the same step during amino acid transport (e.g., a ΔlivJK mutant to eliminate both binding proteins, a ΔlivHM mutant to eliminate both permeases, and a ΔlivGF mutant to eliminate both ATPases). We also constructed a mutant containing an in-frame deletion of ilvE, which encodes a transaminase predicted to be involved in a reversible reaction for conversion of oxopentanoate precursors to the branched-chain amino acids leucine, isoleucine, and valine required for biosynthesis or utilization of these amino acids (26).
We confirmed that deletion of specific liv genes from the C. jejuni chromosome did not cause polar defects in expression of downstream liv genes. As representatives for this type of analysis, we analyzed expression of liv genes in wild-type C. jejuni and the C. jejuni ΔlivJ, ΔlivK, and ΔlivJK mutants. No significant alterations in expression of the downstream genes livH, livM, livF, and livG were detected in the ΔlivJ, ΔlivK, or ΔlivJK mutant (Fig. 1B). Additional characterization of strains revealed that all liv and ilvE mutants demonstrated wild-type motility in semisolid agar (data not shown).
Growth of C. jejuni wild-type and mutant strains in liquid medium.
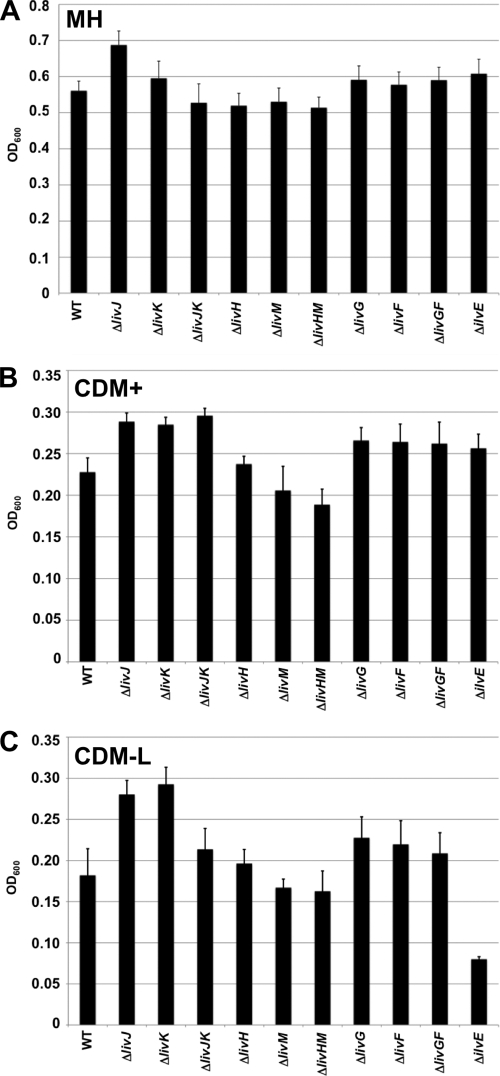
To assay the in vitro growth rates of wild-type and mutant strains in Mueller-Hinton (MH) broth or a Campylobacter chemically defined liquid medium containing all amino acids (CDM+), C. jejuni strains were grown in liquid medium over a 24-h period. The final optical densities of cultures revealed that all strains grew equally well in MH broth, a complex medium routinely used to grow C. jejuni strains, and less robustly in CDM+ (Fig. 2A and B). However, none of the liv mutants displayed an appreciable growth defect relative to wild-type C. jejuni in these media. When strains were grown in CDM lacking leucine (but containing all other amino acids [CDM-L]), all liv mutants grew equally as well as or even better than wild-type C. jejuni (Fig. 2C). These results suggest that if the C. jejuni LIV system transports leucine, C. jejuni has the capacity to synthesize leucine when the amino acid is not exogenously supplied. Therefore, we analyzed growth of a C. jejuni ΔilvE mutant, which is predicted to render C. jejuni incapable of synthesizing leucine from an oxopentanoate precursor. As expected, the ΔilvE mutant was not defective for growth in MH broth or CDM+, which contains leucine, but the mutant was severely impaired for growth in CDM-L (Fig. 2). These results combined suggest that C. jejuni has a system to acquire a branched-chain amino acid such as leucine but also has the capacity to synthesize leucine when it is not exogenously supplied.
Fig. 2.
Analysis of in vitro growth rates of wild-type C. jejuni and mutant strains in different liquid media. C. jejuni strains were grown in three different types of liquid media over 24 h at 37°C under microaerobic conditions. The liquid media included Mueller-Hinton (MH) broth (A), Campylobacter defined medium supplemented with all amino acids (CDM+) (B), or Campylobacter-defined medium supplemented with all amino acids except leucine (CDM-L) (C). Final OD600 measurements were taken at 24 h postinoculation. Strains include wild-type C. jejuni 81-176 Smr (DRH212) and in-frame deletions of each liv gene or pairs of liv genes encoding proteins with similar functions. Growth of 81-176 Smr ΔilvE (SMS270) was also measured. Data are reported as the averages of three experiments ± standard errors.
