Abstract
hSMG-1 is a member of the phosphoinositide 3 kinase-like kinase (PIKK) family with established roles in nonsense-mediated decay (NMD) of mRNA containing premature termination codons and in genotoxic stress responses to DNA damage. We report here a novel role for hSMG-1 in cytoplasmic stress granule (SG) formation. Exposure of cells to stress causing agents led to the localization of hSMG-1 to SG, identified by colocalization with TIA-1, G3BP1, and eIF4G. hSMG-1 small interfering RNA and the PIKK inhibitor wortmannin prevented formation of a subset of SG, while specific inhibitors of ATM, DNA-PKcs, or mTOR had no effect. Exposure of cells to H2O2 and sodium arsenite induced (S/T)Q phosphorylation of proteins. While Upf2 and Upf1, an essential substrate for hSMG-1 in NMD, were present in SG, NMD-specific Upf1 phosphorylation was not detected in SG, indicating hSMG-1's role in SG is separate from classical NMD. Thus, SG formation appears more complex than originally envisaged and hSMG-1 plays a central role in this process.
INTRODUCTION
Cells are exposed to a variety of genotoxic stresses that impact on DNA integrity, gene regulation, subcellular organelles, and metabolic events. The phosphoinositide 3 kinase-like kinase (PIKK), hSMG-1, is an ∼400-kDa protein that plays an important role in cellular viability which is demonstrated by the embryonic lethality observed in hSMG-1-deficient mice (39). In addition, hSMG-1 plays a central role in maintaining mRNA quality through the process of nonsense-mediated mRNA decay (NMD), where it has been demonstrated to be crucial for initiating the signaling cascade through phosphorylation of Upf1 at S1078 and S1096, resulting in degradation of mRNA containing premature termination codons (PTC) (4, 7, 24, 42, 59). PTC-containing mRNAs can be produced through genomic mutations, alternative splicing, or RNA damage, and NMD is responsible for the elimination of aberrant PTC-containing mRNAs which could encode nonfunctional truncated proteins that could interfere with their endogenous counterparts (39, 59). NMD is elicited by recognition of the SURF complex (hSMG-1, Upf1, eRF1, and eRF3) when the termination codon is situated within ∼50bp of the last exon junction complex (EJC) (7, 23, 24). This results in SMG-1 phosphorylating Upf1, leading to NMD-mediated mRNA degradation (24, 42, 58). Recent work has also implicated two cofactors in hSMG-1 regulation: SMG-8 and -9 (58). These proteins form a trimeric complex with hSMG-1 and are required for NMD to occur. SMG-8 acts to inhibit hSMG-1 kinase activity prior to interaction with the EJC. In addition to NMD, mRNAs can be regulated through storage in cytoplasmic stress granules (SG) or by degradation in the related and often associated structures, processing bodies (P bodies) (3, 12, 13, 31). SG are formed in response to cellular stress such as heat shock and oxidative stress that results in the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) (32). SG are composed of accumulated mRNA and their associated proteins, such as TIA-1, eIF4G, and G3BP1 (32, 33). That SG are only transiently formed suggests that they are active sites where individual mRNAs are processed for storage, translation during stress and recovery, or shuttled to the associated structures, PB, for degradation (3, 28, 49).
