Abstract
The peptidoglycan (PG) of Lactobacillus plantarum contains amidated meso-diaminopimelic acid (mDAP). The functional role of this PG modification has never been characterized in any bacterial species, except for its impact on PG recognition by receptors of the innate immune system. In silico analysis of loci carrying PG biosynthesis genes in the L. plantarum genome revealed the colocalization of the murE gene, which encodes the ligase catalyzing the addition of mDAP to UDP-N-muramoyl-d-glutamate PG precursors, with asnB1, which encodes a putative asparagine synthase with an N-terminal amidotransferase domain. By gene disruption and complementation experiments, we showed that asnB1 is the amidotransferase involved in mDAP amidation. PG structural analysis revealed that mDAP amidation plays a key role in the control of the l,d-carboxypeptidase DacB activity. In addition, a mutant strain with a defect in mDAP amidation is strongly affected in growth and cell morphology, with filamentation and cell chaining, while a DacB-negative strain displays a phenotype very similar to that of a wild-type strain. These results suggest that mDAP amidation may play a critical role in the control of the septation process.
INTRODUCTION
Peptidoglycan (PG) is a heteropolymer of glycan strands cross-linked by peptidic stems most often between the fourth and the third amino acids of the donor and the acceptor stem, respectively. The composition of this peptidic stem can vary from one bacterial species to another and, in Lactobacillus plantarum, is composed of l-Ala, d-Glu, meso-diaminopimelic acid (mDAP), and d-Ala. d-Lactic acid is found as the last moiety of the peptidic stem in PG precursors (5).
Different amino acids found in PG have been reported as amidated in various species: d-Glu into d-iso-Gln (5, 9, 14, 21), mDAP into amidated mDAP (3, 5, 21), and d-Asp into d-Asn in species possessing a d-Asp as a cross bridge. Among these PG modifications, only the d-Asp amidation was characterized in Lactococcus lactis (23). d-Asp amidation is catalyzed by an asparagine synthase, AsnH, which is involved in autolysis control and resistance to cationic peptides (23).
We have recently shown that both d-Glu and mDAP are highly amidated (100% and 94%, respectively) in L. plantarum (5). The functional role of these amidations remains poorly understood, except for their impact on PG recognition by the mammalian host innate immune system (2, 11, 14). As a typical prokaryotic structure, PG is sensed by pattern recognition receptors involved in bacterial detection (e.g., Nod1, Nod2, and Toll-like receptor 2[TLR2]), and PG modifications were shown to affect this process (2, 6, 11, 12). Unamidated mDAP is essential for Nod1 detection (2), whereas amidated mDAP was reported to modulate the recognition by TLR2 (11). Despite these important immunomodulatory properties, the genetic determinants of mDAP amidation have never been described, and the functional role of mDAP amidation in bacterial physiology remains completely unexplored.
In this study, we identify the first mDAP amidotransferase and reveal its importance for growth and cell morphology in L. plantarum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. Plasmids were constructed in Escherichia coli MC1061. E. coli was grown in LB medium with shaking at 37°C. L. plantarum was grown in MRS broth (Difco Laboratories Inc., Detroit, MI) at 30°C. When required, erythromycin (250 μg/ml for E. coli, 5 μg/ml for L. plantarum) or chloramphenicol (10 μg/ml for E. coli and L. plantarum) was added to the medium. Solid agar plates were prepared by adding 2% (wt/vol) agar to the medium. Nisin A (Sigma, Bornem, Belgium) was routinely used at a concentration of 20 ng/ml for the induction of genes under the control of the nisA expression signals as previously described (18). For growth rate determinations, cell cultures were inoculated at an initial optical density at 600 nm (OD600) of 0.1, and growth was monitored in MRS medium by measuring the OD600 of cell cultures every 20 min with a Varioskan Flash multimode reader (ThermoFisher Scientific, Zellic, Belgium). The growth rate was calculated from the exponential growth phase.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Lactobacillus plantarum strains | ||
| NZ7100 | WCFS1 lp_0076::nisRK | 22 |
| EB042 | NZ7100 thrA1::P32-cat | This work |
| EB043 | NZ7100 thrA1::pGIEB14 | This work |
| EB044 | EB043 containing pNZ8048 | This work |
| EB045 | EB043 containing pGIEB15 | This work |
| EB046 | EB043 containing pGIEB16 | This work |
| EB047 | NZ7100 dacB::pGIEB17 | This work |
| Escherichia coli strain MC1061 | F− Δ(ara-leu)7697 [araD139]B/r Δ(codB-lacI)3 galK16 galE15 λ− e14−mcrA0 relA1 rpsL150 (strR) spoT1 mcrB1 hsdR2(r− m+) | 8 |
| Plasmids | ||
| pNZ5319 | Cmr Emr; pACYC184 derivative containing the cat gene under the control of the P32 constitutive promoter of Lactococcus lactis (lox66-P32-cat-lox71 cassette) | 16 |
| pNZ8048 | Cmr; shuttle vector containing PnisA promoter and start codon in NcoI site | 15 |
| pGIZ907 | Emr Apr; pUC18Ery with a 0.352-kb insert containing the ldhL expression signals of L. plantarum NCIMB8826 | 13 |
| pRV300 | Emr Apr; pBluescript derivative | 17 |
| pGIEB13 | Emr Apr; pGIZ907 derivative containing a 759-bp fragment from asnB1 | This work |
| pGIEB14 | Emr Apr; pGIZ907 derivative containing a 501-bp fragment from thrA1 | This work |
| pGIEB15 | Cmr; pNZ8048 derivative expressing asnB1 in transcriptional fusion | This work |
| pGIEB16 | Cmr; pNZ8048 derivative expressing thrA1 in transcriptional fusion | This work |
| pGIEB17 | Emr Apr; pRV300 derivative containing a 425-bp fragment from dacB | This work |
Cmr, Apr, and Emr indicate resistance to chloramphenicol, ampicillin, and erythromycin, respectively.