A role for the C. jejuni LIV system in amino acid transport.
To determine the biological role of the liv genes, we tested mutants lacking each gene or lacking two genes encoding proteins with putative similar functions for acquisition of branched-chain amino acids. For these experiments, wild-type and mutant strains were incubated with 3H-labeled amino acids and the level of transport of the amino acids into each strain was calculated by determining the amount of radioactivity associated with the bacteria over time. In initial experiments, strains were grown on MH agar prior to performing the amino acid transport assays. However, we obtained inconsistent measurements of transport from one assay to another (data not shown), possibly due to exogenous unlabeled amino acids remaining from prior growth of strains on MH agar that competed with 3H-labeled amino acids for transport by the LIV system. Subsequent assays in which strains were grown on CDM-LIV agar prior to the transport assays produced more consistent results. Therefore, for all transport experiments, C. jejuni strains were grown on CDM-LIV agar.
As reported below, the levels of amino acid transport that we observed were generally lower than those reported by other laboratories studying other amino acid transport systems of C. jejuni (27, 28, 41). In these other studies, C. jejuni strains were suspended in M9 with lactic acid or sodium lactate rather than M9 with glucose, which we used in our assays. We repeated some of our assays by suspending strains in M9 with lactic acid but did not observe any differences in amino acid transport. Therefore, in all assays described below, data from amino acid transport assays with bacteria in M9 with glucose are reported.
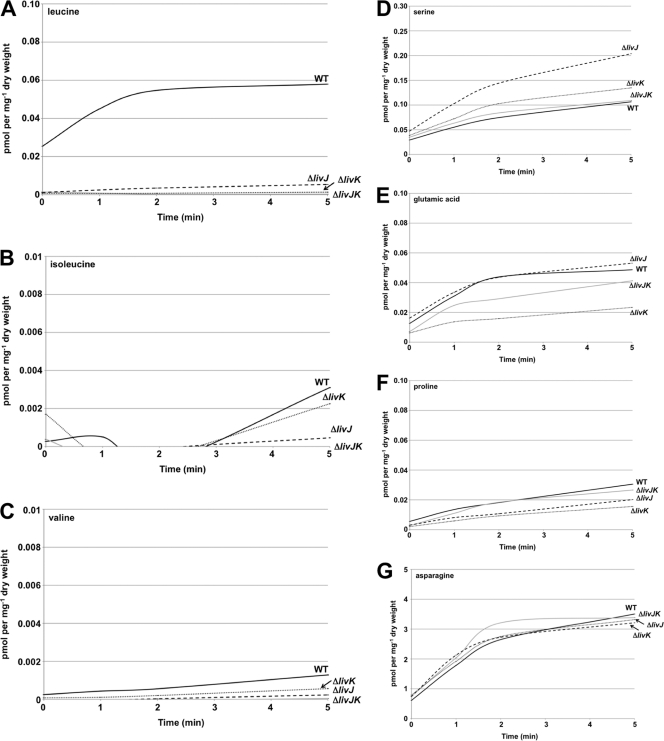
In our experiments, we observed abundant transport of leucine by wild-type C. jejuni that increased linearly up to 2 min before reaching a maximum level of transport (Fig. 3A). In contrast, the C. jejuni ΔlivJ mutant showed a 10-fold reduction in leucine transport, whereas the ΔlivK mutant had a more severe reduction in leucine transport, which was approximately 43-fold lower than that for the wild-type strain (Fig. 3A). When both livJ and livK were deleted, we observed only a minimal transport of leucine, which was over 200-fold lower than that for wild-type C. jejuni (Fig. 3A). These results suggest that LivK and LivJ are involved in leucine transport, with LivK likely having a more prominent role in acquisition of this amino acid than LivJ.
Fig. 3.
Involvement of C. jejuni LivJ and LivK in transport of various amino acids. After growth on CDM-LIV, strains were assayed for transport of 3H-labeled amino acids over a 5-min period by measuring the amount of radioactivity (as pmol per mg [dry weight] of cells) associated with each strain. Each strain was assayed in triplicate. The data presented represent an average of three assays. Standard errors were calculated for each data point and are too small to noticeably appear in graphs. y axis values were adjusted as appropriate for the level of amino acid transported for each graph. Data for transport assays for leucine (A), isoleucine (B), valine (C), serine (D), glutamic acid (E), proline (F), and asparagine (G) are shown. Strains include wild-type C. jejuni 81-176 Smr (DRH212) and ΔlivJ (SMS301), ΔlivK (SMS241), and ΔlivJK (SMS263) mutants.
