Abstract
Leptospira spp. are thin, highly motile, slow-growing spirochetes that can be distinguished from other bacteria on the basis of their unique helical shape. Defining the mechanisms by which these bacteria generate and maintain this atypical morphology should greatly enhance our understanding of the fundamental physiology of these pathogens. In this study, we showed that peptidoglycan sacculi from Leptospira spp. retain the helical shape of intact cells. Interestingly, the distribution of muropeptides was different from that in the Escherichia coli model, indicating that specific enzymes might be active on the peptidoglycan macromolecule. We could alter the shape of Leptospira biflexa with the broad-spectrum β-lactam antibiotic penicillin G and with amdinocillin and aztreonam, which are β-lactams that preferentially target penicillin-binding protein 2 (PBP2) and PBP3, respectively, in some species. Although genetic manipulations of Leptospira spp. are scarce, we were able to obtain mutants with alterations in genes encoding PBPs, including PBP3. Loss of this protein resulted in cell elongation. We also generated an L. biflexa strain that conditionally expresses MreB. Loss of the MreB function was correlated with morphological abnormalities such as a localized increased diameter and heterogeneous length. A prolonged depletion of MreB resulted in cell lysis, suggesting that this protein is essential. These findings indicate that important aspects of leptospiral cell morphology are determined by the cytoskeleton and the murein layer, thus providing a starting point for a better understanding of the morphogenesis in these atypical bacteria.
INTRODUCTION
Shape determination and maintenance are fundamental issues in biology (60). This is particularly true for prokaryotic cells, which need to maintain their shape to preserve their integrity. Bacteria can take on a variety of shapes, but for most of the well-described bacteria, these shapes are created by manipulating a common essential component, peptidoglycan (PG), a polymer of glycosaminopeptides that forms a rigid exoskeleton (15). Among bacteria with a peculiar morphology are the helix-shaped Leptospira organisms, which are 10 to 20 μm long with a cell diameter of approximately 0.15 μm. These cells belong to the phylum Spirochetes, an evolutionarily and structurally unique group of bacteria (43). This group also comprises flat wave- or helix-shaped bacteria like Borrelia and Treponema, which include the agents of Lyme disease and syphilis, respectively. Pathogenic Leptospira species are responsible for leptospirosis, a zoonotic disease which has a worldwide distribution but is particularly prevalent in impoverished regions (28). Leptospirosis is transmitted to humans through contact with water that is contaminated with animal (usually rodent) urine. Leptospira then disseminates in the host and causes a systemic infection. It has been shown that Leptospira moves faster in viscous environments than other bacteria (12). Although it has never been experimentally proven, this corkscrew motility is undoubtedly an advantage during the infectious process. Leptospira motility depends on the presence of two endoflagella (or periplasmic flagella), each arising at one end of the bacterium. Leptospira biflexa flaB mutants cannot form functional endoflagella, but their cell bodies remain intact and helical (45). The endoflagella are therefore not responsible for dictating the helical shape of the cell body in Leptospira spp., as they are in Borrelia burgdorferi (35).
It has been shown in other bacterial species, albeit with a different morphology (rod or crescent shaped), that several factors can be involved in acquiring and maintaining their shape (9, 10). Penicillin-binding proteins (PBPs) are involved in PG biosynthesis. PBPs are divided into two categories: high molecular weight (HMW) and low molecular weight (LMW). HMW PBPs comprise 2 classes of proteins, A and B. Escherichia coli cells need two proteins of the latter class, PBP2 and PBP3, to divide and elongate. PBP2 (encoded by pbpA) is involved in lateral PG synthesis, and PBP3 (encoded by ftsI) is involved in septal PG synthesis. A pbpA mutant becomes spherical, whereas an ftsI mutant produces filaments (51). Antibiotics that affect the function of these PBPs, such as amdinocillin and aztreonam, induce the same phenotypes (19, 40). LMW PBPs also participate in shape maintenance by modifying the preexisting cell wall (38, 39). The role of PBPs is closely interconnected with proteins of the cytoskeleton, like the tubulin homolog FtsZ and the actin homolog MreB, which are also important in bacterial shape determination (11, 42). Other types of cytoskeletal proteins are involved in cell morphology, such as crescentin, an intermediate filament responsible for the crescent shape of Caulobacter crescentus (8).
Five PBPs have been identified after subcellular fractionation in Leptospira interrogans (22), and functional homologs of E. coli ponA (PBP1a) and ftsI (PBP3) have been isolated from this bacterium (7). Leptospira organisms have a Gram-negative-like cell envelope with their PG layer associated with the cytoplasmic membrane (23). Peptidoglycan from L. biflexa apparently conforms to the A1γ chemotype, with the disaccharide pentapeptide GlucNAc-(β 1→4)-MurNAc-l-Ala-d-Glu-(γ)-mDAP-d-Ala-d-Ala as the basic monomeric subunit (59), but its precise composition is not known. This chemotype is the most common among Gram-negative bacteria, but genera of the family Spirochaetaceae are an exception and contain ornithine instead of diaminopimelate (DAP) as the di-amino acid at position 3 of the peptide moiety (59).
To explore Leptospira morphology, we further investigated the PG composition of saprophytic and pathogenic Leptospira strains. We also examined the role of different PBPs that might be involved in maintaining the shape of these bacteria. We then showed that MreB is involved in maintaining certain features of Leptospira morphology.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The sequenced L. biflexa serovar Patoc strain Patoc 1 and L. interrogans serovar Lai strain Lai 56601 (National Reference Center for Leptospirosis, Institut Pasteur, Paris, France) as well as L. interrogans serovar Manilae strain L495 (Monash University, Melbourne, Australia) were grown in EMJH liquid medium (17, 26) at 30°C with agitation or on plates containing 1% Noble agar. For conjugation experiments, E. coli strain Pi1 was used as the host strain for plasmid constructions and E. coli strain β2163 (14) was used as the donor to mobilize plasmids into L. biflexa. E. coli was grown in Luria-Bertani (LB) medium. Diaminopimelate (DAP) and thymidine (dT) were used at a final concentration of 0.3 mM. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to cultures at a final concentration of 1 mM. When appropriate, 50 μg/ml spectinomycin, 50 μg/ml kanamycin, or 100 μg/ml ampicillin was added to culture media.
MICs.
MICs were determined by broth microdilution testing as previously described (36). Briefly, after incubation with 2-fold serial dilutions of the antibiotics, alamarBlue (Trek Diagnostics, Cleveland, OH), which is an oxidation-reduction indicator, was added to each well following the manufacturer's instructions. When cells grow, the blue color indicator turns pink. The MIC (aztreonam, 0.5 μg/ml; amdinocillin, 16 μg/ml; penicillin G, 0.12 μg/ml; A22, 4 μg/ml) was defined as the lowest antibiotic concentration that inhibited visible growth (no color change) at the second day of incubation.