DNA techniques and electrotransformation.
General molecular biology techniques were performed according to the instructions given by Sambrook and Russell (20). Electrotransformation of E. coli was performed as described by Dower et al. (10). Electrocompetent L. plantarum cells were prepared as previously described (4). PCR was performed with the Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) in a 2400 GeneAmp PCR system (Applied Biosystems, Foster City, CA). The primers used in this study were purchased from Eurogentec (Seraing, Belgium) and are listed in Table S1 in the supplemental material.
Construction of mutants by single crossover (SCO) recombination.
Internal fragments of asnB1 (lp_0980) and thrA1 (lp_0979) were amplified by PCR using primer pairs intAsnB1pstI/intAsnB1SpeI and intthrA1pstI/intthrA1SpeI, respectively (see Table S1 in the supplemental material). Both PCR products were restricted by PstI and SpeI and then cloned between NsiI and XbaI restriction sites of the pGIZ907 suicide vector, resulting in the disruption plasmids pGIEB13 and pGIEB14, respectively. An internal fragment of dacB (lp_1010) was amplified by PCR using primers intdacB_1/intdacB_2 (see Table S1) and then cloned into the SmaI restriction site of the suicide vector pRV300, resulting in the disruption plasmid pGIEB17. The thrA1 and dacB disruptions by chromosomal integration of pGIEB14 and pGIEB17, respectively, were successfully achieved. The genotypes of the resulting mutant strains EB043 (thrA1::pGIEB14) and EB047 (dacB::pGIEB17) were validated by PCR using primers flanking the sites of recombination (see Table S1).
Construction of a stable thrA1 mutant.
Construction of the thrA1 deletion mutant was performed as previously described (16). A double crossover (DCO) gene replacement strategy was used to replace the open reading frame (ORF) of the thrA1 gene by a chloramphenicol resistance cassette lacking a transcriptional terminator (P32-cat). Briefly, the upstream and downstream flanking regions of thrA1 were amplified by PCR using primer pairs Uthra1/Uthra2 and Dthra1/Dthra2, respectively (see Table S1 in the supplemental material). Subsequently, amplicons were, respectively, cloned in the SwaI and SmaI restriction sites of the suicide vector pNZ5319 (16). The mutagenesis plasmids were transformed in L. plantarum WCFS1, and colonies displaying a chloramphenicol-resistant and erythromycin-sensitive phenotype represent candidate double crossover gene replacements. The genotype of the resulting EB042 mutant strain (thrA1::P32-cat) was confirmed by PCR by using primers flanking the site of recombination (see Table S1).
Construction of complementation vectors.
The asnB1 and thrA1 ORFs and their associated ribosomal binding sites (RBSs) were amplified by PCR using primer pairs 5′AsnB1lp-SDpstI/3′AsnB1lpXbaI and 5′Thra1lp-SDpstI/3′Thra1lpXbaI, respectively (see Table S1 in the supplemental material). Both PCR amplicons were restricted by PstI and XbaI and then cloned between PstI and SpeI of vector pNZ8048 (15), leading to expression plasmids pGIEB15 and pGIEB16, respectively. The integrity of the asnB1 and thrA1 genes was verified by DNA sequencing (see primers in Table S1). The two generated transcriptional fusions are under the control of PnisA, which allows their induction in the presence of nisin (18). These two vectors were electroporated in the EB043 strain for complementation trials.
Purification and structural analysis of PG.