Deletion of any other liv gene either individually (e.g., ΔlivH, ΔlivM, ΔlivG, or ΔlivF) or as paired double mutants (e.g., ΔlivHM or ΔlivGF) essentially abolished transport of leucine, reducing it to levels seen with the C. jejuni ΔlivJK mutant (data not shown). These results indicate that both permeases and both ATPases are required for transport, similarly to what has been found in the E. coli LIV system (1). The C. jejuni ΔilvE mutant transported leucine at a level equivalent to that of wild-type C. jejuni (data not shown), confirming that IlvE does not have a role in leucine transport but rather a function in leucine biosynthesis, since this mutant demonstrated a severe growth defect in the absence of leucine (Fig. 2C).
We also found that the C. jejuni LIV proteins are involved in transport of other branched-chain amino acids but at more modest levels than what we observed for leucine. For both isoleucine and valine, wild-type C. jejuni demonstrated roughly 19- and 45-fold less transport than that of leucine, respectively (Fig. 3B and C). When the ΔlivK and ΔlivJ mutants were examined, we observed that the ΔlivK mutant was reduced 2- to 3-fold for transport of isoleucine and valine compared to wild-type C. jejuni. However, the ΔlivJ mutant demonstrated an approximately 7-fold decrease in acquisition of both amino acids, suggesting that LivJ may have a more prominent role for isoleucine and valine transport than LivK. As seen for leucine, deletion of both livJ and livK as well as both permeases or ATPases resulted in abolishment of transport of isoleucine and valine (Fig. 3B and C; data not shown). Based on these results, we conclude that C. jejuni has a bona fide LIV system responsible for acquisition of branched-chain amino acids, especially leucine and to a lesser extent isoleucine and valine.
In E. coli, LivJ has been shown to be involved in the transport of alanine, serine, and threonine, albeit at reduced levels compared to branched-chain amino acids (38). The metabolism of many C. jejuni strains is largely dependent on amino acids rather than sugars as carbon sources (17, 25, 27, 31, 32, 41, 44). In metabolic studies, C. jejuni was shown to primarily utilize serine, aspartic acid, asparagine, glutamic acid, and, to a lesser extent, glutamine, threonine, and proline when grown in complex medium, CDM+, or minimal medium with single amino acids exogenously supplied (11, 17, 25, 44). Considering this information, we tested wild type or mutants lacking livJ, livK, or both livJ and livK for an expanded role in transport of preferably utilized amino acids in the metabolism of C. jejuni.
Previous research suggests that serine is a readily utilized amino acid in C. jejuni metabolism, with serine transporters and utilization enzymes produced by the bacterium (25, 40, 41, 44). Similar to previous observations, serine acquisition was robust across all strains, with even some enhanced transport observed in the ΔlivJ, ΔlivK, and ΔlivJK mutants compared to wild-type C. jejuni (Fig. 3D) (41). In contrast, we observed that glutamic acid transport in a ΔlivK mutant was reduced approximately 2-fold compared to that for wild-type C. jejuni (Fig. 3B). We also found a slight reduction in proline transport (approximately 33 to 50% less) in the ΔlivJ, ΔlivK, and ΔlivJK mutants compared to wild-type C. jejuni, but asparagine transport was not defective in any mutants (Fig. 3F and G). We were unable to observe any transport of threonine and aspartic acid by wild-type or mutant strains of C. jejuni 81-176 (data not shown).
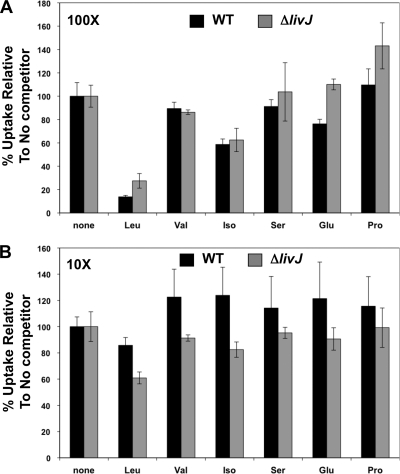
Because leucine appeared to be the predominant amino acid transported in an LIV-dependent manner, we performed competition experiments to verify the specificity of leucine transport by the C. jejuni LIV system. For these experiments, we analyzed only wild-type C. jejuni and the ΔlivJ mutant, which still had observable leucine acquisition through LivK. It was not possible to analyze the ΔlivK mutant in competition experiments to examine the specificity of LivJ-mediated transport due to the extremely low levels of leucine transport observed in this mutant. We performed competition assays by adding unlabeled leucine, valine, isoleucine, serine, glutamic acid, and proline at 10- and 100-fold-higher concentrations than that of 3H-labeled leucine and then measuring the amount of leucine associated with C. jejuni strains after 2 min. When added at a 100-fold excess, the only unlabeled amino acids that reduced the amount of [3H]leucine associated with wild-type C. jejuni were leucine and, to a small extent, isoleucine and glutamic acid (Fig. 4A). Serine or proline did not appear to compete with leucine for transport through the LIV system in wild-type C. jejuni. The fact that proline did not compete with leucine for transport suggests that the small amount of proline transport that we observed to be dependent upon LivJ or LivK is likely not specific (Fig. 3F). With a 10-fold excess of unlabeled amino acids, only leucine competed with 3H-labeled leucine for transport through the LIV system (Fig. 4B).