Brumbaugh et al. (7) and Gewandter et al. (19) demonstrated that hSMG-1 is a genotoxic stress-activated protein kinase that displays some functional overlap with the related kinase, ATM. Expression of hSMG-1 was required for optimal activation of p53 in response to ionizing radiation (IR) and small interfering RNA (siRNA) depletion of hSMG-1 caused constitutive activation of p53 and Chk2, leading to an increased sensitivity to IR (7). As in the case of NMD, Upf1 was shown to be a substrate for hSMG-1 in response to radiation damage. hSMG-1 has also been shown to regulate the G1/S checkpoint in response to prolonged oxidative stress by p53 activation and p53-independent proteolysis of p21 (18). hSMG-1 also plays a role in telomere stability. Telomeric repeats are transcribed into noncoding RNA known as TERRA. hSMG-1 negatively regulates TERRA association with telomeres, and hSMG-1 depletion increased the number of TERRA-positive chromosomes and resulted in telomere destabilization (6, 9). In addition, depletion of hSMG-1 in tumor cells markedly increased the extent and accelerated the rate of apoptosis induced by tumor necrosis factor alpha (TNF-α) (46). Furthermore, hSMG-1 was demonstrated to be required for granzyme B-mediated apoptosis in a primary tumor cell line (41). Inactivation of smg-1 has also been shown to increase the life span of Caenorhabditis elegans, which appears to be related to resistance to oxidative stress (38). hSMG-1 has also been shown to negatively regulate hypoxia-inducible factor 1α (HIF-1α) in part by blocking mitogen-activated protein (MAP) kinase activation (10). Thus, it is evident that hSMG-1 is a key player not only in NMD but also functions in the DNA damage response, oxidative stress response, hypoxia, and apoptosis.
We describe here a new role for hSMG-1 in the formation of SG. This role appears to be separate from its role in active NMD, since although we demonstrated that Upf1 localized to SG, Upf1 was not phosphorylated on residues known to play a key role in NMD. hSMG-1 colocalized with a number of SG-specific markers, and knockdown by siRNA prevented SG formation. Inhibition of PIKK activity by wortmannin treatment reduced SG formation similar to hSMG-1 knockdown, but overexpression of kinase dead hSMG-1 did not prevent SG formation. Our data point to a novel and complex role for hSMG-1 in SG formation as part of the stress response.
MATERIALS AND METHODS
Cell culture conditions.
Cells were cultured in RPMI 1640 (lymphoblastoid cell lines [LCLs]) or Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (Invitrogen), 100 U of penicillin (Invitrogen)/ml, and 100 U of streptomycin (Invitrogen)/ml. Primary normal foreskin fibroblasts (NFF) were established from primary tissue (Glen Boyle, Queensland Institute of Medical Research). Approximately 11 different NFF strains, obtained from separate donors, were used during this study. Primary keratinocytes were supplied by N. Saunders (University of Queensland), and human kidney proximal tubular cells were supplied by C. Percy (University of Queensland). Primary melanocytes were also supplied by G. Boyle. The cell lines used were HeLa, U2OS, and A549 and LCLs. All cells were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
IR treatment of cells.
Irradiation of cells was performed at room temperature using a 137Cs source delivering gamma rays at a dose rate of 1.096 Gy/min (MDS Gammacell irradiator; Nordion, Ottawa, Ontario, Canada).
Hydrogen peroxide treatment of cells.
Adherent cell lines were washed twice in serum-free medium. The cells were resuspended in 1× original culture volume of serum-free medium containing H2O2 (at the indicated concentrations) and returned to the incubator.
Heat shock treatment of cells.
Adherent cells were placed in a 45°C water bath and allowed to equilibrate for 5 min. Cells were held at this temperature for 1 h prior to fixation.
Kinase inhibitor treatment of cells.
Wortmannin, AMA-37, and Ku55933 (Calbiochem, La Jolla, CA) were dissolved in dimethyl sulfoxide (DMSO). Adherent cells were grown to the desired density (∼70% confluent), and inhibitors were added directly into the cell culture medium at final concentrations of 10 μM for wortmannin and Ku55933 and 30 μM for AMA-37. Rapamycin (a gift from Dianne Watters, Griffith University) was used at 20 μM. Cells were returned to the incubator for 2 h. Inhibitor treatment was continued in conjunction with any second agent.
NaAs treatment of cells.
Sodium arsenite (NaAs) was dissolved in water to create a 1 M stock solution. NaAs was added directly into the cell culture medium at a final concentration of 1 mM for up to 1 h.
Antibodies.