PG from L. plantarum strains was prepared as described previously (9), with some modifications. DNase (50 μg/ml) and RNase (50 μg/ml) treatments were applied before hydrofluoric acid treatment. PG was digested with mutanolysin from Streptomyces globisporus (Sigma), and the resulting muropeptides were analyzed by reversed-phase high-performance liquid chromatography (RP-HPLC) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as previously reported (9). For tandem mass spectrometry (MS-MS) structural analysis, muropeptides were desalted on a Betasil C18 column (4.6 by 250 mm; Thermo Electron Corporation) with the acetonitrile/formic acid buffer system and dried with a speed vacuum. Samples were solubilized in 2% acetonitrile and 0.1% formic acid in Milli-Q water (1 μl for 1 mAU [10−3 absorbance unit] detected at 214 nm in the previous HPLC system). Each purified muropeptide was injected and analyzed at a flow rate of 0.2 μl/min on the mass spectrometer (LTQ-Orbitrap; Thermo Fisher) located on the PAPPSO platform (INRA, UMR1319 Micalis, France; http://PAPPSO.inra.fr). The injected muropeptide was first fragmented by source-induced dissociation (SID), leading to the loss of GlcNAc. The resulting ion was selected and fragmented in the LTQ ion trap; fragments were analyzed in the Orbitrap analyzer (accuracy of 10 ppm with external calibration).
Microscopy observations.
Microscopy analyses were performed using an Axio observer Z1 inverted microscope (Carl Zeiss). FM4-64 (Molecular Probes, Leiden, The Netherlands) and DAPI (4′,6-diamidino-2-phenylindole) (Sigma, Bornem, Belgium) staining was performed as previously reported (1). Analyses of micrographies were performed using the AxioVision 4.8. software (Carl Zeiss).
RESULTS AND DISCUSSION
The asnB1-thrA1-murE operon contains putative gene candidates for mDAP amidation.
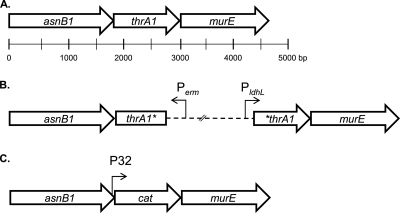
Based on the hypothesis that genes involved in the same biosynthetic pathway are often colocalized in a genome sequence, we examined all possible loci carrying PG biosynthetic genes in the genome of L. plantarum WCFS1 in order to identify a candidate amidotransferase that could be involved in mDAP amidation. Among these loci, the putative asnB1-thrA1-murE (accession numbers and locus tags are NP_784689 and lp_0980, NP_784688 and lp_0979, and NP_784687 and lp_0977, respectively) operon was especially relevant, since the murE gene codes for the well-conserved MurE ligase catalyzing the addition of mDAP to the UDP-N-muramoylalanyl-d-glutamate PG precursor (Fig. 1 A). The asnB1 and thrA1 genes are predicted to code for a potential asparagine synthase with an N-terminal amidotransferase domain and a putative diaminopimelate-sensitive aspartokinase, respectively. Intriguingly, Cahyanto et al. (7) have shown that ThrA1 was unable to phosphorylate l-Asp and that this enzymatic reaction is performed by ThrA2 in L. plantarum. By analogy to AsnH, which is the amidotransferase responsible for d-Asp amidation in L. lactis (23), we hypothesized that AsnB1 could be responsible for mDAP amidation in L. plantarum.
Fig. 1.
Genetic organization of the asnB1-thrA1-murE operon in the L. plantarum wild type (A), SCO thrA1 mutant (B), and DCO thrA1 mutant (C). The putative operon is composed of three genes (white arrows), encoding AsnB1, a putative amidotransferase; ThrA1, a putative aspartokinase; and MurE, a muramoyl-tripeptide synthetase. The dotted line represents a pGIEB14 insertion in the SCO thrA1 mutant. Perm and PldhL represent promoters carried by pGIEB14 allowing expression of the erythromycin resistance gene and genes located downstream of the insertion, respectively. The insertion of the P32-cat cassette in the DCO thrA1 mutant results from the allelic exchange of thrA1 with P32-cat. The murE gene is under the transcriptional control of P32 due to the absence of a transcriptional terminator at the 3′ end of cat.
Peptidoglycan from a thrA1 mutant obtained by single crossover recombination shows a defect in mDAP amidation.