Fig. 4.
Specificity of leucine-mediated transport by the LIV system of C. jejuni. Competition assays were performed to measure the specificity of leucine transport by adding unlabeled amino acids in excess to potentially compete for 3H-labeled leucine for transport via the LIV system. Unlabeled amino acids were added at 100 (A)- or 10 (B)-fold-higher concentrations than 3H-labeled leucine. Assays were stopped after 2 min, and the total [3H]leucine associated with C. jejuni was measured. The amount of transport of 3H-labeled leucine in samples with competing unlabeled amino acid is reported as a percentage relative to the amount of 3H-labeled leucine transported in respective strains without competing unlabeled amino acid. Competition experiments were performed with wild-type C. jejuni 81-176 Smr (DRH212; black bars) and ΔlivJ strain (SMS301; gray bars). Bars represent standard errors of data points.
We also performed competition assays with the C. jejuni ΔlivJ mutant to monitor the specificity of LivK-specific leucine transport. Results from these assays were mostly similar to those performed with wild-type C. jejuni in that at a 100-fold excess, the only amino acid that competed with 3H-labeled leucine transport was leucine, and there was some modest inhibition by isoleucine (Fig. 4A). However, glutamic acid did not compete with leucine for LivK-mediated transport, suggesting that the low level of transport of glutamic acid by LivK that we initially observed is likely not specific (Fig. 3E). At a 10-fold excess, unlabeled leucine was able to reduce the amount of 3H-labeled leucine transport, suggesting that the LivK-mediated transport of leucine is specific.
Cumulatively, our data suggest that amino acid transport by the C. jejuni LIV system is largely limited to leucine, isoleucine, and valine. Transport of these amino acids is entirely dependent on both LIV permeases and LIV ATPases. Both LivJ and LivK are involved in transport of individual branched-chain amino acids. However, LivJ has more appreciable activity in isoleucine and valine transport than does LivK, whereas LivK appears to mediate a higher level of leucine transport.
Analysis of expression of the liv operon and ilvE.
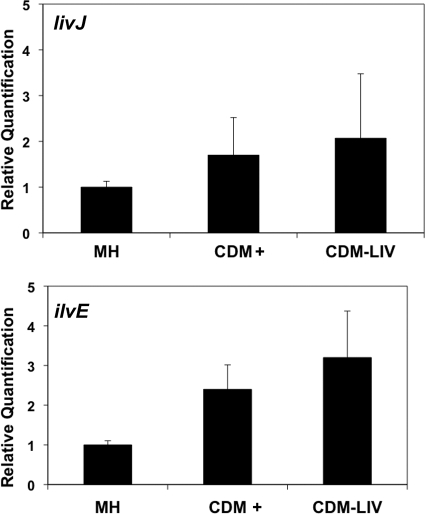
In E. coli, expression of the liv operon is repressed in the presence of leucine by the leucine-responsive protein (Lrp) (12). This repression is relieved upon leucine limitation. A homologous Lrp does not appear to be encoded in the C. jejuni genome. To determine if a leucine-responsive mechanism for controlling liv expression is present in C. jejuni, we grew C. jejuni strains on MH agar, CDM+, and CDM-LIV and examined the levels of expression of livJ, the first gene of the liv operon, and ilvE. We did not detect any significant leucine-dependent changes in livJ or ilvE expression between strains grown in MH, CDM+, or CDM-LIV, suggesting that leucine-dependent repression of liv gene expression likely does not exist in C. jejuni (Fig. 5).
Fig. 5.
Expression of liv operon upon growth of C. jejuni in the presence or absence of leucine. Quantitative real-time RT-PCR was performed on RNA isolated from C. jejuni grown on Mueller-Hinton (MH) agar or Campylobacter defined medium with all amino acids (CDM+) or without LIV (CDM-LIV). Expression of livJ or ilvE is reported relative to expression of these genes in wild-type C. jejuni 81-176 Smr (DRH212) grown on MH agar and has been set to a baseline of 1. Bars represent standard errors of data points.
