Abstract
Under conditions of nutrient limitation and high population density, the bacterium Bacillus subtilis can initiate a variety of developmental pathways. The signaling systems regulating B. subtilis differentiation are tightly controlled by switch proteins called Raps, named after the founding members of the family, which were shown to be response regulator aspartate phosphatases. A phr gene encoding a secreted pentapeptide that regulates the activity of its associated Rap protein was previously identified downstream of 8 of the chromosomally encoded rap genes. We identify and validate here the sequence of an atypical Phr peptide, PhrH, by in vivo and in vitro analyses. Using a luciferase reporter bioassay combined with in vitro experiments, we found that PhrH is a hexapeptide (TDRNTT), in contrast to the other characterized Phr pentapeptides. We also determined that phrH expression is driven by a promoter lying within rapH. Unlike the previously identified dedicated σH-driven phr promoters, it appears that phrH expression most likely requires σA. Furthermore, we show that PhrH can antagonize both of the known activities of RapH: the dephosphorylation of Spo0F and the sequestration of ComA, thus promoting the development of spores and the competent state. Finally, we propose that PhrH is the prototype of a newly identified class of Phr signaling molecules consisting of six amino acids. This class likely includes PhrI, which regulates RapI and the expression, excision, and transfer of the mobile genetic element ICEBs1.
INTRODUCTION
In response to nutritional deprivation and high population density, the Gram-positive soil bacterium Bacillus subtilis can initiate several developmental pathways: sporulation, genetic competence, biofilm formation, and cannibalism (5–7, 15). In B. subtilis, the complex regulatory circuits controlling sporulation and competence initiation are highly connected and share important regulators, notably Spo0A and AbrB. Despite this, these two pathways are mutually exclusive, limiting the cells to a single differentiation process at any given time (9, 29).
RapH is a member of the Rap protein family that can control both sporulation and competence by acting on two distinct response regulator proteins: Spo0F and ComA, respectively (29). At the molecular level, RapH exhibits two distinct modes of action. It represses sporulation by dephosphorylating the intermediate response regulator Spo0F, modulating phosphate flow through the phosphorelay and ultimately the level of Spo0A phosphorylation (29). RapH also inhibits competence, by sequestering ComA, preventing it from binding to its target promoters (29). In fact, RapH was shown to be involved in the temporal separation of late stage competence and sporulation gene expression (29).
An additional level of complexity controlling B. subtilis differentiation derives from the modulation of Rap protein activity by specific peptides encoded by the phr genes (25). Most of the rap genes, with the exception of rapB, rapD, and rapJ (22), are transcriptionally coupled with a phr gene encoding a propeptide that regulates the associated Rap protein activity (26). It is known that once produced in the cell, the propeptides enter an export-import circuit. Proteolytic processing generates the mature pentapeptides (31), which are internalized by an oligopeptide permease system (18). Once in the cytosol, the mature pentapeptides can bind their associated Rap proteins (e.g., PhrA binds to RapA) and regulate their activities.
phr genes encoding putative or identified secreted peptides that regulate the cognate Rap protein activities are found downstream from 8 of the 11 chromosomal rap genes of B. subtilis. With the exception of phrA and phrH, all of these previously identified phr genes are known to be read from σH promoters embedded in their cognate rap genes, as well as from a promoter located upstream of the associated rap (19). Although PhrH activity has been demonstrated in vivo (29), the mature peptide itself has not been identified or characterized. RapH overexpression strongly inhibits the transcription of rapA, which is activated by ComA∼P and induces the transcription of abrB, which is repressed by Spo0A∼P (29). These observations are consistent with the RapH activities described above—the dephosphorylation of Spo0F∼P and the sequestration of ComA. The transcription of genes dependent on ComA and Spo0F (via Spo0A∼P) for their transcription therefore provides a readout for the interaction of RapH with ComA and Spo0F∼P. Simultaneous overexpression of PhrH with RapH partially restores abrB expression, suggesting that PhrH is able to counteract RapH activities (29).
In this report, we use B. subtilis luciferase reporter bioassays to monitor phrH expression or detect PhrH activity in vivo. We show that a promoter within rapH drives phrH expression and that in both sporulation and competence-inducing media, PhrH peptide is secreted in supernatants of growing B. subtilis cultures. Using alanine-scanning mutagenesis and synthetic peptides, we demonstrate that PhrH is a hexapeptide (TDRNTT). In vivo and in vitro approaches show that the synthesized hexapeptide antagonizes RapH dephosphorylation of Spo0F and confirm that TDRNTT modulates the RapH activity toward ComA. These studies demonstrate that Phr peptides are not exclusively pentapeptides and that the TDRNTT hexapeptide is not only able to inhibit the dephosphorylation of Spo0F by RapH but can also modulate its sequestration of ComA.
MATERIALS AND METHODS
Strains and plasmids.
B. subtilis strains (see Table S1 in the supplemental material) were constructed by transformation into BD630 (his leu met), and all of the strains are therefore isogenic. The details of strain and plasmid constructions are presented in the supplemental material. When it was desired to combine the constructs described below, this was performed by transformation, with selection for the appropriate antibiotic resistance marker. For transformation, competent cultures were prepared and incubated in competence medium (CM) with transforming DNA (1 μg/ml) for 30 min at 37°C (1).
The overexpression of rapH, phrH, or rapH and phrH was achieved by cloning each open reading frame amplified by PCR generated with the oligonucleotide pairs (RapH-SalI-FWD and RapH-SphI-REV, PhrH-SalI-FWD and PhrH-SphI-REV, and RapH-SalI-FWD and PhrH-SphI-REV, respectively) into the SalI and SphI sites of pDR111 (kindly provided by David Rudner) downstream of the Phyperspank promoter. The plasmids were then introduced by a double-crossover event into the ectopic amyE locus of the B. subtilis chromosome.
Media and growth conditions.
The media used in all of the in vivo experiments (luciferase assay) were either DSM (28) or competence medium (1) supplemented, when necessary, with 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Construction of deletions.
To inactivate B. subtilis genes, we replaced them cleanly with antibiotic cassettes without using a vector. This method was used for the knockouts of the rghR, rapH, and phrH genes. All of the PCR primers used in the present study are listed in Table S2 in the supplemental material. We first amplified 1-kb fragments upstream and downstream of the gene. These fragments are each flanked with one restriction site at the junctions with the start and the stop codons of the gene. In parallel, we amplified an antibiotic cassette flanked with the same restriction sites. The three fragments were then digested and ligated together. The ligated DNA was then purified through a QIAquick column (Qiagen) and the desired product, produced by ligation of the three fragments, was purified from an agarose gel. This fragment was then amplified by PCR using the outside primers previously used to amplify the upstream and downstream fragments. After further purification on QIAquick columns, the full fragment (upstream + antibiotic cassette + downstream) was used to transform B. subtilis, yielding a double-crossover event between the chromosome and the region of homology, replacing the gene with the antibiotic cassette.
Construction of luciferase promoter fusion strains.