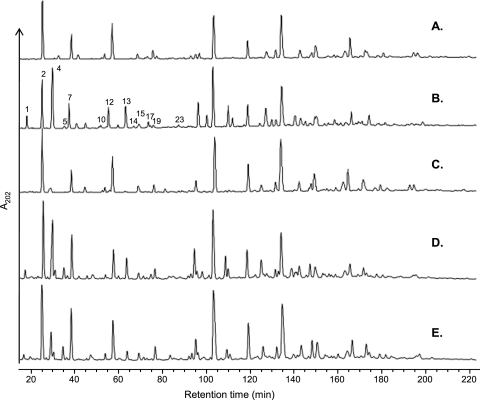
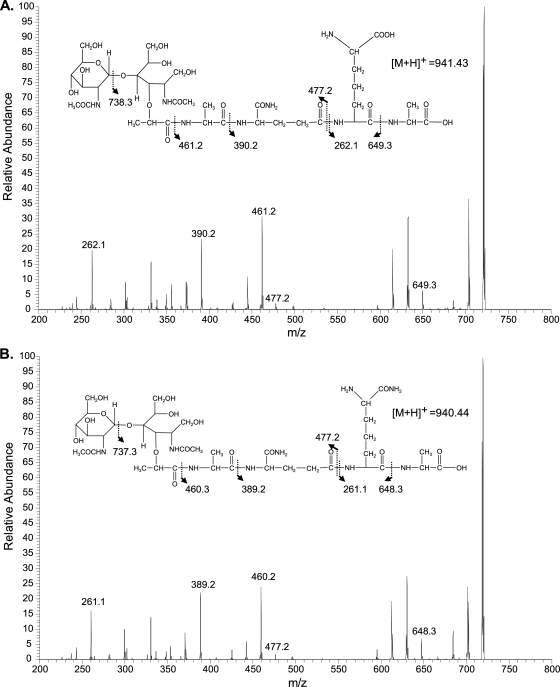
In order to assign a role to asnB1 and thrA1, we attempted to disrupt each of them by SCO homologous recombination using derivatives of the pGIZ907 suicide vector. This vector was chosen since it contains a PldhL promoter that allows the constitutive expression of the essential murE gene after chromosomal integration (Fig. 1). thrA1 was successfully disrupted (Fig. 1B), while despite several attempts, asnB1 inactivation was never obtained, suggesting that asnB1 plays an essential role in L. plantarum. The PG of wild-type NZ7100 and EB043 (SCO thrA1 mutant) strains was purified and digested by mutanolysin, and the resulting muropeptides were separated by RP-HPLC. Notably, muropeptides lacking amidation on mDAP in the control strain NZ7100 dramatically increased in the SCO thrA1 mutant (∼20-fold in monomers) (Fig. 2 A and B, peaks 1, 4, 13; Table 2; see also Table S2 in the supplemental material). Tandem mass spectrometry (MS-MS) on the muropeptides present in peaks 4 and 13 showed the presence of unamidated mDAP in the molecules, as illustrated for peak 4 in Fig. 3 A. The mass fragmentation of the muramoyl tetrapeptide derived from the disaccharide tetrapeptide yielded a fragment of 262.17 Da corresponding to unamidated mDAP linked to d-Ala. Other fragments of interest showed the loss of the unamidated mDAP (172.08 Da) between the muramoyl tripeptide (649.33 Da) and its derived dipeptidic form (477.25 Da). Similar fragments were obtained from the molecule of peak 7, corresponding to an amidated disaccharide tetrapeptide used as a control (Fig. 3B). In this case, the ending dipeptide has a mass of 261.17 Da, and the difference between the muramoyl tripeptide and dipeptide displayed a mass of 171.08 Da, which corresponds to an amidated mDAP.
Fig. 2.
RP-HPLC separation of muropeptides from L. plantarum PG. Wild-type NZ7100 (A), SCO thrA1 mutant (EB043) (B), DCO thrA1 mutant (EB042) (C), SCO thrA1 mutant complemented with thrA1 (EB046) (D), and SCO thrA1 mutant complemented with asnB1 (EB045) (E). Peak numbers refer to those in Table 2. Peaks 4, 7, 13, and 17 were analyzed by MS-MS.
Table 2.