Plasmid and strain construction.
The PflgB promoter was amplified from B. burgdorferi chromosomal DNA using primer pair FLG5 and FLG3 (4) and cloned in pCR2.1 (Invitrogen) to create pPflgB. lacI was amplified using primers LI1 (5′-CATATGGTGAATGTGAAACCAG-3′) and LI2 (5′-TCAAACCAGATCAATTCGCG-3′) and chromosomal DNA from E. coli as a template. The amplification product was cloned between the NdeI and NsiI restriction sites of plasmid pPflgB to generate plasmid pLacIEc. Transcriptional fusion PflgB-lacIEc, amplified from pLacIEc with primer pair Lic1 (5′-TTTGGCGCGCCTAATACCCGAGCTTCAAGGAA-3′) and Lic2 (5′-TTTGGCGCGCCTCAAACCAGATCAATTCGCG-3′), was inserted in the AscI site of vector pSW29T-TKS2 (44) to create plasmid pTK-LacIEc.
The mreB coding sequence was amplified from L. biflexa chromosomal DNA using primers mreB1 (5′-GGAATTCCATATGATATTTGATAACCTTTATG-3′) and mreB2 (5′-CTAGTCTAGAAAAATGGGAAACCTCGGAG-3′) and cloned between the NdeI and XbaI sites of plasmid pPhsp10-LacO (2) to generate plasmid pPhsp10-LacO-mreB, which carries a transcriptional fusion between an IPTG-inducible promoter and the mreB gene. A PCR fragment corresponding to the region upstream from mreB, amplified from L. biflexa chromosomal DNA using primers mreB3 (5′-TCGGGCCCGTTTCGAATGATATAATAAC-3′) and mreB4 (5′-CGGAATTCATCAGTTCCTGGTGAAAG-3′), and a spectinomycin resistance cassette, amplified using primers SpcEco3′ (5′-ACGGAATTCAACGCGTAAAGTAAG-3′) and SpcNot5′ (5′-ATAAGAATGCGGCCGCAACGCGTCCCGAGC-3′), were cloned between the NotI and ApaI sites of pPhsp10-LacO-mreB, creating plasmid pMreB::Spc-Phsp10-LacO. All of these fragments were then cloned between the SmaI and XbaI sites of pSW29T after digestion of pMreB::Spc-Phsp10-LacO with the same enzymes, thus generating plasmid pPind-mreB.
Strain LS Pind-mreB, which conditionally expresses mreB, was constructed in the following manner. E. coli β2163 cells harboring pTK-LacIEc were used as donor cells to mobilize the vector into L. biflexa as described previously (44). Exconjugants were selected on kanamycin, and the insertion site of the transposon carrying Phsp10-lacI was verified in a few of these. The lic2 strain with the transposon inserted at position 3446875 of the large chromosome between LEPBIa3341 and LEPBIa3342 was selected as a recipient for conjugation with E. coli β2163 harboring pPind-mreB as well as pLacIEc to prevent mreB transcription in E. coli. Conjugation was carried out as described, except that IPTG was added to the medium. Exconjugants were selected on spectinomycin. Replacement of the native mreB promoter with the spectinomycin resistance cassette and the inducible Phsp10-LacO promoter by allele exchange was verified by PCR and sequencing.
Random insertion mutagenesis was carried out in L. biflexa serovar Patoc strain Patoc 1, L. interrogans serovar Manilae strain L495, and L. interrogans serovar Lai strain Lai 56601 with a kanamycin-resistant Himar1 transposon as previously described (37). Among the kanamycin transformants, we identified mutants with an insertion into LA1045, LEPBIa1432, LA3698, and LA3692 (Table 1).
Table 1.
Penicillin-binding proteins present in all sequenced Leptospira strains
| Name | Gene no. |
||
|---|---|---|---|
| L. interrogans | L. borgpetersenii | L. biflexa | |
| Class A | |||
| PBP1a | LA3692a | LBL_0402 | LEPBIa0847 |
| PBP1a | LA1221 | LBL_2123 | LEPBIa1010 |
| PBP1a | LA1009 | LBL_0498 | LEPBIa0637 |
| PBP1a | LA1045a | LBL_2793 | LEPBIa1432a |
| PBP1c | LA2187 | LBL_1576 | LEPBIa0534 |
| Class B | |||
| PBP2 | LA2755 | LBL_1957 | LEPBIa3075 |
| PBP3 | LA3698a | LBL_2647 | LEPBIa0257 |
Mutants harboring a transposon in these genes are available. The locations of the Himar1 insertion are as follows: LA3692 (808 amino acids [aa]), insertion after aa 66; LA1045 (827 aa), insertion after aa 122; LA3698 (602 aa), insertion after aa 96; and LEPBIa1432 (850 aa), insertion after aa 437.
Plasmid and chromosomal DNAs were prepared using Qiagen and Promega Maxwell cell purification kits, respectively.
Real-time qRT-PCR.
Real-time quantitative reverse transcriptase PCR (qRT-PCR) experiments were carried out as previously described (2). The specific primers for L. biflexa mreB (5′-GGTATGGTGATTGCCGAGTC-3′ and 5′-TCTGCTGTTCTTTCCCCAAC-3′) and L. interrogans ftsI (5′-GAACCGAAAGGTGGAACTCA-3′ and 5′-AACCGAAATCCCAAGAGGTC-3′) and LA3697 (5′-TTTAGTTTGTGGGGCTGGTC-3′ and 5′-CCTGGGGATCAACTGAGAAA-3′), as well as specific rpoB primers (5′-GCAAAATGAAAATCGCTGGT-3′ and 5′-CAGCATCGAGCATTACCTCA-3′ for L. biflexa and 5′-ATGGAGCGGAACGTGTAGTC-3′ and 5′-CTTCGTTCGTTCCATGTCCT-3′ for L. interrogans), were used for PCR. Relative RNA levels were determined using the comparative threshold cycle (CT) method, also called the 2−ΔΔCT method (31).
Peptidoglycan preparation for visualization and analysis.
L. biflexa or L. interrogans cells were inoculated in 200 ml EMJH and grown until stationary phase and harvested at 5,000 × g for 10 min at room temperature. Cells were resuspended in 3 ml phosphate-buffered saline (PBS) and dropped into 6 ml of a boiling 6% SDS solution being stirred with a magnetic bar. The mixture was stirred for 4 h in a boiling water bath and then overnight at room temperature as previously described (16). Sacculi were washed clean of SDS by four successive centrifugations and resuspensions in water using a Beckman TL100 centrifuge equipped with a TLA100.3 rotor at 300,000 × g for 10 min at 18°C.