A 1-kb fragment ending with the initiating codon of the gene of interest and containing the promoter was amplified by PCR from the B. subtilis chromosome using the primers spoIIGA1 and spoIIGA2 for PspoIIG, rapH7 and rapH8 for PrapH, srfA1 and srfA2 for PsrfA, and phrH7 and phrH8 for PphrH. A single nucleotide was inserted in the primer to restore the correct reading frame. The primers are listed in Table S2 in the supplemental material. In the case of PspoIIG and PsrfA, the 1-kb fragment was cut by KpnI/NcoI in sites present at the extremities of the primers used for the amplification. In parallel, the luciferase gene was cut from the pGL3 plasmid (Promega) by NcoI/BamHI digestion. A three-fragment ligation was then carried out between the promoter of interest, the luciferase gene, and plasmid pUC18Cm digested with KpnI and BamHI. The resulting plasmid, pCU18Cm-promoter::luc, which cannot replicate autonomously in B. subtilis was used to transform B. subtilis where it integrated, by a single crossover. This event reconstructs the “normal” regulatory region in front of the fusion and a complete copy of the gene of interest, downstream of the fusion. In the case of PrapH and PphrH, the 1-kb fragment was cut by EcoRI/NcoI in sites present at the extremities of the primers used for the amplification. In parallel, the luciferase gene was cut from the pGL3 plasmid (Promega) by NcoI/BamHI digestion. A three-fragment ligation was then carried out between the promoter of interest, the luciferase gene and plasmid pDR111 (Spc) digested with EcoRI and BamHI. The resulting plasmid, pDR111-promoter::luc was used to transform B. subtilis, where it integrated at the amyE locus, by a double crossover.
Luciferase assay.
For the detection of luciferase activity, strains were first grown in LB medium to an optical density at 600 nm (OD600) of 2. The cells were then centrifuged and resuspended in fresh sporulation medium (DSM [28]) or CM (1), adjusting all of the cultures to an OD600 of 2. These precultures were then diluted 20-fold in fresh DSM or competence medium and 200 μl was distributed into each of two wells in a 96-well black plate (Corning). Then, 10 μl of luciferin was added to each well to reach a final concentration of 1.5 mg/ml (4.7 mM). The cultures were incubated at 37°C with agitation in a Perkin-Elmer Envision 2104 multilabel reader equipped with an enhanced sensitivity photomultiplier for luminometry. The temperature of the clear plastic lid was maintained at 38°C to avoid condensation. The relative luminescence units and OD600 were measured at 1.5-min intervals. Each curve is representative of at least three independent experiments performed in duplicate.
mRNA extraction and rapH or phrH transcription start mapping.
To obtain freshly growing cells from the strains BD5510 (PrapH::luc) or BD5509 (PphrH::luc), overnight cultures grown at 30°C in LB were diluted 20-fold into fresh CM. After 2 h (for BD5509) or 3 h (for BD5510) of growth, 15-ml samples were taken, rapidly chilled, pelleted by centrifugation for 10 min at 4°C, and resuspended in 1 ml of RNApro solution (MPbio). RNA was then extracted by using FastRNA Pro Blue Kit (MPbio). 5′-RACE (5′ rapid amplification of cDNA ends) PCR was carried out using a 5′-RACE kit (Invitrogen). Sequences of the luciferase specific primers (GSP1-luc and GSP2-luc) used for the mapping are shown in Table S2 in the supplemental material. The final PCR products obtained with the 5′-RACE kit were separated by gel electrophoresis, purified and sequenced, using the primer seq+1-luc (see Table S2 in the supplemental material).
Protein expression and purification, Spo0F labeling, and in vitro phosphatase assays were performed as described previously (21), except that the final reaction contained: 6.5 to 1,300 μM TDRNTT or 1,300 μM DRNTT, 6.5 μM RapH, 6.0 μM radiolabeled Spo0F∼P, 24 μM Spo0F, 2.85 μM KinA, 14.55 mM Tris (pH 8.0), 50 mM EPPS (pH 8.5), 0.1 mM EDTA, 100 mM KCl, 23 mM MgCl2, 3 mM dithiothreitol (DTT), 11.6% glycerol, 0.04 μM [γ-32P]ATP, and 1 mM ATP.
Purification of ComA.
ComA was overexpressed and purified from E. coli strain BL21(DE3)pLysS as an N-terminal, hexahistidine fusion protein (pET28b; Novagen). Briefly, cells were lysed in 20 mM Tris (pH 8.0), 300 mM NaCl, 5 mM MgCl2, 10 mM β-mercaptoethanol, and 5% glycerol supplemented with 20 mM imidazole, 20 μg of DNase I/ml, and 20 mM phenylmethylsulfonyl fluoride, and insoluble material was pelleted at 22,000 rpm. Cleared lysates were purified by Ni-affinity chromatography using a gradient of 25 to 500 mM imidazole, followed by anion-exchange chromatography (MonoQ HR5/5; Pharmacia) using a linear gradient of 50 to 850 mM NaCl. ComA-containing fractions were pooled, concentrated by ultracentrifugation (molecular mass cutoff, 10 kDa; Amicon), and stored at −80°C.
Peptide synthesis.
The synthesized peptides—TDRNTTY, TDRNTT, DRNTTY, TDRNT, and DRNTT—were obtained from LifeTein. The peptides were resuspended in 10 mM Tris (pH 7.6). The concentrations were adjusted by resuspending the lyophilized peptides (weight provided by LifeTein) in the appropriate buffer volume.
Electrophoretic mobility shift assay (EMSA).
A 323-bp DNA fragment comprising 308 bp from the srfA promoter region (from −185 to +123) (20) and unique EcoRI and HindIII restriction sites was amplified by using the primers pSrfAA-HindIII-F and pSrfAA-EcorI-R and Phusion polymerase. The resulting fragment was digested with EcoRI, end labeled with T4 polynucleotide kinase (NEB) and [γ32P]ATP (MP Biomedicals), and purified from unincorporated label by centrifugation through a Micro Biospin 30 column (Bio-Rad). Samples containing either (i) ComA, (ii) ComA and RapH, or (iii) ComA, RapH, and synthesized peptides were incubated in binding buffer [25 mM HEPES (pH 7.5), 25 mM KCl, MgCl2, 1 mM DTT, 1 mM EDTA, 10% glycerol, 0.2 mg of poly(dI-dC) (Sigma)] for 15 min at 4°C. The radiolabeled PsrfA probe was then added to a final concentration of 0.1 or 1 nM as indicated, and the samples were incubated at room temperature for 15 min. Glycerol was then added to each sample to a final concentration of 10%, and the complexes were separated on a nondenaturing 5% polyacrylamide gel (Bio-Rad) running in 0.5× Tris-borate-EDTA (TBE) buffer at 300 V for 3 min and then 150 V for 65 min. The gels were then visualized by phosphorimager analysis.
RESULTS
phrH is transcribed from two promoters.