Monomer composition of PG from L. plantarum wild-type NZ7100, SCO thrA1 (EB043) mutant, EB043 complemented by asnB1 (SCO thrA1/asnB1 mutant; EB045), and EB043 complemented by thrA1 (SCO thrA1/thrA1 mutant; EB046)
| Peaka | Proposed structureb | Calculated mass [M+Na]+c | % of all peaksd |
|||
|---|---|---|---|---|---|---|
| NZ7100 (wild type) | EB043 (SCO thrA1 mutant) | EB045 (SCO thrA1/asnB1 mutant) | EB046 (SCO thrA1/thrA1 mutant) | |||
| 1 | Tri missing NH2 | 892.37 | 0.36 | 1.52 | 0.45 | 0.84 |
| 2 | Tri | 891.39 | 10.38 | 6.41 | 9.86 | 8.91 |
| 4 | Tetra missing NH2 | 963.41 | 0.47 | 10.12 | 3.20 | 8.47 |
| 5 | Di | 720.29 | 1.06 | 0.51 | 0.95 | 1.08 |
| 7 | Tetra | 962.42 | 3.30 | 3.39 | 5.87 | 5.18 |
| Tri missing NH2 (Ac) | 934.39 | 0 | 0.66 | 1.04 | 0.76 | |
| 10 | Tri (OAc-M) | 933.40 | 0.96 | 0.39 | 0.31 | 0.225 |
| 12 | Tri (OAc-M) | 933.40 | 6.75 | 3.04 | 5.10 | 3.69 |
| 13 | Tetra missing NH2 (Ac) | 1,005.42 | 0.11 | 3.77 | 1.12 | 2.78 |
| 14 | Tri (OAc-G) | 933.40 | 1.09 | 0.46 | 0.94 | 0.85 |
| 15 | Di (Ac) | 762.30 | 0.51 | 0.46 | 0.40 | 0.40 |
| 17 | Tetra (Ac) | 1,004.44 | 1.33 | 0.83 | 1.68 | 1.36 |
| 19 | Tetra (Ac) | 1,004.44 | 0.22 | 0.25 | 0.135 | 0.105 |
| 23 | Tri (2Ac) | 975.41 | 1.19 | 1.53 | 1.19 | 1.72 |
| Monomers | 27.7 | 33.4 | 32.3 | 36.4 | ||
| Monomers with amidated mDAP | 25.2 | 16.1 | 25.1 | 22.0 | ||
| Monomers with nonamidated mDAP | 0.9 | 16.3 | 5.8 | 12.8 | ||
| Ratio amidated/nonamidated | 28.0 | 1.0 | 4.3 | 1.7 | ||
| Tripeptide chain | 20.7 | 14.0 | 18.9 | 17.0 | ||
| Tetrapeptide chain | 5.4 | 18.4 | 12.0 | 17.9 | ||
| Ratio of tripeptide/tetrapeptide | 3.8 | 0.8 | 1.6 | 0.95 | ||
Tri, disaccharide tripeptide [l-Ala-d-iGln-mDAP(NH2)]; tetra, disaccharide tetrapeptide [l-Ala-d-iGln-mDAP(NH2)-d-Ala]; penta, disaccharide pentapeptide [l-Ala-d-iGln-mDAP(NH2)-d-Ala-D-Lac]; disaccharide, GlcNAc-MurNAc; (NH2), amidation; Ac, acetylation on MurNAc or GlcNAc; OAcM, O-acetyl MurNAc; OAcG, O-acetyl GlcNAc. Muropeptides missing one amidation are indicated in bold.
Calculated masses are those of sodiated molecular ions that were the most abundant ions on MALDI-TOF mass spectra for all muropeptides.
The percentage of each peak was calculated as the ratio of the peak area over the sum of areas of all the peaks identified on the corresponding chromatogram.
Fig. 3.
MS-MS analysis of disaccharide tetrapeptides contained in peak 4 (Fig. 2 and Table 2) (A) and peak 7 (Fig. 2 and Table 2) (B). Fragmentation was performed on the [M+H]+ ions at m/z 738.3 (A) and 737.3 (B), which result from the loss of GlcNAc by source-induced dissociation (SID). Indicated m/z values correspond to ions obtained by cleavage of peptide bonds, as represented on the chemical structures.
Intriguingly, the ratio between disaccharide tripeptides and disaccharide tetrapeptides in the pool of monomers is affected by the level of mDAP amidation (Table 2). While the disaccharide tripeptide (e.g., peak 2) is the major monomer among the fully amidated muropeptides in wild-type NZ7100 (Fig. 2A), the disaccharide tetrapeptide (e.g., peak 4) dominates in monomers lacking mDAP amidation in the SCO thrA1 mutant (Fig. 2B). The global quantification of monomers confirmed that tripeptidic forms were almost 4-fold more represented than the tetrapeptidic forms in the wild type, while this ratio was slightly below 1 in the SCO thrA1 mutant (Table 2).
Importantly, these data demonstrate that the asnB1-thrA1-murE locus is involved in mDAP amidation and suggest that mDAP amidation plays an important role in the control of the l,d-carboxypeptidase activity in this species.
ThrA1 is not involved in mDAP amidation.