For electron microscopy, sacculi were observed directly (raw) or after chymotrypsin digestion to remove contaminating protein. In the latter case, once SDS was removed, sacculi were digested overnight at 37°C with 100 μg/ml chymotrypsin in PBS. After digestion, SDS was added to the suspension to a final concentration of 1%, and sacculi were incubated for 2 h in a boiling water bath and then cleaned free of SDS as described above. Sacculi were then resuspended into 100 μl of water, and carbon-Formvar-coated copper electron microscopy (EM) grids (200 mesh) were floated onto drops of this suspension. After 10 min, the grids were dried with filter paper and, after a few minutes of air drying, were floated onto water drops and washed 6 times. Grids were then floated onto 1% uranyl acetate in water for 2 min. After removing excess uranyl acetate, grids were washed once again very briefly with water, dried, and either observed directly or subjected to carbon-platinum shadowing at a 10° angle. Observations were made in a JEOL JEM1010 at 60 KV, equipped with a TemCam-F416 (TVIPS, Germany) digital camera.
For composition analysis, PG was digested overnight at 37°C with 40 μg/ml muramidase (Cellosyl; Hoechst), reduced with NaBH4 to avoid anomerization of sugars, and subjected to high-performance liquid chromatography (HPLC) to separate and purify the muropeptides as previously described (20, 44). Individual components were collected after HPLC separation and subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Autoflex; Bruker Daltonics) to determine the molecular mass of the components. To confirm the proposed structures, most muropeptides were further analyzed by electrospray ion-trap mass spectrometry (LCQ Classic; Thermo-Finnigan) to define their amino acid and amino sugar sequences.
For preparation of the mreB mutant sacculi, LS Pind-mreB cells grown until midexponential-phase in the presence of IPTG were harvested, the pellet was washed once in EMJH to remove IPTG, and growth was resumed in EMJH for 16 h. Preparation of sacculi was then pursued as described above.
Scanning electron microscopy.
For the analysis of Leptospira morphology, bacteria in exponential growth phase (optical density at 420 nm [OD420] of ∼0.2) were incubated with various compounds (aztreonam, 0.2 μg/ml; amdinocillin, 20 μg/ml; penicillin G, 0.1 μg/ml; A22, 5 μg/ml). Cells were then harvested at 4, 8, 12, 20, or 24 h by centrifugation, washed once in PBS, and resuspended in cacodylate buffer (0.1 M; pH 7.2) supplemented with 2.5% glutaraldehyde. Samples were then processed for electron microscopy experiments as described previously (49). For cell length measurements, 50 to 100 cells were randomly chosen in the micrographs, and the distance between two ends was manually calculated using the ImageJ program (http://rsb.info.nih.gov/ij/).
RESULTS
Sacculi from Leptospira retain a helical shape.
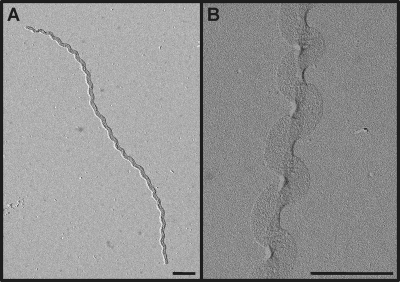
To study the morphology of Leptospira, we first wanted to examine the shape of their murein sacculi. The PG macromolecules were purified and subjected to electron microscopy as described in Materials and Methods. The micrograph in Fig. 1 illustrates that Leptospira sacculi present a helical shape similar to the shape of the live microorganisms. This shape is intrinsic to the PG macromolecule since they must be devoid of protein after the chymotrypsin treatment to which they were subjected (Fig. 1B). The darker area along the longitudinal axis on Fig. 1B is probably due to the flattening of the sacculi. Purified sacculi of L. biflexa have a helix pitch of 470 ± 40 nm, a helix diameter of 326 ± 12 nm, and a sacculus width of 175 ± 10 nm (data from 12 distinct sacculi).
Fig. 1.
Electron microscopy of L. biflexa sacculi. L. biflexa cells were grown in EMJH at 30°C, and samples were processed as described in Materials and Methods. (A) Untreated sacculus. (B) Chymotrypsin-digested sacculus. The bars represent 0.5 μm.
Alteration of cell shape by antibiotics or mutagenesis targeting PBPs.
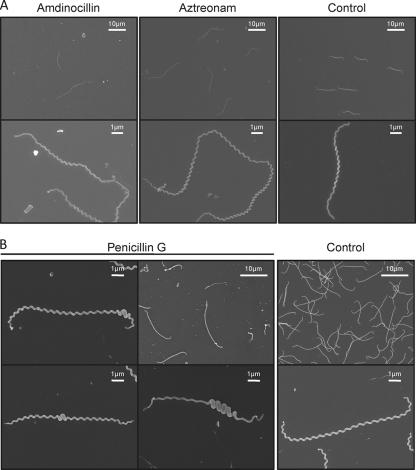
To understand the role of enzymes involved in peptidoglycan synthesis and modification, we first listed the PBPs found in the genome of all sequenced Leptospira strains. Seven PBPs were identified: 5 class A PBPs, including four PBP1a proteins, and 2 class B PBPs (Table 1). No low-molecular-weight PBPs were identified. Transposon mutants carrying genes encoding two PBP1a protein variants were subjected to microscopic analysis. No morphological alterations were noted for these mutants compared to the wild-type strain (data not shown). This might be due to a functional redundancy in the Leptospira genomes. We then subjected L. biflexa cells to the action of inhibitors of these PG enzymes. Different antibiotics were chosen according to their spectrum of action in E. coli. Aztreonam preferentially inhibits PBP3, involved in septal peptidoglycan synthesis. Amdinocillin preferentially inhibits PBP2, involved in lateral peptidoglycan synthesis. Penicillin G inhibits all PBPs. In E. coli, aztreonam induces the formation of elongated cells, whereas addition of amdinocillin leads to round cells.
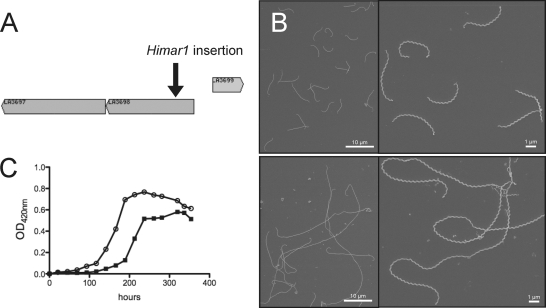
The micrographs on Fig. 2A show that the majority (∼87%) of L. biflexa cells incubated with 0.2 μg/ml aztreonam, which is a concentration below the MIC (0.5 μg/ml), were longer than the control sample (19.21 ± 5.60 μm versus 10.72 ± 2.27 μm in the treated and untreated samples, respectively). The growth rate was comparable to that of the control cells (doubling time of 4 h) until 8 h after the addition of the antibiotic. Shape changes appeared after 4 h, and the growth rate decreased to 70% of the control until the end of the experiment. Furthermore, a mutant with a transposon insertion in LA3698 (ftsI) encoding PBP3 in L. interrogans serovar Manilae strain L495 (Fig. 3A) presented an elongated-cell phenotype (25.11 ± 11.74 μm, in comparison to 6.44 ± 0.93 μm for the parental strain), with ∼16% of the cells being aberrantly elongated (≥40 μm) (Fig. 3B). The mutant showed poor growth in EMJH, compared to the parental strain (Fig. 3C), but was viable even after prolonged incubation times. In other bacteria, with the exception of cyanobacteria (34), ftsI is included in a gene cluster involved in cell division (3, 56). Similar to the organization in other Leptospira species, ftsI in L. interrogans is not included in such a cluster and is adjacent to a gene encoding a protein of unknown function (LA3697) and to a gene encoding an arsenate reductase (LA3699). The mRNA level of LA3697 was decreased in the mutant, in comparison to the level in the parental strain (data not shown), suggesting that disruption of ftsI exerted a polar effect on the downstream gene and that we cannot rule out an effect of this gene in the morphological alterations observed.