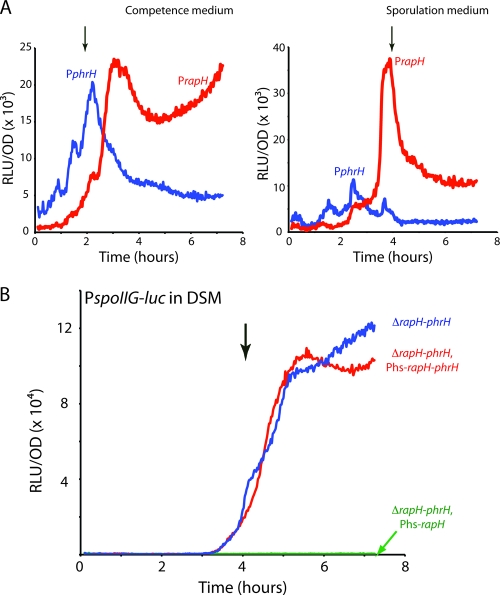
In order to study the phrH transcription profile, we used the luciferase reporter gene from the firefly Photinus pyralis. Although phrH was known to exist in a transcription unit coupled with rapH (K. Fujii, K. Asai, and N. Ogasawara, unpublished results, quoted in reference 10), we investigated the possibility that a second promoter may initiate the transcription of this downstream gene. For this, we studied the transcription profile from both a 1-kb fragment corresponding to the promoter upstream of the rapH gene (BD5510, PrapH-luc) and a 700-bp fragment directly upstream of the phrH gene (BD5509, PphrH-luc). In both competence and sporulation-inducing media the transcription rate from the PrapH-luc fusion increased in the middle of the exponential phase with a decrease beginning about 2 h after the transition between the exponential and stationary phases (Fig. 1 A and B). Transcription was also detected from the phrH fusion, confirming the existence of a second promoter. The expression pattern of PphrH was different from that of PrapH, increasing earlier and reaching a maximum at the entrance to stationary phase, followed by a decrease and then by a sustained expression until the end of the experiment (Fig. 1A and B). The patterns in the two media are similar, except that the ratio of transcription from PrapH to that of PphrH was higher in DSM (sporulation medium) than in competence medium. Further evidence for the independent regulation of rapH and phrH, and hence for the existence of two promoters, was obtained by the introduction of a rghR knockout. RghR (10) is known to repress the transcription of rapH and, indeed, the inactivation of rghR resulted in an increase of the transcription rate from PrapH in both DSM and competence medium, but had no effect on the transcription rate from PphrH (see Fig. S2 in the supplemental material).
Fig. 1.
Transcription from the PrapH, PphrH, and PspoIIG promoters. (A) The blue curve shows the relative luminescence readings corrected for the OD for the PphrH promoter (BD5509) and the red curve shows the same for the PrapH promoter (BD5510). (B) Effect of rapH or rapH plus phrH overexpression on expression of the PspoIIG promoter. The relative luminescence readings corrected for the OD for the PspoIIG promoter are presented when rapH and phrH are knocked out and rapH alone (BD5190 in green), rapH and phrH (BD5263 in red) or none of them (BD5035 in blue) are overexpressed. T0, the transition between growth and stationary phase, is indicated by the downward-pointing arrows. Phs stands for the IPTG-inducible hyperspank promoter (kindly provided by D. Z. Rudner).
Using 5′-RACE as described in Materials and Methods, we mapped the transcription start of the rapH promoter, which is downstream from a likely SigA promoter (see Fig. S1A in the supplemental material). Inspection of the DNA sequence upstream of phrH revealed no obvious σH-dependent promoter, and the expression pattern of PphrH was unaffected by inactivation of sigH (see Fig. S1B in the supplemental material). Instead, putative −35 and −10 hexanucleotide core elements characteristic of σA-dependent promoters (11) were noted upstream from the phrH coding sequence (see Fig. S1A in the supplemental material). However, using the 5′-RACE procedure, we were unable to determine the phrH start site. This may reflect a lower abundance of the phrH transcript.
We conclude that rapH and phrH are both transcribed from the SigA-driven rapH promoter (10), although phrH is also transcribed from a putative SigA-driven promoter embedded within the rapH gene sequence.
An in vivo assay for RapH and PhrH activity.
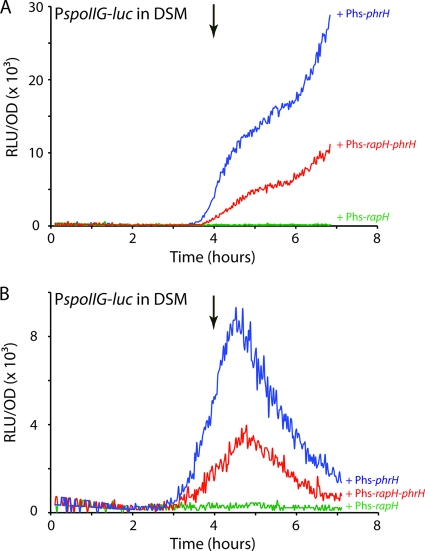
To identify PhrH and determine whether it is a secreted signal involved in cell-cell signaling, we developed an in vivo assay for PhrH activity using the P. pyralis luciferase gene as a reporter under the control of the Spo0A-driven promoter PspoIIG (Fig. 1B). As expected, the expression of PspoIIG in DSM was undetectable during the three first hours of growth in the strain deleted for both rapH and phrH, and it increased at the transition between exponential and stationary phase because it requires a high level of Spo0A∼P (8). Expression was identical in a wild-type background (not shown).
The overexpression of rapH from the Phyperspank (Phs) promoter in the presence of IPTG completely abolished PspoIIG-luc expression (Fig. 1B) (21). This was expected because RapH dephosphorylates Spo0F, draining phosphoryl groups from Spo0A∼P. When both rapH and phrH were overexpressed from Phs, the rate of PspoIIG-luc transcription was restored to about the level exhibited in the absence of overexpression, confirming that PhrH was produced and active (Fig. 1B).
PhrH is exported to the extracellular environment.
We next determined whether PhrH is exported into the medium and works in trans to inhibit the activity of RapH in cells that are mutant for phrH. For this, we conducted mixing experiments in which “donor” strains lacking a luc reporter and either expressing or not expressing phrH were cocultured with a “recipient” strain (BD5135) that carries the PspoIIG-luc reporter, a knockout of phrH and overproduces RapH.
When the donor strain did not express phrH (BD5190), the overexpression of rapH in the recipient strain inhibited PspoIIG transcription, as expected (Fig. 2 A). In contrast, when rapH was overexpressed in the recipient strain and rapH and phrH or phrH alone were expressed in the donor strain (BD5191 and BD5192, respectively), the inhibitory effect of the peptide on RapH activity could be detected in the recipient strain as a partial restoration of PspoIIG-luc transcription (Fig. 2A). In this experiment, PspoIIG expression was greater when phrH was expressed alone from the Phs promoter than when coexpressed with rapH. This may be due to the greater expression of phrH when it is proximal to the Phs promoter. The maximal restoration of PspoIIG-luc expression detected was ∼4-fold less than that in Fig. 1, although the expression in Fig. 2A was still increasing when the experiment was terminated, and a 2-fold decrease was expected because only half the cells were expressing the reporter. These results confirm that PhrH is matured and exported outside the cells. However, PhrH could either be exported and diffuse from donor to recipient cells or could be localized on the donor cell surface where it could be sensed by cell-cell contact.
Fig. 2.