LivJ and LivK are required for optimal commensal colonization of the chick ceca.
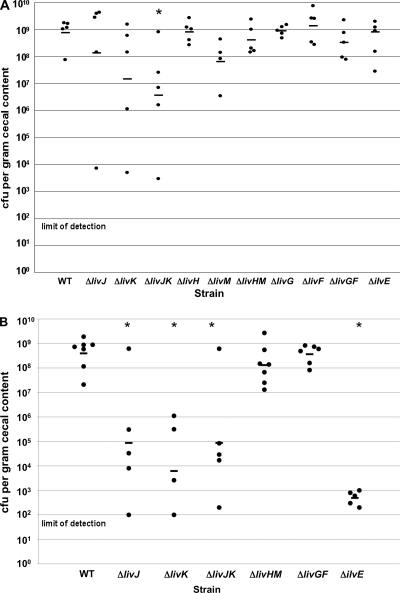
The identification of a mutant with a transposon insertion in livJ that showed a reduced colonization capacity suggested that livJ and possibly other downstream liv genes are required for wild-type levels of commensal colonization of the chick ceca. To determine which component(s) of the LIV system functions in commensal colonization, we tested each mutant lacking a single liv gene or mutants lacking pairs of liv genes encoding LIV components with similar functions for the ability to promote commensal colonization in a 1-day-old chick model of infection. Chicks were orally gavaged with approximately 104 or 100 CFU of wild-type C. jejuni 81-176 Smr or mutant strains lacking one or more liv genes. At day 7 postinfection, chicks were sacrificed and the levels of wild-type C. jejuni or mutant strains were enumerated in the chick ceca.
Upon inoculation with 104 CFU of wild-type C. jejuni, four out of five chicks had levels of C. jejuni greater than 109 CFU per gram of cecal content, with one chick colonized just below 108 CFU per gram of cecal content (Fig. 6A). The ΔlivJ and ΔlivK mutants as a group demonstrated defective colonization levels averaging 5- and 50-fold lower than that of wild-type C. jejuni, respectively (Fig. 6A), but the differences were not statistically significant. However, in each group infected with the ΔlivJ or ΔlivK mutants, chicks with 1,000- to 10,000-fold-lower C. jejuni loads than those of chicks infected with wild-type C. jejuni were evident. In contrast, the C. jejuni ΔlivJK mutant averaged a 200-fold-lower colonization capacity than did wild-type C. jejuni, which was statistically significant. These results suggested that LivJ and LivK are important commensal colonization determinants. In contrast, none of the other liv mutants appeared to be necessary for optimal commensal colonization of chicks (Fig. 6A). Deletion of each putative permease or ATPase component, or deletion of pairs of these components, did not hinder the colonization capacity of C. jejuni, with the possible exception of livM. However, the observed colonization defect of the livM mutant was not statistically significantly different from the colonization capacity of wild-type C. jejuni and was not recapitulated in the ΔlivHM mutant.
Fig. 6.
Chick colonization phenotypes of wild-type C. jejuni and isogenic liv or ilvE mutants. One-day-old chicks were orally inoculated with C. jejuni strains with approximately 104 CFU (A) or 100 CFU (B). At 7 days postinfection, chicks were sacrificed and the C. jejuni loads in the ceca of each chick were determined as the number of CFU per gram of cecal content. Each circle represents the number of C. jejuni bacteria from one chick. Bars represent the geometric means of bacterial loads from chicks infected with specific strains. Asterisks indicate statistically significant differences in colonization of mutants compared to wild-type C. jejuni using the Mann-Whitney U test (P < 0.05). Strains include wild-type C. jejuni 81-176 Smr (DRH212) and in-frame deletions of each liv gene or pairs of liv genes encoding proteins with similar functions. The colonization capacity of 81-176 Smr ΔilvE (SMS270) is also included.
Because we observed potentially reduced colonization capacities with the single ΔlivJ or ΔlivK mutants, we performed a second colonization experiment by lowering the inoculum to 100 CFU. With this inoculum, the wild-type strain reached a level of colonization between 108 and 109 CFU per gram of cecal content at day 7 postinfection (Fig. 6B). In contrast, the levels of colonization in the C. jejuni ΔlivJ and ΔlivK mutants were dramatically reduced, with colonization capacities that averaged 5,000- to 60,000-fold lower than that of wild-type C. jejuni, differences that were statistically significant (Fig. 6B). This decreased colonization ability was also seen in the ΔlivJK mutant. Mutants lacking both putative LIV permeases (LivH or LivM) or ATPases (LivG or LivF) did not show defects in colonization compared to wild-type C. jejuni (Fig. 6B). Since the LIV permeases and ATPases were required for LIV transport but not for colonization, these data suggest that LivJ and LivK may have other functions besides acquiring branched-chain amino acids that are important for promoting commensal colonization of poultry.