In order to confirm a direct role of ThrA1 in the amidation of mDAP, a stable thrA1 mutant was constructed by double crossover (DCO) recombination, resulting in the allelic exchange of thrA1 by a P32-cat cassette (Fig. 1C). Surprisingly, comparative PG analysis between NZ7100 and EB042 (thrA1::P32-cat) strains revealed a very similar muropeptide profile (Fig. 2A and C). This observation shows that the presence of muropeptides lacking mDAP amidation in the PG of the SCO thrA1 mutant (thrA1::pGIEB14) was not due to thrA1 inactivation as such but was more likely due to a polar effect resulting from plasmid integration. One possible explanation is the antisense orientation of the erm resistance marker after pGIEB14 integration (Fig. 1B), which could decrease the expression of the upstream asnB1 gene due to the leaky transcriptional terminator of the erm gene (P. Hols, unpublished data).
The asnB1 gene is able to complement the mDAP amidation defect of the SCO thrA1 mutant strain.
In order to clarify the phenotype of the SCO thrA1 mutant (EB043), two complementation vectors carrying asnB1 and thrA1 were constructed. Both genes are under the control of PnisA, which allows their induction in the presence of nisin. Notably, the complementation with asnB1 (EB045) was able to partially restore the muropeptide profile observed in the control strain NZ7100 (Fig. 2E; Table 2), while the complementation with thrA1 (EB046) has only a minor impact on peptidoglycan composition (Fig. 2D; Table 2). These data confirm that the phenotype observed in the SCO thrA1 mutant (EB043) was due to a polar effect on asnB1 expression and that asnB1 encodes the amidotransferase responsible for mDAP amidation in L. plantarum.
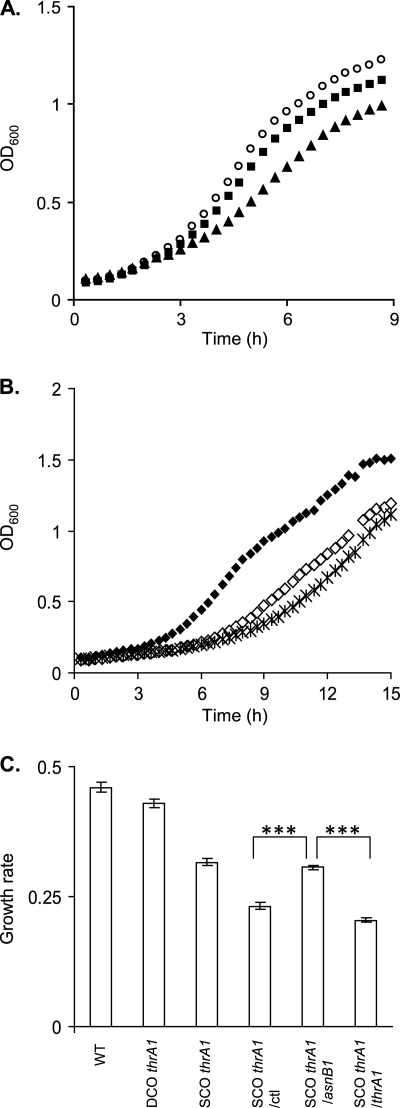
Defect in mDAP amidation has a negative impact on growth.
In order to evaluate the impact of mDAP amidation on the growth rate of the different mutant strains, the OD600 of cell cultures performed in MRS medium was monitored. The stable thrA1 mutation (strain EB042) had no significant effect on growth compared to that of the NZ7100 control strain, while thrA1 inactivation by SCO (EB043) negatively affects the growth rate (Fig. 4 A and C). We then monitored growth of the EB045 and EB046 complemented strains for comparison with that of the EB043 control strain harboring the empty expression vector (EB044) in the presence of antibiotics and nisin. The additional presence of nisin and chloramphenicol is responsible for a decrease in growth rate of the EB044 control strain compared to that of EB043. However, the complementation of strain EB043 with asnB1 (EB045) was able to significantly increase the growth rate compared to that of the EB044 control strain, while the complementation with thrA1 has no effect (EB046) (Fig. 4B and C). The growth rates observed above corroborate the levels of defect in mDAP amidation of the different mutant strains.
Fig. 4.
Effect of mDAP amidation on growth in MRS medium. (A and B) Growth curves of NZ7100 (wild type [WT]; open circles), DCO thrA1 mutant (EB042; black squares), SCO thrA1 mutant (EB043; black triangles), SCO thrA1 mutant carrying the empty plasmid pNZ8048 (SCO thrA1/ctl mutant, EB044; open diamonds), and SCO thrA1 mutant complemented with asnB1 (SCO thrA1/asnB1, EB045; black diamonds), and SCO thrA1 mutant complemented with thrA1 (SCO thrA1/thrA1 mutant, EB046; crosses). (C) Effect of mDAP amidation on growth rate of all constructed mutants. Mean values of one of two independent experiments (with 6 repetitions for each). Significance based on Student's t test; ***, P value of <0.001.