PhrH activity is released in the growth medium. (A) Effect of the overproduction of PhrH from a donor strain on the PspoIIG promoter in a recipient strain in cocultures. The relative luminescence readings corrected for the OD for the PspoIIG promoter in the recipient strain (BD5135) is presented while growing with a donor strain overexpressing rapH alone (BD5190), rapH and phrH (BD5191), or phrH alone (BD5192) in green, red, or blue, respectively. (B) Effect of donor strain supernatants on the PspoIIG promoter in a recipient strain. The relative luminescence readings corrected for the OD are presented for the PspoIIG promoter in the recipient strain (BD5135) growing in the presence of 1:1 diluted supernatants from stationary-phase cultures of a donor strain overexpressing rapH alone (BD5190), rapH and phrH (BD5191), or phrH alone (BD5192) in green, red, and blue, respectively. T0, the transition between growth and stationary phase, is indicated by the downward-pointing arrows. Phs stands for the IPTG-inducible “hyperspank promoter.”
To determine whether PhrH is secreted into the medium, we grew the recipient strain (BD5135) in fresh DSM mixed with equal volumes of culture supernatants prepared from different “donor” strains (BD5190, BD5191, and BD5192) also grown in DSM (Fig. 2B). Consistent with the data in Fig. 2A, we could only detect a restoration of PspoIIG expression in conditioned media from cultures expressing PhrH. In Fig. 2 the rates of PspoIIG transcription obtained with the three donor strains are ranked in the same order (Phs-phrH > Phs-rapH-phrH > Phs-rapH), but the kinetics differ, probably reflecting the exhaustion or proteolysis of PhrH in the conditioned medium. Although we cannot rule out the possibility that cell-cell contacts contribute in the wild-type situation, we conclude from these results that the PhrH peptide is exported into the extracellular environment and probably imported back into the recipient strain, where it antagonizes RapH.
PhrH may not be a pentapeptide.
The phr genes encode Phr precursors that are generally small proteins of approximately 40 amino acids with typical structural features of exported proteins, i.e., N-terminal signal peptide sequences consisting of a few positively charged residues, followed by a hydrophobic region (27). Most of the characterized matured Phr peptides consist of five residues derived from the C terminus of the precursor protein, but in a number of cases the pentapeptide is derived from an internal portion of the precursor peptide, e.g., PhrE (13) and PhrK (24). In each case, the second amino acid of the pentapeptide is positively charged (K or R) (24).
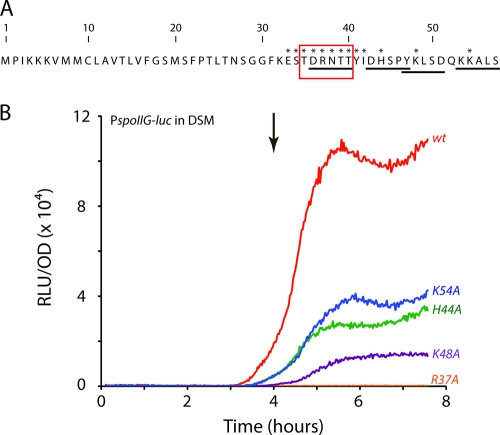
Based on this last consideration, four potential pentapeptides (DRNTT, YKLSD, DHSPY, and KKALS) were identified within the PhrH propeptide sequence (Fig. 3 A). To determine whether any of these candidate peptides might correspond to the mature PhrH, each of the second position residues was changed to alanine and the resulting PhrH mutants were tested for their ability to counteract the inhibitory effect of RapH overexpression on PspoIIG transcription (Fig. 3B). The parent strain (BD5263) for these constructions carried PspoIIG-luc, a deletion of both rapH and phrH and Phs-rapH-phrH in the ectopic amyE locus. The mutations were introduced into this ectopic copy of phrH. As shown above (Fig. 2A), when both rapH and phrH were overexpressed, the wild-type mature peptide was able to inhibit RapH activity, allowing Spo0A phosphorylation and ultimately PspoIIG expression. If the mutation that we introduced in PhrH inactivated the mature peptide, PspoIIG transcription would remain low. As shown in Fig. 3B, the R37A mutation completely inactivated PhrH activity, suggesting that DRNTT might be the sequence of the mature PhrH or at least that the biologically relevant sequence is located near the R37 residue. Effects on processing of the propeptide or on its in vivo stability could explain the reduction in activity due to the K54A, H44A, and K48A mutations. Taken together, these results suggest that, like PhrE and PhrK, PhrH does not correspond to the five C-terminal residues but may be generated from an internal sequence within phrH.
Fig. 3.
In vivo tests of potential pentapeptides for PhrH activity. (A) Sequence of the PhrH propeptide. The potential pentapeptides (DRNTT, YKLSD, DHSPY, and KKALS) are underlined. Asterisks indicate the amino acids that were individually mutated. (B) Effects of single mutations in the conserved second amino acid of the four potential pentapeptides on the PhrH activity. The control curve (wt) represents the relative luminescence readings corrected for the OD for the PspoIIG promoter when wild-type rapH and phrH are both overexpressed (BD5263, in red). The remaining curves show the relative luminescence readings corrected for OD for the PspoIIG promoter when the R37 (in orange), K48 (in violet), H44 (in green), or K54 (in blue) residues are changed to alanine. T0 is indicated by the downward-pointing arrow.
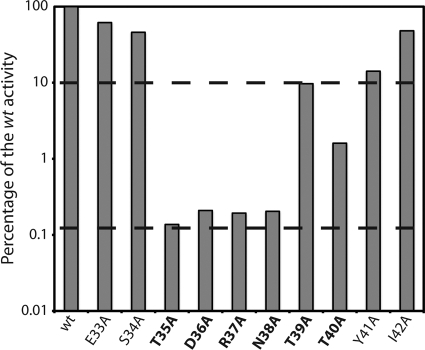
To further pinpoint the mature PhrH peptide boundaries, we extended the alanine scanning to the entire region around the DRNTT sequence. In addition to each amino acid within this sequence, we also mutated the three residues immediately upstream from the N terminus and the two residues downstream from the C terminus of the putative pentapeptide (Fig. 3A) and analyzed their abilities to counteract the effect of RapH overproduction on PspoIIG transcription (Fig. 4). The semilogarithmic histogram in Fig. 4 shows that mutation of residues 35 to 41 reduces the activity to ca. 10% or less of the wild-type peptide. Mutations of residues 35 to 38 reduced PhrH activity to the background level (ca. 0.1% of the wild-type activity). These data show that the DRNTT residues are indeed important to regulate RapH activity, as are residues that flank this sequence. The effects of the flanking mutations may be due to interference with processing, or these flanking residues may be part of the mature PhrH peptide.
Fig. 4.
Alanine scanning mutagenesis of the putative PhrH peptide coding sequence. The wild-type (wt) reference shows the level of the PspoIIG promoter activity after 5.5 h of growth in DSM when rapH and phrH are both overexpressed (BD5263). The effect of each mutation is presented as a percentage of the wild-type activity. The single mutations that depress the PspoIIG activity to less than 10% of the wild-type reference are highlighted in boldface type. The upper and lower horizontal dashed lines show the 10% activity and the background (zero activity) level, respectively, when only rapH is overexpressed. For the full curves of expression see Fig. S3 in the supplemental material.