Fig. 2.
Shape alterations of L. biflexa induced by antibiotics. L. biflexa cells were grown in EMJH at 30°C until midexponential phase, antibiotics were added, cultures were resumed, and samples were taken at different time points for electron microscopy observation. Cells with no additive are the control sample. (A) Cells grown with aztreonam (0.2 μg/ml) or amdinocillin (20 μg/ml) for 12 h. (B) Cells grown with penicillin G (0.1 μg/ml) for 20 h. This experiment is representative of two independent experiments.
Fig. 3.
L. interrogans ftsI mutant cells are elongated. (A) Genetic organization of the ftsI locus (LA3798) in L. interrogans. The insertion site of Himar1 in the chromosome of the ftsI mutant is indicated. (B) Scanning electron microscopy of L. interrogans wild-type strain (top panel) and isogenic ftsI mutant (bottom panel). (C) Growth kinetics of the wild-type (open circles) and ftsI mutant (filled squares) strains.
Contrary to E. coli, L. biflexa cells incubated with amdinocillin at concentrations around the MIC level also presented an elongated phenotype (22.38 ± 7.22 μm). Morphological changes appeared after 4 h. The growth rate was comparable to that of the control cells until 8 h after addition of the antibiotic, and then the cells stopped growing and started lysing 12 h after addition of amdinocillin.
Interestingly, when penicillin G was added to the culture at concentrations around the MIC level, cells presented a different morphology. There was heterogeneity in cell length but also a localized increase of cell diameter at various points along the longitudinal cell axis in ∼43% of the cells (Fig. 2B). Shape changes appeared after 12 h. The growth rate was comparable to that of the control cells until 8 h after addition of the antibiotic; it then decreased to 75% of that of the control and was 50% at the end of the experiment.
MreB is involved in maintaining the diameter and length of the cells.
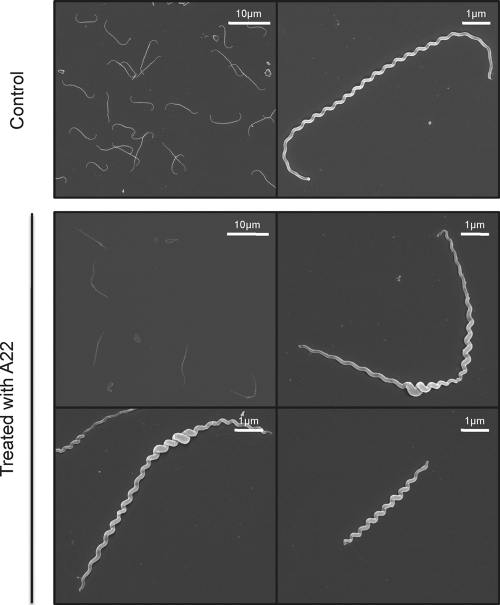
We then examined the role of MreB, a protein involved in maintaining the proper morphology of several rod-shaped bacteria. MreB is present in all sequenced Leptospira strains. In the pathogens, but not in the saprophyte L. biflexa, mreB is genetically linked to the mreC and mreD genes, as is also the case in E. coli. L. biflexa MreB shares 50 to 60% identity with MreB from E. coli or C. crescentus. To understand the role of MreB, we first used the A22 compound, which has been shown to affect the function and to mimic loss of this actin-like homologue in several Gram-negative microorganisms (5, 20, 41). Figure 4 shows that cells treated with A22 at concentrations around the MIC level presented a phenotype similar to what we observed when they were subjected to penicillin G. Morphological changes appeared after 8 h. The growth rate was comparable to that of the control cells until 8 h after addition of the compound, and then the cells stopped growing and started lysing 12 h after addition of A22. Cells were of different lengths, and there was a localized increase in cell diameter in various points along the longitudinal cell axis in ∼95% of the cells. A22-treated cells seem to present a less homogeneous helix pitch than the control cells. However, the cells remained helical.
Fig. 4.
A22-induced morphological alterations of L. biflexa. L. biflexa cells were grown in EMJH at 30°C until early exponential phase, and A22 was added at a final concentration of 5 μg/ml. The control sample was supplemented with ethanol, the solvent used for A22 solubilization, in the same proportion as for the culture with this compound. Cultures were resumed, and samples were taken at different time points for electron microscopy observation. The samples presented here were harvested 24 h after addition of A22. This experiment is representative of three independent experiments.
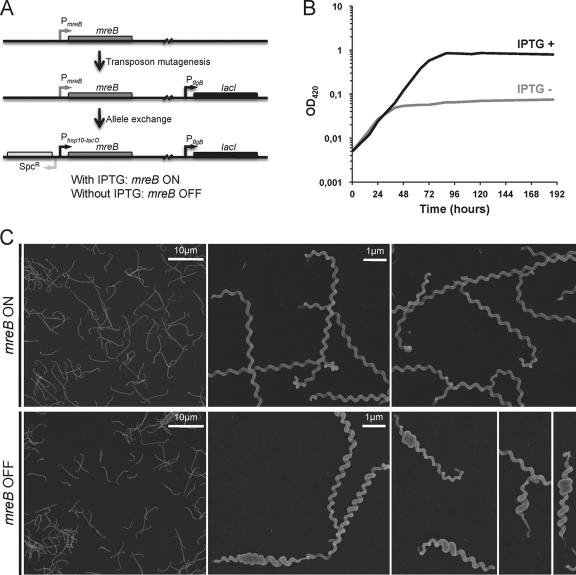
Since we did not succeed in disrupting the mreB gene by allelic exchange (our unpublished data), we constructed strain LS Pind-mreB carrying lacI from E. coli and mreB under the control of a promoter containing LacO operator sequences (Fig. 5A). In the presence of IPTG, the mreB mRNA level of LS Pind-mreB is comparable to that of the wild-type strain. In the absence of IPTG, the LacI regulator binds to LacO and represses transcription of mreB. The transcript level of mreB decreased approximately 14-fold 1 h after IPTG had been removed from the culture compared to the mreB mRNA level in cells grown with IPTG (data not shown). The growth curve presented in Fig. 5B shows that in the absence of IPTG, the culture reached a plateau at an OD420 of ∼0.05 in 36 h, whereas the cells grew until they reached stationary phase at an OD420 of ∼1 after 90 h in the presence of IPTG. The length of most of the cells grown without IPTG was shorter (5.92 ± 1.84 μm versus 8.58 ± 1.1 μm in the absence of IPTG), and 16% of the cells were smaller than 4 μm. Thirty-eight percent of the cells of the mutant also exhibited an increased diameter in localized regions along the longitudinal axis (Fig. 5C), as was observed with A22-treated cells. Furthermore, after a longer incubation period, cells started lysing. This observation and the fact that we could not disrupt the mreB gene suggest that MreB is essential for the growth of L. biflexa under standard laboratory conditions.