Proper cell septation in L. plantarum requires amidated mDAP.
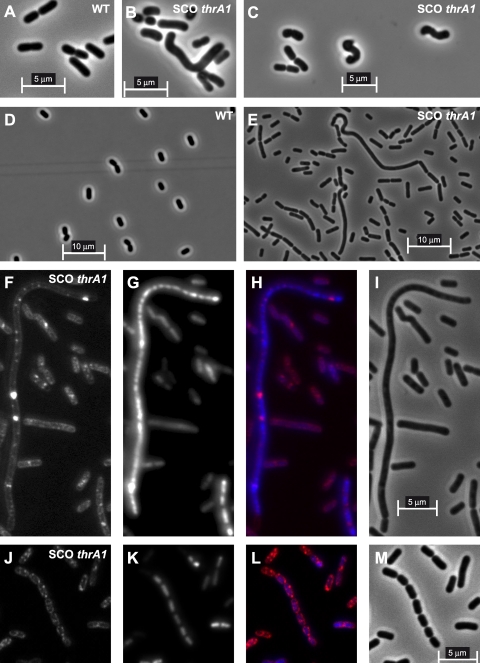
Since the defect in mDAP amidation of PG could result in altered cell morphology, microscopy analyses were performed on thrA1 mutant cells (EB042 and EB043) for their comparison to wild-type cells (NZ7100). The cell morphology of the DCO thrA1 mutant (EB042) is very similar to that of the wild-type strain at all growth stages (data not shown), while the SCO thrA1 mutant (EB043) shows multiple morphological alterations (Fig. 5). In the early exponential phase, curved cells and some short filaments are observed (Fig. 5B and C). The morphological defects are more pronounced in the late exponential phase, with an increased filamentation and cell chaining (Fig. 5E to M). At this growth stage, ∼21% (n = 197) of cells from the SCO thrA1 mutant harbored these morphotypes, while only one filamentous cell (n = 304) was found in the wild type and none (n = 661) in the DCO thrA1 mutant. Remarkably, DAPI staining showed that up to 39 separated nucleoids could be found in one filament (Fig. 5G and K), while only four distinct cells were observed by membrane staining with FM4-64 (Fig. 5F and J). LIVE/DEAD assays with Syto9/propidium iodide staining (LIVE/DEAD BacLight kit; Invitrogen) showed that more than 99% of the bacterial cell population was alive, including the cell subpopulation with a strongly altered morphology (data not shown).
Fig. 5.
Micrographs of L. plantarum wild type and SCO thrA1 mutant cells grown in MRS medium. WT NZ7100 (A) and SCO thrA1 mutant (EB043) (B and C) cells from early exponential phase; WT NZ7100 (D) and SCO thrA1 mutant (E) cells from late exponential phase; FM4-64 staining (F and J), DAPI staining (G and K), merge of FM4-64 and DAPI staining (H and L), and phase-contrast (I and M) of SCO thrA1 cells from late exponential phase.
Since PG analysis of the SCO thrA1 mutant revealed that the lack of mDAP amidation also results in a higher proportion of muropeptides carrying a tetrapeptide, the observed morphological alterations could be due to a modification of the l,d-carboxypeptidase activity. In order to test this hypothesis, the dacB gene (lp_1010), coding for the unique l,d-carboxypeptidase (9) identified in the genome of L. plantarum WCFS1, was inactivated by SCO as described in Materials and Methods. PG analysis of the SCO dacB mutant showed that the tetrapeptide/tripeptide ratio was dramatically changed (see Fig. S1 and Table S3 in the supplemental material), with a small amount of muropeptides with a tripeptide acceptor chain and a much higher proportion of muropeptides with a tetrapeptide acceptor chain than in the wild type (see Table S3 in the supplemental material). Interestingly, the morphology of dacB mutant cells is very similar to that of the wild type, without filamentation aberrations (see Fig. S2 in the supplemental material). All together, these results suggest that the morphological alterations observed in the SCO thrA1 mutant could be linked to the decrease of mDAP amidation levels and that mDAP amidation may play a key role in the septation process of L. plantarum.
Concluding remarks.