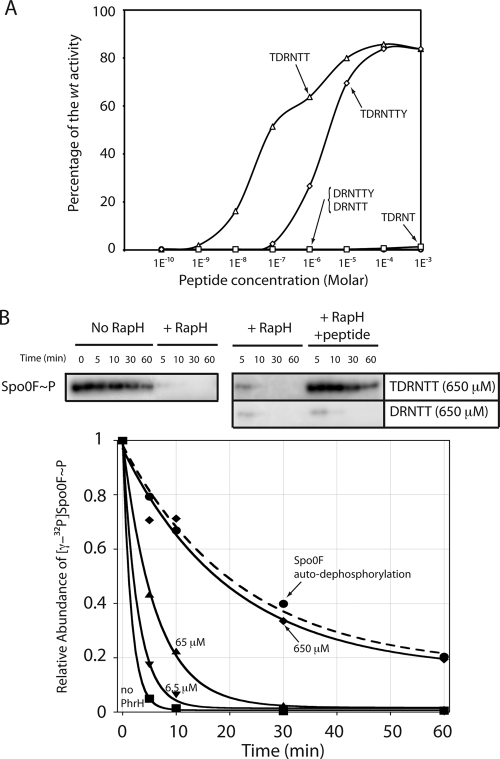
To test the first possibility, we examined the effect of synthetic DRNTT in vivo and in vitro (Fig. 5). When added to growth medium at a concentration as high as 1 mM, this peptide could not counteract the effect of RapH overproduction on PspoIIG expression (Fig. 5A). The synthesized DRNTT pentapeptide was also tested in vitro for its ability to inhibit the dephosphorylation of Spo0F∼P (Fig. 5B). Again, no inhibition was noted even at a concentration of 650 μM. These results strongly suggested that, unlike the other characterized Phr peptides, PhrH might not be a pentapeptide.
Fig. 5.
In vivo and in vitro effects of the synthesized peptides. (A) In vivo effects. The effects on PspoIIG promoter activity in DSM, when only rapH is overexpressed, of the indicated molar concentrations of synthesized TDRNTTY (diamonds), TDRNTT (triangles), DRNTTY (circles), and DRNTT (squares) are presented as a percentage of the wild-type activity. The wild-type reference is the level of PspoIIG promoter activity after 5.5 h of growth when both rapH and phrH are overexpressed. For the full curves of expression, see Fig. S4 in the supplemental material. (B) In vitro inhibition of RapH-mediated Spo0F dephosphorylation by synthesized peptides. 6 μM 32P-labeled Spo0F∼P, 6.5 μM RapH, and PhrH at the indicated concentrations were incubated at 25°C. The autodephosphorylation of Spo0F, in the absence of RapH (•) (labeled Spo0F autodephosphorylation) was similarly measured at 25°C. Aliquots were removed from the reaction at the indicated time points and visualized by phosphorimaging. The concentrations of TDRNTT used were as follows: 0 μM (labeled no PhrH) (▪), 6.5 μM (▾), 65 μM (▴), and 650 μM (⧫). Phosphatase activity data were fit to exponential curves using KaleidaGraph software (Synergy Software). The dashed curve shows autodephosphorylation. Gels depicting Spo0F dephosphorylation in the absence of RapH, the presence of RapH, or the presence of RapH plus peptide are also presented. The additional gels used to construct this figure are shown in Fig. S5 in the supplemental material.
The PhrH hexapeptide TDRNTT inhibits RapH dephosphorylation of Spo0F both in vivo and in vitro.
We initially hypothesized that the loss-of-function displayed by PhrH-T35A and PhrH-Y41A in vivo resulted from inefficient proteolytic cleavage. However, when we subsequently showed that the DRNTT pentapeptide does not possess PhrH activity using an in vitro phosphatase assay, we considered the possibility that PhrH was the pentapeptide TDRNT or in fact not a pentapeptide but rather a hexapeptide (TDRNTT or DRNTTY) or a heptapeptide (TDRNTTY) (Fig. 5A; see also Fig. S4 in the supplemental material). Indeed, the synthetic TDRNTT hexapeptide counteracts the inhibitory effect of RapH on PspoIIG activity in vivo, reaching nearly 100% of the activity observed when rapH and wild-type phrH are overexpressed together (Fig. 5A and see Fig. S4 in the supplemental material). Practically no activity was detected with the DRNTTY hexapeptide or the TDRNT pentapeptide (Fig. 5A). Finally, the TDRNTTY heptapeptide was 10- to 100-fold less active than TDRNTT. We concluded from these results that the PhrH sequence was TDRNTT and that this hexapeptide had an apparent 50% effective concentration (EC50) of 100 nM.
To determine whether the TDRNTT peptide directly inhibits RapH function, we measured its ability to inhibit RapH catalyzed dephosphorylation of Spo0F∼P in vitro. In the absence of TDRNTT, after 5 min 5% of radiolabeled Spo0F∼P remained in the presence of RapH compared to 80% in the no-RapH autodephosphorylation control reaction (Fig. 5B). However, low micromolar concentrations of TDRNTT detectably inhibited RapH-mediated Spo0F∼P dephosphorylation (see Fig. S5 in the supplemental material) and, in the presence of 650 μM TDRNTT, the reaction containing RapH was nearly indistinguishable from the no RapH control (Fig. 5B and see Fig. S5 in the supplemental material).
The PhrH hexapeptide TDRNTT inhibits RapH sequestration of ComA both in vivo and in vitro.
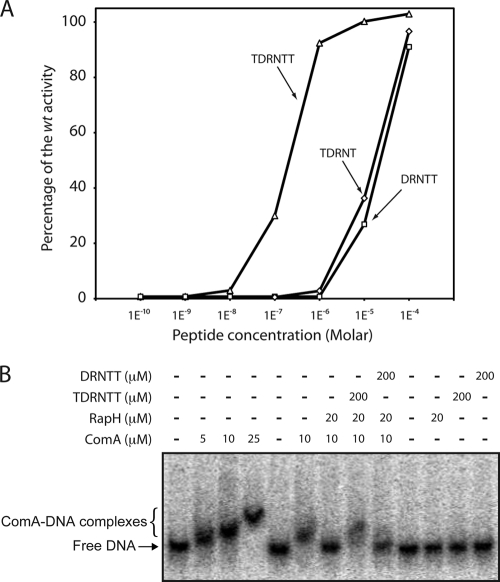
We next investigated whether the TDRNTT and DRNTT peptides could also modulate competence by interfering with the ability of RapH to inhibit ComA activity. During the development of competence, the transcription of the srfA operon is activated by the interaction of ComA∼P with PsrfA (20). The srfA operon encodes the anti-adaptor protein ComS that prevents degradation of the competence master regulator ComK (32). We therefore used a PsrfA-luc reporter to measure the ability of the two synthetic peptides to reverse the RapH inhibition of ComA binding to PsrfA analogously to the use of PspoIIG-luc as a reporter for the reversal of RapH activity toward Spo0F∼P. First, however, we performed a coculture experiment identical in design to the one shown in Fig. 2A, except that the PsrfA-luc reporter was used (not shown). In the recipient strain, the expression from PsrfA-luc was totally inhibited by the overexpression of rapH alone (BD5177). When cocultured with donor strains that were deleted for the rapH-phrH locus and overproducing either RapH alone (BD5190), RapH and PhrH (BD5191) or PhrH alone (BD5192), the PsrfA expression was only restored by the two strains that were wild type for phrH (not shown). This experiment verified that a producing strain secreted enough PhrH to prevent RapH inhibition of PsrfA-luc expression (not shown).