Fig. 5.
Effect of mreB depletion on L. biflexa morphology. (A) Details of the MreB-depleted strain construction. (B) Growth curves of the LS Pind-mreB strain in the absence (gray line) or presence (black line) of IPTG, which was added at the time of inoculation. (C) L. biflexa cells were grown in EMJH at 30°C until midexponential phase with IPTG. Cells were washed, and half the culture was supplemented with IPTG. Cultures were resumed, and samples were taken at different time points for electron microscopy observation. The samples presented here were harvested 24 h after broth change. This experiment is representative of three independent experiments.
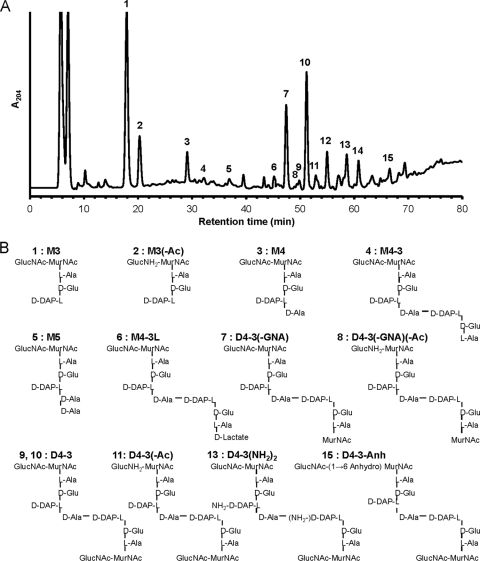
Composition of L. biflexa peptidoglycan.
The morphological peculiarities of L. biflexa—in particular its extremely small diameter and helical geometry—might well impose constraints on the organization of the sacculus. Therefore, the muropeptide composition of its PG was determined as previously done for other microorganisms (21, 47). The HPLC elution pattern obtained is shown in Fig. 6A. Each peak should correspond to a specific PG subunit. The results presented in Table S1 in the supplemental material allowed for the unambiguous identification of the major components resolved by HPLC and whose structures are displayed in Fig. 6B. Identified peaks accounted for more than 80% of the total peak area, as calculated from the integration of the HPLC results shown in Fig. 6A. The major components, peaks 1 and 10, were identified as the disaccharide tripeptide GlucNAc-MurNAc-l-Ala-d-Glu-(meso)-DAP (M3) and the cross-linked dimer GlucNAc-MurNAc-l-Ala-d-Glu-(meso)-DAP-d-Ala→(meso)-DAP-d-Glu-l-Ala-MurNAc-GlucNAc (D4-3), respectively. An unusual feature of the HPLC elution pattern was the identification of two well-separated peaks (9 and 10) as the same component, D4-3. The components in both peaks had virtually identical masses (see Table S1). Furthermore, liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis supported identical sequences for both (data not shown). Peak 5 corresponded to the disaccharide pentapeptide GlucNAc-MurNAc-l-Ala-d-Glu-(meso)-DAP-d-Ala-d-Ala (M5) the canonical basic subunit of A1γ peptidoglycan (50), and peak 4 is the same component without the terminal d-Ala, which is usually the major muropeptide in Gram-negative bacteria. We identified 1→6 anhydromuramic acid residues in peak 15. These residues are important because they occupy the C-1 terminal position on the glycan chain, and their relative abundance is an indication of the average length of PG strands (57). Most remarkable was the presence of a series of muropeptides (Fig. 6, peaks 4, 6, 7, 8, 11, and 13) derived from D4-3, which might represent intermediate steps of a specific PG remodeling process. M4-3, M4-3L, D4-3(−GNA) and D4-3(−GNA)(−Ac) (Fig. 6, peaks 4, 6, 7, and 8, respectively) are seldom detected in macromolecular PG in other species. [For definitions of “(−GNA)” and “(−Ac),” see the legend to Fig. 6.] Muropeptides in peaks 2, 8, 11, and 13 were also quite interesting because of specific chemical modifications: deacetylation of the GlucNAc residue in M3(−Ac), D4-3(−Ac) and D4-3(−GNA)(−Ac); and amidation of the DAP residues in muropeptide D4-3(NH2)2. Neither modification is particularly rare, but both require specific enzymatic activities.
Fig. 6.
L. biflexa muropeptides. (A) A sample of purified and muramidase-digested L. biflexa PG was subjected to HPLC. The eluate was monitored measuring A204. Peaks were collected and subjected to MS for further analyses. Peak numbers correlate with Fig. 6B (and see Table S1 in the supplemental material). (B) Composition and structure of L. biflexa muropeptides as deduced from HPLC and MS data presented in panel A and Table 1. The proposed abbreviations are defined as follows. M represents the monomer, and D represents the dimer. The first number indicates the number of amino acids in the donor stem peptide, and the second number indicates the number of amino acids in the acceptor stem peptide. (−GNA), lacking a residue of N-acetylglucosamine; (−Ac), lacking an acetyl group at position 2 of N-acetylglucosamine; (NH2), amidated at the d-carboxyl of DAP; -Anh, with a residue of (1→6) anhydro muramic acid.
Once the nature of the major muropeptides was defined, it was possible to quantify the relative abundance of each in molar terms (Table 2). Taking the well-known PG of E. coli as a comparative standard, L. biflexa PG is as cross-linked (31.6%) as the PG of E. coli (30 to 35%) and other Gram-negative bacteria. Based on anhydro muropeptide proportion, L. biflexa PG seems made of longer glycan strands, averaging 125 disaccharides/strand, than E. coli PG, which is made of 30 to 40 disaccharides/strand (21).
Table 2.