Since acquisition of leucine, isoleucine, and valine by the LIV system did not appear to be required for in vivo growth of C. jejuni, we tested if LIV biosynthesis is required for colonization of chicks. For these experiments, we tested the ability of the ΔilvE mutant to promote commensal colonization. At an inoculum of 104 CFU, the C. jejuni ΔilvE mutant was found at levels similar to that of wild-type C. jejuni at 7 days postinfection (Fig. 6A). However, at an inoculum of 100 CFU, the C. jejuni ΔilvE mutant clearly demonstrated a severe colonization defect, which was statistically significant (Fig. 6B). These results suggest that at a low inoculum, an ΔilvE mutant is hindered in initiating or maintaining infection of the chick ceca.
DISCUSSION
Due to the prevalence and persistence of C. jejuni in poultry, the bacterium has evolved a metabolism well suited for conditions and components present within the avian intestinal tract. For instance, C. jejuni tolerates well an elevated growth temperature of 42°C, which is the body temperature of many avian species, and has developed a microaerobic metabolism likely reflecting the low oxygen tensions present in the chick gut (43). Other than a fucose uptake and utilization system present in some C. jejuni strains, many C. jejuni strains appear to rely on amino acids rather than sugars as carbon sources (11, 32, 39, 41). Investigations have found that serine, aspartic acid, glutamic acid, and proline are depleted from media during in vitro growth of C. jejuni (11, 25, 31, 44). Corroborating the importance of amino acid utilization by C. jejuni, genome analysis as well as both genetic and biochemical studies has revealed that C. jejuni possesses the necessary catabolic pathways to convert these amino acids to intermediaries used in other biological processes (8, 34). Studies have verified the importance of a serine transporter (SdaC), a serine deaminase (SdaA), an aspartase (AspA), and an aspartate:glutamate aminotransferase (AspB) for both in vitro growth and commensal colonization of avian species by C. jejuni (11, 41). In addition, these amino acids are found most abundantly in chick feces, indicating that these nutrients are likely available for C. jejuni in infected hosts (35). Other amino acid transporters such as the Paq glutamine transport system have been shown to alter the virulence properties of the bacterium (28).
Upon identification of a component of the putative LIV amino acid transport system as an important commensal colonization determinant for C. jejuni (14), we initially hypothesized that C. jejuni may require one or more branched-chain amino acids to be supplied by the host for in vivo growth. In E. coli, the LIV system functions to transport branched-chain amino acids such as leucine, isoleucine, and valine (and to a lesser extent alanine, serine, and threonine) into the cytoplasm in an ATP-dependent manner (2, 3, 38, 42). The six proteins of the E. coli LIV system that function as the amino acid-binding, permease, and ATPase components are conserved in the C. jejuni LIV system. By analyzing an extensive collection of mutants lacking individual liv genes or combinations of liv genes, we were able to thoroughly analyze a role for the LIV system in in vitro growth, amino acid transport, and commensal colonization of chicks.
Whereas both LIV permeases and ATPases of C. jejuni were required for acquisition of leucine, isoleucine, and valine, we noticed differences in the involvement of LivJ and LivK for transport of these amino acids. We found that both LivJ and LivK were required for substantial levels of leucine transport, with LivK possessing a more prominent role. We also found that transport of isoleucine and valine was reduced to baseline levels in the absence of both livJ and livK, but the levels of transport of these amino acids were lower than that of leucine. A majority of the isoleucine and valine transport into C. jejuni appeared to be due to LivJ rather than LivK. In drawing analogies to the E. coli LIV system, C. jejuni appears to possess a similar LS system with LivK being fairly specific for transport of leucine and a LIV-I system with LivJ having a broader range of amino acids that can be transported than LivK, including leucine, valine, and isoleucine. We initially found that C. jejuni LivK may be responsible for transport of a small amount of isoleucine, valine, glutamic acid, and proline. However, competition experiments to assess the specificity of transport by LivK revealed that only isoleucine could compete to a small extent with leucine for transport. Thus, LivK is most likely dedicated to transport of leucine among all the amino acids tested. We were unable to demonstrate that the LIV system of C. jejuni can transport to a substantial level amino acids previously reported to be preferred substrates in C. jejuni metabolism.