Constructs encoding fragments of hSMG-1 fused to glutathione S-transferase (GST; antibody 1 [Ab1] against amino acids [aa] 24 to 101, Ab2 against aa 155 to 222, and Ab3 against aa 2458 to 2851) were generated in pGEX5. Recombinant protein production and immunization of sheep was performed as described previously (15). Antibodies were purified from sheep sera by affinity chromatography. Sera were diluted 1/10 in phosphate-buffered saline (PBS) and passed over a GST column to deplete the anti-GST antibodies. Eluted sera were then passed over GST–hSMG-1 columns to isolate anti-hSMG-1 antibodies (the columns were produced using the same protein fragments used for immunization or a fragment including the N-terminal region detected by Ab1 and Ab2, aa 24 to 222). hSMG-1-specific antibodies were eluted, buffer exchanged, and concentrated into a final buffer of PBS, 50% glycerol, and 0.1% sodium azide. hSMG-1 antibodies were used for immunofluorescence at a 1/100 to 1/600 dilution. hSMG-1 Ab3 was used for Western blotting at 1/1,000. Upf1 was detected using anti-Upf1 antibody (rabbit serum) and Upf2 detected with anti-Upf2 (affinity-purified rabbit antibody [a gift from L. E. Maquat, University of Rochester, Rochester, NY]). The phospho-UPF1 antibody that recognizes pS1078 and pS1096 was supplied by S. Ohno and has been described previously (59). The phosphorylated (S/T)Q motif antibody (rabbit serum) at a dilution of 1/100 to 1/400 was generated in Robert. T. Abraham's Laboratory at the Burnham Institute for Medical Research (La Jolla, CA). Two batches of this antibody were used; although the staining intensity differed between the batches of antibody, the overall pattern of staining was consistent. The commercial antibodies and the respective dilutions used for immunofluorescence were as follows: goat polyclonal TIA-1 antiserum (C20, Sc-1751; Santa Cruz Biotechnology), 1/100; G3BP1 antibody clone 23 (catalog no. 611126; BD Biosciences), 1/100; rabbit anti-eIF4G antibody (Santa Cruz catalog no. SC11373, H300), 1/500; and mouse anti-γH2AX (Upstate Biotechnology). HA was detected by using mouse anti-hemagglutinin (anti-HA; Covance) at 1/2,000. The antibodies used for Western blotting were mouse anti-ATM (Genetex) at 1/2,000, rabbit anti-phospho-1981 ATM (R&D Systems) at 1/2,000, mouse anti-DNA-PKcs (Oncogene/Merck) at 1/1,000, and rabbit anti-phospho-2056 DNA-PKcs (Abcam) at 1/2,000.
Conditions for immunofluorescent detection of hSMG-1.
Cells were fixed in 1% paraformaldehyde–PBS, followed by permeabilization in 0.1% Triton X-100 and PBS. Nonspecific sites were blocked using 10% fetal calf serum in PBS. All primary antibodies diluted in 1% newborn calf serum in 0.1% Triton X-100 and PBS. Alexa Fluor 488- or Alexa Fluor 594-conjugated (Molecular Probes) secondary antibodies were used at 1/200 to 1/1,000. DNA was visualized using Hoechst 3339 or DAPI (4′,6′-diamidino-2-phenylindole). Images were captured using a digital camera (Zeiss AxioCam MRm) attached to a fluorescence microscope (Zeiss Axiovert 2 Mot Plus) with a ×63/1.4 Zeiss Plan-Apochromat oil lens or using a DeltaVision fluorescence microscopy system (Applied Precision) with ×60/1.4 Olympus Plan-Apochromat oil lens, and images were analyzed with Softworx imaging software. Deconvolution was performed for five cycles, using a conservative setting with high noise filtering (Applied Precision). All images were acquired at room temperature. Colocalization analysis was performed using Softworx imaging software. Line profiles were determined by drawing a line through the cytoplasm of SG-positive fibroblasts. Pearson correlation coefficients (PCCs) were determined for a defined region of the cytoplasm of SG-positive cells. Between five and ten measurements were performed for each of these analyses, and representative data are shown.
Mammalian expression vectors.