In this study, we show that the asnB1-thrA1-murE locus encodes the determinants of mDAP amidation in L. plantarum. Our genetic analysis by gene disruption and complementation revealed the key contribution of the asnB1 gene to this modification, which indicates that it encodes the first-described mDAP amidotransferase. In contrast, we were not able to assign any functional role to the thrA1 gene in our growth conditions. Its colocalization with asnB1 and murE suggests that it may play a role in mDAP amidation in unknown conditions, possibly through the biosynthesis of the substrate that acts as the amino group donor. This work also revealed the important role played by mDAP amidation of PG for the survival of L. plantarum. Despite several attempts, we were not able to either disrupt or delete the asnB1 gene. The attenuated mutant (SCO thrA1 mutant) strongly deficient in mDAP amidation displayed a major growth defect, suggesting that this PG modification could be essential in this species. Nevertheless, the expression of the downstream murE gene in the SCO thrA1 mutant is under the control of the constitutive PldhL promoter, which could also contribute to a certain extent to the growth defect of the mutant. A detailed genetic analysis using the construction of conditional mutants will be required to definitively conclude on the essentiality of mDAP amidation in this species. PG structural analysis revealed that PG with lowered mDAP amidation levels displays a higher tetrapeptide/tripeptide ratio than the highly amidated wild-type PG, which suggests that the unamidated mDAP-containing tetrapeptide is a poor substrate compared to its amidated counterpart for the L. plantarum l,d-carboxypeptidase DacB. The septation defect leading to filamentation observed for the mutant deficient in mDAP amidation (SCO thrA1 mutant) is reminiscent of the filamentation phenotype of L. lactis grown in the presence of methicillin that inhibits the septal penicillin-binding protein Pbp2X (19). Thus, mDAP amidation could be critical for septal PG biosynthesis in L. plantarum. Future work will be dedicated to better understand the contribution of mDAP amidation to the control of septation in time and space in this species as well as its importance in live cells for its recognition by the innate immune system.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Guillot (PAPPSO platform, UMR1319 Micalis, INRA, France) for helpful advice for MS-MS.
The work of M.-P.C.-C. was supported by INRA (Jeune Equipe grant). The work of P.H. was supported by the National Foundation for Scientific Research (FNRS), the Université Catholique de Louvain (Fonds Spéciaux de Recherche), and the Research Department of the Communauté Française de Belgique (Concerted Research Action). E.B. was the recipient of a Marie Curie fellowship for Early Stage Research Training (EST) of the FP6 LabHealth project (MEST-CT-2004-514428). T.R. held a doctoral fellowship from FRIA. P.H. is a research associate of the FNRS.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Andre G., et al. 2011. Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem. Biol. 6:366–376 [DOI] [PubMed] [Google Scholar]
- 2. Asong J., Wolfert M. A., Maiti K. K., Miller D., Boons G. J. 2009. Binding and cellular activation studies reveal that Toll-like receptor 2 can differentially recognize peptidoglycan from Gram-positive and Gram-negative bacteria. J. Biol. Chem. 284:8643–8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atrih A., Bacher G., Allmaier G., Williamson M. P., Foster S. J. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aukrust T. W., Brurberg M. B., Nes I. F. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201–208 [DOI] [PubMed] [Google Scholar]
- 5. Bernard E., et al. 2011. Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286:23950–23958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boneca I. G., et al. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 104:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cahyanto M. N., Kawasaki H., Nagashio M., Fujiyama K., Seki T. 2006. Regulation of aspartokinase, aspartate semialdehyde dehydrogenase, dihydrodipicolinate synthase and dihydrodipicolinate reductase in Lactobacillus plantarum. Microbiology 152:105–112 [DOI] [PubMed] [Google Scholar]
- 8. Casadaban M. J., Cohen S. N. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207 [DOI] [PubMed] [Google Scholar]
- 9. Courtin P., et al. 2006. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an l,d-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188:5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dower W. J., Miller J. F., Ragsdale C. W. 1988. High-efficiency transformation of Escherichia coli by high-voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Girardin S. E., et al. 2003. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300:1584–1587 [DOI] [PubMed] [Google Scholar]
- 12. Girardin S. E., et al. 2003. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278:41702–41708 [DOI] [PubMed] [Google Scholar]
- 13. Goffin P., et al. 2005. Lactate racemization as a rescue pathway for supplying d-lactate to the cell wall biosynthesis machinery in Lactobacillus plantarum. J. Bacteriol. 187:6750–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kraus D., et al. 2007. Muropeptide modification-amidation of peptidoglycan d-glutamate does not affect the proinflammatory activity of Staphylococcus aureus. Infect. Immun. 75:2084–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuipers O. P., de Ruyter P. G. G. A., Kleerebezem M., de Vos W. M. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 16. Lambert J. M., Bongers R. S., Kleerebezem M. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leloup L., Ehrlich S. D., Zagorec M., Morel-Deville F. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pavan S., et al. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perez-Nunez D., et al. 2011. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol. Microbiol. 79:759–771 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Schleifer K. H., Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Serrano L. M., et al. 2007. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb. Cell Fact. 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Veiga P., et al. 2009. Identification of the asparagine synthase responsible for d-Asp amidation in the Lactococcus lactis peptidoglycan interpeptide crossbridge. J. Bacteriol. 191:3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.