Unlike the E. coli LIV system, we were not able to detect leucine-dependent repression of liv gene expression in C. jejuni. Consistent with this observation, we were unable to identify a gene encoding a protein similar to Lrp that may mediate repression of expression of the liv operon in the presence of leucine. However, we cannot rule out the possibility that the LIV system is regulated by an additional mechanism. With the data acquired so far, we hypothesize that the liv operon may be constitutively expressed.
Comparing the requirements of each LIV component for amino acid transport and for colonization of the chick ceca, we uncovered evidence for a more complex role of specific LIV components beyond acquisition of branched-chain amino acids. All components of the LIV system were required for wild-type levels of transport of leucine, isoleucine, and valine, but only LivJ and LivK were required for wild-type levels of commensal colonization of chicks. Neither the LIV permease mutants nor ATPase mutants were defective for colonization. If the sole functions of LivJ and LivK were to bind amino acids to be eventually transported into the cytoplasm in a manner dependent on the LIV permeases and ATPases, then we would expect the livH, livM, livG, and livF mutants to possess defects in colonization. However, these mutants were able to colonize chicks at levels similar to that of the wild-type strain. These findings suggest that LivJ and LivK likely have additional functions besides branched-chain amino acid transport that are required by C. jejuni during in vivo growth in the ceca of chicks. Furthermore, these results suggest that acquisition of branched-chain amino acids that may be supplied by the avian hosts is not a requirement of C. jejuni for promoting commensalism.
A few hypotheses of various degrees of validity can be proposed for these potentially alternative functions of LivJ and LivK. Some evidence exists that the periplasmic binding proteins of transport systems of other bacteria are able to use noncognate permeases and ATPases for transport of an amino acid into the cytoplasm of a bacterium (16, 18). It is possible that LivJ and LivK may interact with alternative permeases and ATPases for transport of branched-chain amino acids during in vivo growth. However, in our amino acid transport assays, the amounts of leucine, isoleucine, or valine transported into C. jejuni were reduced to the same baseline levels in the absence of the LIV permeases (ΔlivHM) or LIV ATPases (ΔlivGF) as in the ΔlivJK mutants. Thus, we could not find evidence that LivJ or LivK uses noncognate permeases or ATPases to transport branched-chain amino acids. It is possible that LivJ and LivK may bind an amino acid other than branched-chain amino acids whose transport is required for optimal in vivo growth. However, an examination of acquisition of the most preferred amino acids acquired by C. jejuni and used in its metabolism during in vitro growth did not reveal substantial transport of any of these amino acids in our assays that was dependent on LivJ or LivK. Alternatively, it is tempting to speculate that LivJ and LivK may bind any of a number of possible factors present in the chick gut that are essential for in vivo growth of C. jejuni and colonization of the chick ceca. We may be able to address these possibilities by determining if LivJ or LivK can bind components found in the cecal lumen of chicks. In summary, our work has revealed a surprising involvement of only specific components of an amino acid transport system for commensal colonization by C. jejuni and has provided further insight into the interactions between the bacterium and its natural avian host.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI065539 and by National Research Initiative grants 2006-35201-17382 and 2009-35201-05039 from the USDA Cooperative State Research, Education, and Extension Service Food Safety Program.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Adams M. D., et al. 1990. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J. Biol. Chem. 265:11436–11443 [PubMed] [Google Scholar]
- 2. Anderson J. J., Oxender D. L. 1977. Escherichia coli transport mutants lacking binding protein and other components of the branched-chain amino acid transport systems. J. Bacteriol. 130:384–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson J. J., Quay S. C., Oxender D. L. 1976. Mapping of two loci affecting the regulation of branched-chain amino acid transport in Escherichia coli K-12. J. Bacteriol. 126:80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beery J. T., Hugdahl M. B., Doyle M. P. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bingham-Ramos L. K., Hendrixson D. R. 2008. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76:1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black R. E., Levine M. M., Clements M. L., Hughes T. P., Blaser M. J. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472–479 [DOI] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention. 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:418–422 [PubMed] [Google Scholar]
- 8. Fouts D. E., et al. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedman C. R., et al. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3):S285–S296 [DOI] [PubMed] [Google Scholar]
- 10. Guardiola J., De Felice M., Klopotowski T., Iaccarino M. 1974. Multiplicity of isoleucine, leucine, and valine transport systems in Escherichia coli K-12. J. Bacteriol. 117:382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guccione E., et al. 2008. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69:77–93 [DOI] [PubMed] [Google Scholar]