We then repeated this experiment without the donor strains but instead added the synthesized TDRNTT hexapeptide to the growth medium. The PsrfA expression was restored with the same range of peptide concentrations that restored PspoIIG expression (compare Fig. 5A and 6), although the apparent EC50 for the PsrfA-luc reporter was slightly higher than for PspoIIG-luc. Interestingly, when the DRNTT or TDRNT pentapeptides were added, some activity was detected, albeit at concentrations 100-fold higher than with TDRNTT. At the highest and probably nonphysiological concentration used (100 μM), DRNTT counteracted the effect of rapH overexpression, restoring the PsrfA activity to a level comparable to that obtained in the absence of Phs-rapH induction. As described below, existing data suggest that B. subtilis may not in fact produce DRNTT (17).
Fig. 6.
In vivo and in vitro effects of synthesized TDRNTT and DRNTT peptides on PsrfA-luc expression and srfA promoter fragment binding to ComA in the presence or absence of RapH. (A) PsrfA promoter activity in competence medium is shown after 1.9 h of growth when only rapH is overexpressed. The effects of several TDRNTT (▵) or DRNTT (□) concentrations are presented as a percentage of the wild-type activity at the same time. The wild-type reference is the level of the PsrfA-luc promoter activity when rapH and phrH are both overexpressed. The full expression curves are shown in Fig. S6 in the supplemental material. (B) EMSA performed as described in Materials and Methods using 0.1 nM radiolabeled srfA promoter fragment. A dose-response experiment of TDRNTT on the ComA DNA-binding in the presence of RapH is presented in Fig. S7 in the supplemental material.
Finally, we used an EMSA to determine whether the TDRNTT peptide directly antagonizes the interaction of RapH and ComA. Consistent with previous studies, we found that ComA bound stably to the PsrfA promoter and that RapH inhibited this interaction (Fig. 6 B) (29). However, when the reaction included the TDRNTT hexapeptide, ComA bound to PsrfA even in the presence of RapH (Fig. 6B). In contrast, the DRNTT pentapeptide did not disrupt the formation of RapH-ComA complexes at 200 or 500 μM concentrations (Fig. 6B and data not shown). Based on the extensive in vivo and in vitro analysis presented above, we conclude that the TDRNTT hexapeptide is active in antagonizing RapH activity toward both Spo0F∼P and ComA and that TDRNTT is a biologically important signal.
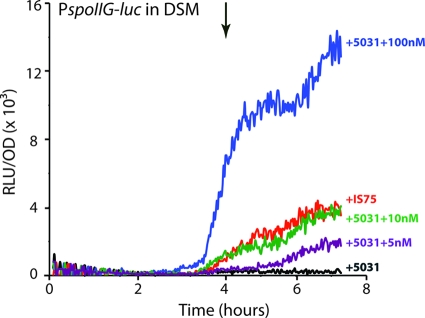
Concentration of secreted PhrH in B. subtilis cultures.
The previous experiments (Fig. 5 and 6) suggest that PhrH is active in vivo in the low nanomolar range even when RapH is overproduced. To evaluate the PhrH concentration in wild-type B. subtilis (IS75) culture supernatants, we conducted additional experiments using cocultured strains. PhrH activity was readily detected when the recipient strain (BD5135) was mixed with IS75 as the donor strain, which produces PhrH from its natural locus (Fig. 7). These results proved that the peptide produced and exported from IS75 could be detected in the recipient strain and that this level of activity could be used as a reference when synthesized peptide was added. For this, the recipient strain BD5135 was cocultured with a strain that was isogenic with IS75 but was deleted for rapH and phrH (BD5031). This “neutral” strain was used to mimic the growth conditions in the experiment just described in which IS75 was cocultured with BD5135. These mixed cultures were grown in the presence of IPTG to induce the rapH expression in BD5135, and to each culture a different amount of synthetic TDRNTT was added. As shown in Fig. 7, the kinetics and level of luminescence observed with 10 nM peptide was nearly identical to that obtained with the “natural producer,” IS75, suggesting that in DSM, the mature PhrH peptide is found in B. subtilis cultures at concentrations in the range of 10 to 20 nM. This concentration is similar to estimates made for other Phr peptides (23).
Fig. 7.
Determination of the PhrH concentration in B. subtilis supernatants. We compared the activity of the PspoIIG promoter in a recipient strain (BD5135) when mixed with IS75 (used as the donor strain, red curve) or when synthesized peptide (TDRNTT) is added at the indicated concentrations. When peptide was added the recipient strain was mixed with a strain carrying a knockout of both rapH and phrH (BD5031) to maintain the same concentration of cells. IPTG (0.25 mM) was added to all of the cultures to induce rapH in the recipient culture. T0, the transition between growth and stationary phase, is indicated by the downward-pointing arrows.
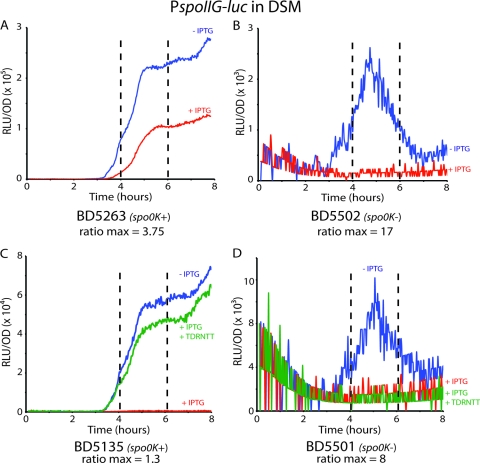
The Spo0K oligopeptide permease is required for PhrH activity.
The apparent greater size of PhrH in comparison to the previously characterized Phr pentapeptides raises the question whether the Spo0K ABC transporter is necessary for its activity. To test this, we reproduced the experiment shown in Fig. 1B in a spo0K background (Fig. 8). As shown in Fig. 1B and 8A, when rapH and phrH were both overexpressed in the spo0K+ background (strain BD5263), PhrH was able to antagonize RapH activity and allow the partial restoration of PspoIIG expression (Fig. 8A). In the spo0K background (BD5502), there was no detectable PspoIIG expression when rapH and phrH were induced (Fig. 8B). It is notable that in the absence of induction, PspoIIG expression is much lower in the spo0K mutant than in the wild-type (Fig. 8A and B), a finding consistent with the role of the oligopeptide permease in the uptake of Phr peptides other than PhrH, which antagonize the dephosphorylating activities of their cognate Rap proteins acting on Spo0F (RapA, RapB, RapE, and RapJ) (14, 21, 25). Comparison of the ratios of the induced and noninduced curves in the wild-type and spo0K backgrounds shows that without the permease, PhrH is not active. In other experiments, the addition of the synthesized TDRNTT peptide to a strain overexpressing rapH and lacking spo0K did not result in a detectable increase in spoIIG-luc expression (Fig. 8C and D). These experiments confirm that the oligopeptide permease is required for the activity of PhrH, most likely by transporting the hexapeptide.
Fig. 8.