Muropeptide composition of peptidoglycan purified from Leptospira strains
| Muropeptidea | Molar fraction (×100)b |
|||
|---|---|---|---|---|
|
L. biflexa |
L. interrogans (wild type) | |||
| Control | A22 | MreB | ||
| Individual peptides | ||||
| M3 | 42.6 | 33.8 | 58.7 | 69.2 |
| M3(−Ac) | 11.1 | 7.5 | 7.8 | 12.6 |
| M4 | 4.4 | 15.6 | 1.7 | 2 |
| M4-3 | 2.0 | 4.2 | 3.4 | 3.3 |
| M5 | 1.7 | 1.1 | 4.9 | 1.6 |
| M4-3L | 2.2 | 0.8 | 2.8 | 0.5 |
| D4-3(−GNA) | 9.6 | 9.7 | 4.0 | 1.5 |
| D4-3(−GNA)(−Ac) | 0.2 | 0.0 | 0.0 | 0 |
| D4-3 | 3.0 | 1.3 | 2.8 | 0 |
| D4-3 | 14.0 | 12.9 | 9.9 | 4.9 |
| D4-3(−Ac) | 1.8 | 1.3 | 0.4 | 0 |
| X1 | 2.3 | 7.1 | 1.2 | 2.9 |
| D4-3(NH2)2 | 2.1 | 2.7 | 1.2 | 0.7 |
| X2 | 2.1 | 1.9 | 0.7 | 0.4 |
| D4-3-Anh | 0.8 | 0.8 | 0.6 | 0.5 |
| Groupsc | ||||
| Monomers | 64.0 | 63.0 | 79.2 | 89.1 |
| Dimers | 31.6 | 28.7 | 18.9 | 7.6 |
| Anhydro | 0.8 | 0.8 | 0.6 | 0.5 |
| Amidase | 2.0 | 3.8 | 3.4 | 3.3 |
| Etherase | 2.2 | 0.7 | 2.8 | 0.5 |
| Glucosaminidase | 9.8 | 9.7 | 4.0 | 1.5 |
| Amidation | 2.1 | 2.7 | 1.2 | 0.7 |
| Deacetylase | 13.0 | 8.8 | 8.1 | 12.6 |
About 29% of the total muropeptides exhibited some alteration over the basic structure of M3 and D4-3. Most abundant were deglucosaminidated and deacetylated muropeptides, which accounted for about 10 and 13% of total muropeptides, respectively. The presumptive products of MurNAc-l-Ala amidases and etherases, M4-3 and M4-3L, accounted for about 2% of the total each, a rather significant fraction in structural terms. Such a large-scale postinsertional modification should certainly have consequences on PG properties and suggests an elaborate processing of macromolecular PG in L. biflexa.
The proposed structures can be explained as naturally derived from the basic disaccharide pentapeptide subunit as the consequence of known biosynthetic and degradative enzyme activities. However, we could not suggest a sound structure even after MS/MS for X1 and X2, which compose peaks 12 and 14, which seem to be modified muropeptides as both contain residues of GlucNAc and MurNAc.
Treatment of L. biflexa PG with pronase-E had no detectable effect on the HPLC elution pattern (data not shown), suggesting absence of covalently PG-bound proteins equivalent to Braun's lipoprotein of E. coli (21).
We also investigated PG composition of a representative pathogenic strain, L. interrogans serovar Manilae strain L495. The results show that overall PG composition of L. interrogans was similar to that of L. biflexa. However, some differences were noted. L. interrogans showed a low proportion of cross-linked muropeptides (D4-3) and consequently of the muropeptides that derive from D4-3. There is also a lower proportion of 1→6 anhydromuramic acid residues, which indicates that the glycan chains might be longer in L. interrogans than in L. biflexa.
Effect of impaired MreB functionality on L. biflexa PG composition.
To study whether the morphological modifications observed on cells subjected to alterations of the function or level of MreB could be associated with specific changes in PG composition, sacculi isolated from A22-treated cells or from a strain depleted of MreB were subjected to HPLC analysis. The results presented in Table 2 show that the presence of A22 caused a dramatic rise in the proportion of M4 and a concomitant drop in M3. In addition, A22 treatment caused a strong reduction in the proportion of deacetylated muropeptides, which dropped to 60% of the control level, and a moderate decrease in the proportion of cross-linked muropeptides.
Decrease of MreB level showed both similarities and important differences in the PG composition of these cells compared to cells treated with A22. The effect on cross-linking was more pronounced than that of A22 and was accompanied by a marked reduction in D4-3(−GNA) muropeptides. Deacetylated muropeptides were reduced to a similar proportion as in the drug-treated cells. The more intriguing observation was the effect on the relative levels of M4 and M3, which was the opposite to the effect of A22 causing a further reduction of M4 instead of an accumulation (Table 2). It has to be noted that while A22 acts fast and inhibits all MreB molecules simultaneously, MreB depletion is probably a progressive and slow process, in particular in an organism with such a long generation time as Leptospira. This difference could explain why the effects on PG composition were not identical, although the trends were similar.