- 12. Haney S. A., Platko J. V., Oxender D. L., Calvo J. M. 1992. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J. Bacteriol. 174:108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hendrixson D. R., Akerley B. J., DiRita V. J. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214–224 [DOI] [PubMed] [Google Scholar]
- 14. Hendrixson D. R., DiRita V. J. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471–484 [DOI] [PubMed] [Google Scholar]
- 15. Hendrixson D. R., DiRita V. J. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687–702 [DOI] [PubMed] [Google Scholar]
- 16. Higgins C. F., Ames G. F. 1981. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc. Natl. Acad. Sci. U. S. A. 78:6038–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hofreuter D., Novik V., Galan J. E. 2008. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4:425–433 [DOI] [PubMed] [Google Scholar]
- 18. Hosie A. H., Allaway D., Galloway C. S., Dunsby H. A., Poole P. S. 2002. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 184:4071–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joslin S. N., Hendrixson D. R. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191:2656–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kakuda T., DiRita V. J. 2006. Cj1496c encodes a Campylobacter jejuni glycoprotein that influences invasion of human epithelial cells and colonization of the chick gastrointestinal tract. Infect. Immun. 74:4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karlyshev A. V., et al. 2004. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150:1957–1964 [DOI] [PubMed] [Google Scholar]
- 22. Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592–596 [DOI] [PubMed] [Google Scholar]
- 23. Landick R., et al. 1980. Regulation of high-affinity leucine transport in Escherichia coli. J. Supramol. Struct. 14:527–537 [DOI] [PubMed] [Google Scholar]
- 24. Landick R., Oxender D. L. 1985. The complete nucleotide sequence of the Escherichia coli LIV-BP and LS-BP genes. J. Biol. Chem. 260:8257–8261 [PubMed] [Google Scholar]
- 25. Leach S., Harvey P., Wali R. 1997. Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J. Appl. Microbiol. 82:631–640 [DOI] [PubMed] [Google Scholar]
- 26. Lee-Peng F. C., Hermodson M. A., Kohlhaw G. B. 1979. Transaminase B from Escherichia coli: quaternary structure, amino-terminal sequence, substrate specificity, and absence of a separate valine-alpha-ketoglutarate activity. J. Bacteriol. 139:339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leon-Kempis M. D., Guccione E., Mulholland F., Williamson M. P., Kelly D. J. 2006. The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60:1262–1275 [DOI] [PubMed] [Google Scholar]
- 28. Lin A. E., et al. 2009. Atypical roles for Campylobacter jejuni amino acid ATP binding cassette transporter components PaqP and PaqQ in bacterial stress tolerance and pathogen-host cell dynamics. Infect. Immun. 77:4912–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindblom G.-B., Sjorgren E., Kaijser B. 1986. Natural campylobacter colonization in chickens raised under different environmental conditions. J. Hyg. (Lond.) 96:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Makarova O., Kamberov E., Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970–972 [DOI] [PubMed] [Google Scholar]
- 31. Mendz G. L., Hazell S. L. 1996. Utilization of amino acids by Campylobacter jejuni, p. 67–73.In Newell D. G. (ed.), Campylobacters, helicobacters, and related organisms. Plenum Press, New York, NY. [Google Scholar]
- 32. Muraoka W. T., Zhang Q. 2011. Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olson C. K., Ethelberg S., van Pelt W., Tauxe R. V. 2008. Epidemiology of Campylobacter jejuni infections in industrialized nations, p. 163–189.In Nachamkin I., Szymanski C. M., Blaser M. J. (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 34. Parkhill J., et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 35. Parsons C. M., Potter L. M., Brown R. D., Jr 1983. Effects of dietary carbohydrate and of intestinal microflora on excretion of endogenous amino acids by poultry. Poult. Sci. 62:483–489 [DOI] [PubMed] [Google Scholar]
- 36. Penrose W. R., Nichoalds G. E., Piperno J. R., Oxender D. L. 1968. Purification and properties of a leucine-binding protein from Escherichia coli. J. Biol. Chem. 243:5921–5928 [PubMed] [Google Scholar]
- 37. Pokamunski S., Kass N., Borochovich E., Marantz B., Rogol M. 1986. Incidence of Campylobacter spp. in broiler flocks monitored from hatching to slaughter. Avian Pathol. 15:83–92 [DOI] [PubMed] [Google Scholar]
- 38. Rahmanian M., Claus D. R., Oxender D. L. 1973. Multiplicity of leucine transport systems in Escherichia coli K-12. J. Bacteriol. 116:1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stahl M., et al. 2011. L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U. S. A. 108:7194–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Velayudhan J., et al. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274–286 [DOI] [PubMed] [Google Scholar]
- 41. Velayudhan J., Jones M. A., Barrow P. A., Kelly D. J. 2004. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72:260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wood J. M. 1975. Leucine transport in Escherichia coli. J. Biol. Chem. 250:4477–4485 [PubMed] [Google Scholar]
- 43. Woodall C. A., et al. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73:5278–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright J. A., et al. 2009. Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155:80–94 [DOI] [PubMed] [Google Scholar]