PhrH activity requires the Spo0K oligopeptide permease. Panels A and B show the expression from the PspoIIG promoter when rapH and phrH are overexpressed (+IPTG, red) or not overexpressed (−IPTG, blue) in spo0K+ (BD5263, panel A) or spo0K-deficient (BD5502, panel B) backgrounds. In both cases, the maximal ratio between the IPTG plus and minus curves has been determined between the fourth and sixth hours. Panels C and D show the expression from the PspoIIG promoter when rapH is overexpressed in the absence (+IPTG, red) or presence (+IPTG+TDRNTT, green) of the hexapeptide or when not overexpressed (−IPTG, blue) in spo0K+ (BD5263, panel C) or spo0K-deficient (BD5502, panel D) backgrounds. In both cases, the maximal ratio between the +IPTG +TDNTT and −IPTG curves has been determined between the fourth and sixth hours.
DISCUSSION
PhrH is a hexapeptide.
The data presented here allowed us to define the sequence of a peptide with PhrH activity. Our data strongly support the conclusion that the minimal active peptide is not a pentapeptide but instead has the composition TDRNTT. This conclusion is supported by alanine scanning, the activity of the synthetic peptide, and the lack of activity exhibited by DRNTT at physiological concentrations. Moreover, the hexapeptide shows activity not only in vivo but also in vitro, demonstrating that the active form is not a modified derivative of the hexapeptide, formed within the cell following uptake. Thus, PhrH is unique among the characterized Phr peptide signals.
How Phr peptides antagonize their Rap protein targets is unknown. An X-ray crystal structure of a Rap protein in complex with a Phr peptide will reveal the mechanism of this regulation. While we recently showed that RapH is amenable to crystallographic analysis (21), structural studies of RapH-PhrH complexes were impossible without knowing the identity of the mature PhrH peptide. Now that the mature PhrH peptide (TDRNTT) has been identified, both crystallographic and biochemical studies of the RapH-PhrH interaction can be carried out. It will be particularly interesting to define the PhrH binding site and reveal the molecular interactions that enable Phr peptides to regulate their cognate Rap proteins with exquisite specificity despite the fact that the mature Phr peptide signals are only five or six amino acids in length.
The specificity of the known Phr peptides for Rap protein recognition is dependent upon the amino acid sequence of the peptides, and single amino acid substitutions can drastically affect their activity and specificity (23). Positions 2 and 5 of the known Phr pentapeptides are highly conserved as arginine and threonine, respectively. For PhrH, we confirmed the importance of the highly conserved R and T in positions 3 and 6, corresponding to positions 2 and 5 in the Phr pentapeptides, e.g., PhrA and PhrC (see Table S3 in the supplemental material). The residues at positions 1, 3, and 4 of Phr pentapeptides are more variable than those at positions 2 and 5, and they probably determine Rap protein target specificity (see Table S3 in the supplemental material). In the case of PhrA, modification of the fourth position did not completely abolish activity toward RapA (23). Modification of the T in position 5 of TDRNTT, corresponding to position 4 in PhrA, did not result in a complete loss of PhrH function (Fig. 4). Thus, PhrH retains the characteristic features governing the activity of the Phr pentapeptides, with the difference that there is an additional residue at its N terminus (see Table S3 in the supplemental material). The presence of an additional residue at the N terminus of PhrH, and perhaps PhrI as discussed below, means that the most conserved feature of Phr peptides is not a basic residue at the second position from the N terminus but rather a conserved basic residue at the fourth position from the C terminus.
We have shown that Spo0K is required for the activity of the hexapeptide PhrH, most likely by mediating its uptake (Fig. 8). The upper size limit for peptide transport by this permease has not been determined, but it is known to transport peptides ranging in size from 3 to possibly 8 amino acids (16, 33). In other Gram-positive bacteria the size limits for peptide transport by similar permeases is variable. For example, the transcriptional regulator PlcR in Bacillus cereus is regulated by the PapR signaling heptapaptide, which is imported back into the cell via the oligopeptide permease system (4). Streptococcus pneumoniae possesses an oligopeptide permease that functions in the uptake of peptides consisting of 2 to 7 residues (2), and a hexa-heptapeptide permease in Streptococcus gordonii has been identified (12).
Regulation of PhrH activity.
We have confirmed that a phr gene is found downstream from rapH as it is from seven other rap-phr gene pairs. All of these phr genes are transcribed by readthrough from their upstream rap gene promoters. With the apparent exception of phrA and phrH, phr genes are also transcribed from a dedicated SigH-dependent promoter that is internal to the rap gene (19). These results suggest that Rap protein activity can be controlled by the nutritional conditions that modulate SigH expression or activity. We have shown here that phrH is indeed transcribed from its own promoter, in addition to its reported readthrough transcription from PrapH, and that PphrH is uniquely not a SigH promoter but is probably σA dependent. The increase in PphrH-luc expression during growth and its sustained expression during stationary phase suggest that this promoter is regulated and the difference in the timing of expression between PrapH and PphrH (Fig. 1A) suggests that the two promoters are regulated differently. This conclusion is also supported by the differential effects of a rghR mutant on the two promoters.
In both competence and sporulation medium there is a peak of phrH transcription rate near the entrance to stationary phase. rapH transcription increases sharply somewhat later than the increase in phrH transcription and reaches a maximum at a later time. In the case of competence medium, part of this increase in rapH transcription must be due to ComK (29), and this part must be restricted to the competent subpopulation. The earlier expression of phrH in both media suggests that the system is designed to minimize the impact of RapH on its targets during growth, including on Spo0F and ComA. As cells transition to stationary phase, phrH transcription declines and that of rapH continues to increase. This may permit RapH to exert its influence on the rate of Spo0F (and hence Spo0A) phosphorylation and on the activity of ComA as cells enter stationary phase and make developmental decisions (29). The redundancy of rap/phr genes, the fact that the Phr peptides are likely to be secreted and hence shared by all of the cells, and the higher expression of rapH in a subpopulation at least in competence medium points to complexity, which is consistent with a role for these systems in fine-tuning development and developmental decision making.
Other potential regulatory inputs occur during processing and internalization of PhrH. In this respect, PhrH differs in three important ways from other Phr peptides. First, like PhrE and PhrK, PhrH is an internally generated peptide and an additional processing step must take place to generate its C terminus. Second, the final N-terminal processing step does not occur one residue upstream from the conserved R or K residue, and it is possible that a distinct protease is used for this process. Third, the internalization of this hexapeptide may differ from that of the pentapeptides. It may be that the need for an additional processing step causes a delay in the uptake of PhrH and a hexapeptide may have a different affinity for the transporter and may therefore be relatively advantaged or disadvantaged in competition with other Phr peptides. The effects, if any, of these potential differences remain to be investigated.
PhrH and the regulation of RapH activities.
RapH is an important member of the Rap protein family that has been proposed to affect the decision between the development of two of the most studied environmental adaptations in B. subtilis, namely, the entry into spore formation and competence. We have shown here that the hexapeptide TDRNTT can counteract both activities of RapH (i.e., Spo0F∼P dephosphorylation and ComA sequestration) in vivo and in vitro. Furthermore, we have shown that the pentapeptides DRNTT and TDRNT have no detectable effect on spoIIG expression, whereas they do have a distinctly measurable effect on srfA operon transcription, although only at high, nonphysiological concentrations. The in vivo effect of DRNTT and TDRNT on srfA was observed only when their concentrations were at least 2 orders of magnitude greater than TDRNTT, and DRNTT did not disrupt RapH-ComA binding at any concentration tested in vitro. In fact, as we will now describe, it is likely that little or no DRNTT or TDRNT peptide is produced by the cells.