DISCUSSION
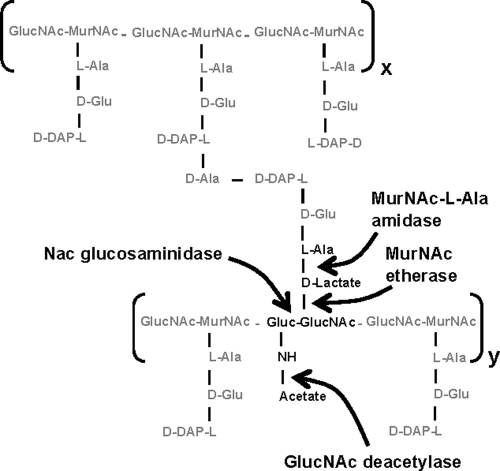
Cell shape determination and maintenance have been studied until now mainly in rod-shaped organisms such as E. coli and Bacillus subtilis or in the crescent-shaped organism C. crescentus. Spirochetes have a unique morphology, ranging from helix- to flat wave-shaped cells (12). It is not clear why some spirochetes form helices and others flat waves. It has been shown that endoflagella is sufficient to determine the flat wave morphology of B. burgdorferi. In contrast to B. burgdorferi, the endoflagella in Leptospira do not cover the entire cell length, and mutants deficient in endoflagella retain their helical morphology (45). The saprophytic and pathogenic Leptospira strains share the same morphological features, except that pathogenic strains are usually shorter than saprophytic strains (18). Our results show that the helical morphology of Leptospira is intrinsic to the PG macromolecule, unlike in B. burgdorferi (35). Similar to Leptospira spp., purified sacculi from Treponema pallidum examined by electron microscopy indicated that the PG contributes to the spiral cell shape (48, 55). Our work shows that although its PG conforms to a common Gram-negative chemotype, the constituents of this macromolecule are peculiar. For example, the fact that muropeptides with terminal d-Ala residues were so scarce in L. biflexa and L. interrogans PG is quite unusual and suggests the presence of specific peptidases, such as the tandem action of dd- and ld-carboxypeptidases or an ld-endopeptidase able to accept M5 as a substrate. Interestingly, a similar occurrence has been documented in the Gram-positive bacterium B. subtilis (1). Detection of M4-3, M4-3L, D4-3(−GNA) and D4-3(−GNA)(−Ac) is rare in the PG of other species. Enzymes potentially able to produce each of them from D4-3 subunits in the sacculus have been described in other systems and are indicated in Fig. 7. Muropeptide M4-3 is normally the product of a MurNAc-l-alanyl-amidase. These enzymes are often found in Gram-negative bacteria, where they play an important role in division, but their products rarely accumulate in macromolecular PG (58). Interestingly amidase activity seems to be upregulated in Salmonella enterica serovar Typhimurium following colonization of nonphagocytic cells (46). Generation of M4-3L from D4-3 would require the activity of an etherase. Such an enzyme designated MurQ has been described in E. coli. However, this enzyme is cytoplasmic and is only active on previously phosphorylated muropeptides in the course of PG recycling (54). Therefore, the putative enzyme responsible for generation of M4-3L might represent a new kind of PG hydrolase. Cleavage of the GlucNAc-MurNAc glycosidic bond by an N-acetylglucosaminidase is the likely origin of muropeptides lacking a GlucNAc residue (peaks 7 and 8). Such enzymes are widespread among bacteria. However, their role in Gram-negative bacteria is often restricted to PG recycling and requires soluble muropeptides as substrates (58). Interestingly, all the unusual muropeptides discussed above require cleavage of preexisting glycan strands or peptide bridges in macromolecular PG and therefore could cause local variations in the physical properties of PG. Peptidoglycan relaxation by hydrolytic enzymes has been found to promote helical shape in Helicobacter pylori (53). A similar process of modification of the organization of the network of peptidoglycan subunits may play a predominant role in the generation of the helical cell shape of Leptospira spp. (24). Subtle and notable differences were found in the muropeptide composition of the PG in the environmental species L. biflexa and the pathogen L. interrogans. The very low cross-linking in L. interrogans may reflect an adaptation to isosmotic conditions.
Fig. 7.
Representation of target sites in macromolecular PG for the enzymes proposed as origins of L. biflexa modified muropeptides. x and y indicate two glycan chains of undefined length.
Genome-wide analysis revealed that several shape-related proteins such as FtsZ, MreBCD, and RodA are present in Leptospira spp. We showed that MreB, one of the most abundant cytoplasmic proteins of L. interrogans, with 2,500 copies per cell (32), is involved in maintaining specific features of Leptospira morphology, such as its length, diameter in localized areas, and homogeneity of the pitch of the helix. This phenotype was similar to what we observed with penicillin G-treated cells. These observations suggest that the role of MreB in Leptospira is linked to at least one PBP. The localized increase in diameter could suggest that there is accumulation of PG synthesis in that area. B. subtilis mreB mutants form elongated and wider cells with bulging poles. It has been proposed that this phenotype was due to mislocalization of PBP1 in these cells and to its accumulation at the poles (27). It would be interesting to examine the localization of MreB and PBPs in healthy cells and in cells treated with various shape-altering compounds.
Among a library of random mutants (37), we identified transposon mutants exhibiting an insertion in genes encoding PBP1a and PBP3 (Table 1). Morphological alteration was only observed in the PBP3 mutant. The numerous HMW PBPs belonging to class A that are present in leptospires may compensate for the loss of PBP1a production. Like in E. coli (13), the L. interrogans PBP3 mutant forms filamentous cells, suggesting its involvement in cell division. However, unlike E. coli, FtsI does not appear to be essential in Leptospira (52). Surprisingly, LMW PBPs that generally act as carboxypeptidases or endopeptidases were not found in the Leptospira genomes, which suggests that other enzymes should undertake these functions, as has recently been shown in H. pylori (6). Among the antibiotics used to assess the role of various PBPs in the morphology of Leptospira, amdinocillin did not induce the expected phenotype, and cells were not affected in lateral peptidoglycan synthesis. It is possible that amdinocillin does not target the same PBP in Leptospira as in E. coli or that this PBP does not have the same role in this species. The antibiotics that we tested did not really affect the overall helical shape of the bacterium. Other proteins might be involved in the determination of this feature. Cytoplasmic filaments have been visualized using tomography in several spirochetes, including Leptospira spp. (29, 30, 32). However, little is known about the function of these elements. Previous findings in Treponema denticola suggest that they might be involved in cell division, structural integrity, motility, and/or chromosome structure and segregation (25). Genome-wide analysis of Leptospira spp. allowed for the identification of several putative coiled-coil proteins (33) that might be involved in shape determination similarly to CreS in C. crescentus.
In conclusion, our data provide the first description of the muropeptide content of Leptospira strains. We also showed that inactivation of mreB or ftsI resulted in mutants with morphological abnormalities such as aberrant diameter or length of the cells, therefore demonstrating that these proteins are important components of the morphology of Leptospira cells. Our results provide a starting point for a better understanding of the involvement of PBPs and cytoskeletal proteins in the morphogenesis of these atypical bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Stéphanie Guadagnini and Marie-Christine Prevost at the Plateforme de Microscopie Ultrastructurale for help with the electron microscopy. We also thank Gustavo Cerqueira for preliminary attempts to inactivate L. biflexa mreB.