Portions (5 μg) of plasmid DNA (pSR-HA-hSMG-1, pSR-HA-hSMG-1-DA, or pSR-HA-Upf1-4SA) (24, 59) were transfected into the indicated cells in a six-well plate using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The following day, transfected cells were replated onto coverslips in a 24-well plate and allowed to adhere for 6 to 8 h prior to treatment with 6 mM sodium butyrate for ca. 16 h prior to stress treatment. At approximately 48 h posttransfection, the cells were either left untreated or subjected to various treatments and then rinsed once with PBS before being fixed in 4% (wt/vol) paraformaldehyde for 10 min. The cells were then permeabilized in 0.5% (vol/vol) Triton X-100 in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2 [pH 6.9]) for 10 min. After a brief rinse in PBS, the cells were blocked in 5% (vol/vol) normal horse serum plus PBS or fetal calf serum plus PBS for 1 h. The remainder of the staining protocol was performed as described above.
siRNA knockdown.
hSMG-1 and Upf1 stealth siRNA duplex oligoribonucleotides were obtained from Invitrogen (hSMG-1-HSS118096-8; Upf1 HSS109171-3). The duplex oligoribonucleotides were resuspended to a concentration of 20 μM. NFF were grown to ca. 70% confluence prior to use. The volumes of reagents listed are per well for a six-well plate. siRNA (120 pmol) was added to final volume of 400 μl of Opti-MEM (Invitrogen) at 37°C, followed by mixing. Concurrently, 20 μl of Lipofectamine 2000 (Invitrogen) was added to 400 μl of Opti-MEM, followed by mixing. Each mixture was incubated at room temperature for 5 min, after which the two mixtures were combined and incubated at room temperature for a further 20 min. While the mixture was incubating, each well was washed twice with serum-free medium. After the final wash, 400 μl of Opti-MEM and 400 μl of transfection mix were added to the well. The plate was returned to the incubator for 6 h, followed by the addition of 2 ml of medium. At approximately 48 h (hSMG-1) or 64 h (Upf1) after the treatment had commenced, the cells were used for assays.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 5 software. The samples were analyzed using a paired two-tailed t test. Statistically significant differences are marked with asterisks in the figures (*, P < 0.05; **, P < 0.01).
Sample preparation for and analysis by MS.
Samples were processed as described previously (35). Briefly, samples were separated by SDS-PAGE, incubated in fixing solution (40% ethanol, 10% acetic acid, 50% H2O), and rebuffered in sensitizing solution (30% ethanol, 6.8% [wt/vol] sodium acetate, 0.5% [wt/vol] sodium thiosulfate), followed by washing. The gel was soaked in silver solution (0.25% [wt/vol] silver nitrate, 0.015% formaldehyde) and briefly washed with developing solution (2.5% [wt/vol] sodium carbonate, 0.0074% formaldehyde). The reaction was terminated by the addition of stop solution (1.46% [wt/vol] EDTA). The bands were excised, and the silver stain was removed prior to tryptic digestion using an adapted form of the method of Gharahdaghi et al. (20); the method for tryptic digestion was modified from that of Shevchenko et al. (50). The pellet returned from tryptic digest was resuspended in 5 μl of 50% acetonitrile and 0.1% trifluoroacetic acid. This was then used for mass spectrophotometric (MS) analysis.
For protein identification, peptides were analyzed using a Microflex MALDI-TOF-PSD (Bruker Daltonics, Bremen, Germany) operated in positive-ion reflectron mode. MS data were acquired using 350 shots of a nitrogen laser at 355 nm with a 20-Hz repetition rate and various intensities. The MS data were calibrated via close external calibration using peptide standards (New England Biolabs) containing angiotensin I (MH+ 1,296.69), neurotensin (MH+ 1,672.92), ACTH (1 to 17 clip, MH+ 2,093.09), ACTH (18 to 39 clip, MH+ 2,465.20), and ACTH (7 to 38 clip, MH+ 3,657.93). The Mascot search engine and the Homo sapiens taxonomic subset of the NCBI nonredundant database were used to identify the MS data. Mass tolerance was set at 150 ppm. Searches took into account carbamidomethylated cysteine and oxidized methionine. For the purposes of protein identification, no other posttranslational modifications were considered.
Cell cycle analysis.