The sequences of known or predicted mature Phr pentapeptides were previously aligned (27), and the identity of the residues in positions −1 to −5 N-terminal to the Phr cleavage site were shown to affect pro-PhrC cleavage (17). In order for pro-PhrH to conform to the pro-Phr peptide consensus sequence, a one-amino-acid gap was introduced in its alignment, and the authors stated that it was difficult at the time to accurately predict the sequence of the mature PhrH peptide (17). Our data suggest that the threonine residue occupying the gap in the alignment is in fact the first residue of the mature PhrH hexapeptide.
In addition, cleavage of pro-PhrH to generate the hexapeptide TDRNTT places phenylalanine at the −4 position (see Table S3 in the supplemental material), which was reported to be highly favorable for pro-PhrC cleavage (17). However, cleavage of pro-PhrH to generate the pentapeptide DRNTT places lysine in the −4 position, and cleavage was found to be most inefficient when lysine occupied the pro-PhrC −4 position (17). Thus, it is unlikely that B. subtilis produces meaningful quantities of DRNTT; rather, mature PhrH is the hexapeptide TDRNTT.
Finally, Lanigan-Gerdes et al. (17) reported that pro-PhrI required the introduction of a single amino acid gap to conform best to the propeptide consensus sequence alignment. Similar to pro-PhrH, cleavage of pro-PhrI to generate a PhrI pentapeptide (DRVGA) places lysine at the −4 position (see Table S3 in the supplemental material). However, cleavage of pro-PhrI to generate a PhrI hexapeptide (ADRVGA) places isoleucine at the −4 position, and pro-PhrC cleavage was shown to be highly efficient with isoleucine in this position (17). The above results suggest that B. subtilis may not efficiently produce the PhrI pentapeptide. Therefore, although synthesized PhrI pentapeptide repressed RapI-dependent excision of ICEBs1 (3), we speculate that the PhrI hexapeptide may have similar activity to the pentapeptide and be more important physiologically. More generally, we propose that PhrH is the prototype of a newly identified class of hexapeptide Phr signaling molecules.
Concluding remarks.
An unresolved question concerns the purpose of the export-import peptide pathway for regulating Rap activity. It was postulated that the peptides accumulate in the medium and act as quorum-sensing pheromones. This has been suggested in the case of PhrC, which has been proposed to serve as a cell density signal for both competence and sporulation in B. subtilis (30). Another theory proposes that the peptide journey through this export-import circuit represents a timing mechanism that allows the Rap activity to persist for a while (23). For example, PhrA is difficult to detect in culture supernatants, and it has been proposed that PhrA is only exported to the immediate environment of the cell, perhaps into the cell wall or periplasm, where a high local concentration can be achieved resulting in autocrine signaling, presumably for the purpose of timing (23). We have shown clearly that PhrH can function in trans between donor and recipient cells, consistent with the quorum-sensing model. However, the low PhrH concentration detected in the supernatants is consistent with the possibility that little of the peptide actually leaves the cell surface. In addition, the trans activity detected in the present study has been identified while RapH was overproduced. It is possible that both models are correct. Producing cells may enjoy an advantage, responding before nonproducers. Thus, cheaters may be punished without denying donor cells any benefits that derive from a population-wide response.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health (NIH) grants AI081736 to M.B.N. and GM057720 to D.A.D. and by a UNCF-Merck Science Initiative Postdoctoral Fellowship to M.D.B.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Albano M., Hahn J., Dubnau D. 1987. Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169:3110–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alloing G., de Philip P., Claverys J. P. 1994. Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the gram-positive Streptococcus pneumoniae. J. Mol. Biol. 241:44–58 [DOI] [PubMed] [Google Scholar]
- 3. Auchtung J. M., Lee C. A., Monson R. E., Lehman A. P., Grossman A. D. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 102:12554–12559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouillaut L., et al. 2008. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 36:3791–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burbulys D., Trach K. A., Hoch J. A. 1991. Initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 6. Chai Y., Chu F., Kolter R., Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67:254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dubnau D., Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 8. Fujita M., Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grossman A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477–508 [DOI] [PubMed] [Google Scholar]
- 10. Hayashi K., Kensuke T., Kobayashi K., Ogasawara N., Ogura M. 2006. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol. Microbiol. 59:1714–1729 [DOI] [PubMed] [Google Scholar]
- 11. Helmann J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jenkinson H. F., Baker R. A., Tannock G. W. 1996. A binding-lipoprotein-dependent oligopeptide transport system in Streptococcus gordonii essential for uptake of hexa- and heptapeptides. J. Bacteriol. 178:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang M., Grau R., Perego M. 2000. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 182:303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang M., Shao W., Perego M., Hoch J. A. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535–542 [DOI] [PubMed] [Google Scholar]
- 15. Kearns D. B., Chu F., Branda S. S., Kolter R., Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739–749 [DOI] [PubMed] [Google Scholar]
- 16. Koide A., Hoch J. A. 1994. Identification of a second oligopeptide transport system in Bacillus subtilis and determination of its role in sporulation. Mol. Microbiol. 13:417–426 [DOI] [PubMed] [Google Scholar]
- 17. Lanigan-Gerdes S., Briceno G., Dooley A. N., Faull K. F., Lazazzera B. A. 2008. Identification of residues important for cleavage of the extracellular signaling peptide CSF of Bacillus subtilis from its precursor protein. J. Bacteriol. 190:6668–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lazazzera B. A., Solomon J. M., Grossman A. D. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell 89:917–925 [DOI] [PubMed] [Google Scholar]
- 19. McQuade R. S., Comella N., Grossman A. D. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis. J. Bacteriol. 183:4905–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakano M. M., Xia L. A., Zuber P. 1991. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 173:5487–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parashar V., Mirouze N., Dubnau D. A., Neiditch M. B. 2011. Structural basis of response regulator dephosphorylation by rap phosphatases. PLoS Biol. 9:e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perego M. 1999. Cell-cell signaling in bacteria, p. 243–258 American Society for Microbiology, Washington, DC [Google Scholar]
- 23. Perego M. 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc. Natl. Acad. Sci. U. S. A. 94:8612–8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perego M., Brannigan J. A. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22:1541–1547 [DOI] [PubMed] [Google Scholar]
- 25. Perego M., et al. 1994. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell 79:1047–1055 [DOI] [PubMed] [Google Scholar]
- 26. Perego M., Hoch J. A. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 93:1549–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pottathil M., Lazazzera B. A. 2003. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front. Biosci. 8:d32–d45 [DOI] [PubMed] [Google Scholar]
- 28. Schaeffer P., Millet J., Aubert J.-P. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smits W. K., et al. 2007. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol. Microbiol. 65:103–120 [DOI] [PubMed] [Google Scholar]
- 30. Solomon J. M., Lazazzera B. A., Grossman A. D. 1996. Purification and characterization of an extracellular peptide factor that affects two developmental pathways in Bacillus subtilis. Genes Dev. 10:2014–2024 [DOI] [PubMed] [Google Scholar]
- 31. Stephenson S., Mueller C., Jiang M., Perego M. 2003. Molecular analysis of Phr peptide processing in Bacillus subtilis. J. Bacteriol. 185:4861–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turgay K., Hahn J., Burghoorn J., Dubnau D. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tynkkynen S., et al. 1993. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 175:7523–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.