This work was supported by the Institut Pasteur, Paris, France, and the French Ministry of Research ANR-08-MIE-018. M.A.D.P. was supported by Ministry of Education and Science, Spain (MEC, BFU2006-04574), and Fundación Ramón Areces.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Atrih A., Bacher G., Allmaier G., Williamson M. P., Foster S. J. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aviat F., Slamti L., Cerqueira G. M., Lourdault K., Picardeau M. 2010. Expanding the genetic toolbox for Leptospira species by generation of fluorescent bacteria. Appl. Environ. Microbiol. 76:8135–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ayala J., Goffin C., Nguyen-Distèche M., Ghuysen J. M. 1994. Site-directed mutagenesis of penicillin-binding protein 3 of Escherichia coli: role of Val-545. FEMS Microbiol. Lett. 121:251–256 [DOI] [PubMed] [Google Scholar]
- 4. Bauby H., Saint Girons I., Picardeau M. 2003. Construction and complementation of the first auxotrophic mutant in the spirochaete Leptospira meyeri. Microbiology 149:689–693 [DOI] [PubMed] [Google Scholar]
- 5. Bean G. J., et al. 2009. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48:4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonis M., Ecobichon C., Guadagnini S., Prevost M. C., Boneca I. G. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol. Microbiol. 78:809–819 [DOI] [PubMed] [Google Scholar]
- 7. Brenot A., Trott D., Saint Girons I., Zuerner R. 2001. Penicillin-binding proteins in Leptospira interrogans. Antimicrob. Agents Chemother. 45:870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabeen M. T., et al. 2009. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28:1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cabeen M. T., Jacobs-Wagner C. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601–610 [DOI] [PubMed] [Google Scholar]
- 10. Cabeen M. T., Jacobs-Wagner C. 2007. Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J. Cell Biol. 179:381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carballido-Lopez R., Formstone A. 2007. Shape determination in Bacillus subtilis. Curr. Opin. Microbiol. 10:611–616 [DOI] [PubMed] [Google Scholar]
- 12. Charon N. W., Goldstein S. F. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47–73 [DOI] [PubMed] [Google Scholar]
- 13. Curtis N. A., Eisenstadt R. L., Turner K. A., White A. J. 1985. Inhibition of penicillin-binding protein 3 of Escherichia coli K-12. Effects upon growth, viability and outer membrane barrier function. J. Antimicrob. Chemother. 16:287–296 [DOI] [PubMed] [Google Scholar]
- 14. Demarre G., et al. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156:245–255 [DOI] [PubMed] [Google Scholar]
- 15. den Blaauwen T., de Pedro M. A., Nguyen-Disteche M., Ayala J. A. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321–344 [DOI] [PubMed] [Google Scholar]
- 16. de Pedro M. A., Quintela J. C., Höltje J. V., Schwarz H. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellinghausen H. C., McCullough W. G. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45–51 [PubMed] [Google Scholar]
- 18. Ellis W. A., Hovind-Hougen K., Möller S., Birch-Andresen A. 1983. Morphological changes upon subculturing of freshly isolated strains of Leptospira interrogans serovar hardjo. Zentralbl. Bakteriol. Mikrobiol. Hyg. 255:323–335 [PubMed] [Google Scholar]
- 19. Georgopapadakou N. H., Smith S. A., Sykes R. B. 1982. Mode of action of aztreonam. Antimicrob. Agents Chemother. 21:950–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gitai Z., Dye N. A., Reisenauer A., Wachi M., Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341 [DOI] [PubMed] [Google Scholar]
- 21. Glauner B., Höltje J. V., Schwarz U. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088–10095 [PubMed] [Google Scholar]
- 22. Haake D. A., et al. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holt S. C. 1978. Anatomy and chemistry of spirochetes. Microbiol. Rev. 42:114–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang K. C., Mukhopadhyay R., Wen B., Gitai Z., Wingreen N. S. 2008. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:19282–19287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Izard J., Samsonoff W. A., Limberger R. J. 2001. Cytoplasmic filament-deficient mutant of Treponema denticola has pleiotropic defects. J. Bacteriol. 183:1078–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson R. C., Harris V. G. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 94:27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawai Y., Daniel R. A., Errington J. 2009. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71:1131–1144 [DOI] [PubMed] [Google Scholar]
- 28. Ko A. I., Goarant C., Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurner J., Frangakis A. S., Baumeister W. 2005. Cryo-electron tomography reveals the cytoskeleton structure of Spiroplasma melliferum. Science 307:436–438 [DOI] [PubMed] [Google Scholar]
- 30. Liu J., et al. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 403:546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) methods. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 32. Malmström J., et al. 2009. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460:762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazouni K., et al. 2006. The Scc spirochetal coiled-coil protein forms helix-like filaments and binds to nucleic acids generating nucleoprotein structures. J. Bacteriol. 188:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyagishima S. Y., Wolk C. P., Osteryoung K. W. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56:126–143 [DOI] [PubMed] [Google Scholar]
- 35. Motaleb M. A., et al. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. U. S. A. 97:10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murray C. K., Hospenthal D. R. 2004. Broth microdilution susceptibility testing for Leptospira spp. Antimicrob. Agents Chemother. 48:1548–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murray G. L., et al. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira spp. Infect. Immun. 77:810–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nelson D. E., Ghosh A. S., Paulson A. L., Young K. D. 2002. Contribution of membrane-binding and enzymatic domains of penicillin binding protein 5 to maintenance of uniform cellular morphology of Escherichia coli. J. Bacteriol. 184:3630–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nelson D. E., Young K. D. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neu H. C. 1983. Penicillin-binding proteins and role of amdinocillin in causing bacterial cell death. Am. J. Med. 75:9–20 [DOI] [PubMed] [Google Scholar]
- 41. Noguchi N., et al. 2008. Anti-infectious effect of S-benzylisothiourea compound A22, which inhibits the actin-like protein, MreB, in Shigella flexneri. Biol. Pharm. Bull. 31:1327–1332 [DOI] [PubMed] [Google Scholar]
- 42. Osborn M. J., Rothfield L. 2007. Cell shape determination in Escherichia coli. Curr. Opin. Microbiol. 10:606–610 [DOI] [PubMed] [Google Scholar]
- 43. Paster B. J., et al. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Picardeau M. 2008. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl. Environ. Microbiol. 74:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Picardeau M., Brenot A., Saint Girons I. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189–199 [DOI] [PubMed] [Google Scholar]
- 46. Quintela J. C., de Pedro M. A., Zollner P., Allmaier G., Garcia-del Portillo F. 1997. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol. Microbiol. 23:693–704 [DOI] [PubMed] [Google Scholar]
- 47. Quintela J. C., Pittenauer E., Allmaier G., Aran V., de Pedro M. A. 1995. Structure of peptidoglycan from Thermus thermophilus HB8. J. Bacteriol. 177:4947–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Radolf J. D., Moomaw C., Slaughter C. A., Norgard M. V. 1989. Penicillin-binding proteins and peptidoglycan of Treponema pallidum subsp. pallidum. Infect. Immun. 57:1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ristow P., et al. 2008. Biofilm formation by saprophytic and pathogenic leptospires. Microbiology 154:1309–1317 [DOI] [PubMed] [Google Scholar]
- 50. Schleifer K. H., Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spratt B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 72:2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Spratt B. G. 1977. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J. Bacteriol. 131:293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sycuro L. K., et al. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141:822–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Uehara T., Suefuji K., Jaeger T., Mayer C., Park J. T. 2006. MurQ etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J. Bacteriol. 188:1660–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Umemoto T., et al. 1981. Chemical and biological properties of a peptidoglycan isolated from Treponema pallidum kazan. Infect. Immun. 31:767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vicente M., Gomez M. J., Ayala J. A. 1998. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell. Mol. Life Sci. 54:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vollmer W., Blanot D., de Pedro M. A. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167 [DOI] [PubMed] [Google Scholar]
- 58. Vollmer W., Joris B., Charlier P., Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259–286 [DOI] [PubMed] [Google Scholar]
- 59. Yanagihara Y., et al. 1984. Chemical compositions of cell walls and polysaccharide fractions of spirochetes. Microbiol. Immunol. 28:535–544 [DOI] [PubMed] [Google Scholar]
- 60. Young K. D. 2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70:660–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.