After indicated treatments, the cells were incubated with propidium iodide (50 μg/ml; Sigma) and RNase A (100 μg/ml; Roche) for 30 min before cell cycle profiles were quantified using a FACScan (BD Biosciences). Analysis was performed using ModFit LT (BectonDickinson).
Western blotting.
Protein samples analyzed on either hand-poured 4.2% polyacrylamide gels or on 4 to 12% Bis/Tris gradient gels (Invitrogen). The proteins were then transferred onto nitrocellulose membrane, and Western blotting was performed as described previously (47).
RESULTS
hSMG-1 characterization.
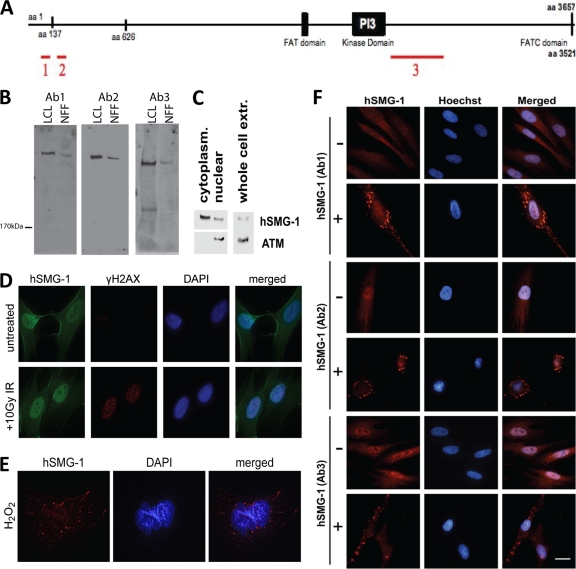
We developed three different antibodies that recognize hSMG-1 with a view to investigating hSMG-1 function in the stress response. The regions of hSMG-1 recognized by the three antibodies are shown in Fig. 1A. Immunoblot analysis revealed that the antibodies recognized a protein of the expected size for hSMG-1 in extracts from NFF and LCLs. For all three antibodies, a single high-molecular-weight band was observed (Fig. 1B), though following further separation multiple isoforms of hSMG-1 could be detected (see Fig. 5A; also data not shown). The bands were confirmed to be hSMG-1 using immunoprecipitation, followed by MS; of the 35 peptides identified from tryptic digests, 25 matched hSMG-1, resulting in 7% protein coverage (data not shown). The specificity of the antibodies was further confirmed by using siRNA knockdown of hSMG-1 that resulted in a loss of signal for both immunoblotting and immunostaining (see Fig. 5A and B). Cellular fractionation followed by immunoblotting confirmed that hSMG-1 is present in both the cytoplasm and the nucleus (Fig. 1C), as demonstrated previously (7, 24). Detection of ATM only in the nuclear fraction demonstrated that nuclei were intact. Immunostaining of primary cells also showed the presence of hSMG-1 in both subcellular compartments (Fig. 1D, untreated). In response to genotoxic stress, other members of the PIKK family, including ATM and ATR (ATM and Rad3 related), localize to nuclear foci at sites of DNA damage (1, 17). Exposure of primary fibroblasts to IR gave rise to γH2AX foci at sites of DNA damage, but there was no evidence that hSMG-1 localized to these sites under these conditions (Fig. 1D). However, in a small percentage of cells (up to 10%) hSMG-1 localized to cytoplasmic granules in response to treatment with the DNA-damaging agent hydrogen peroxide (H2O2) (Fig. 1E). These granules were detected by all three anti-hSMG-1 antibodies after H2O2 treatment (Fig. 1F).
Fig. 1.
hSMG-1 is cytoplasmic and nuclear and localizes to cytoplasmic granules in response to stress. (A) Schematic of the regions recognized by of hSMG-1 antibodies. GST fusion proteins were prepared using the regions indicated (region 1, Ab1; region 2, Ab2; region 3, Ab3) and used to generate polyclonal antibodies in sheep. (B) Specificity of detection of hSMG-1 by immunoblotting. Extracts were prepared from LCLs and NFF, and proteins were separated by SDS-PAGE prior to immunoblotting with the three different hSMG-1 antibodies. (C) hSMG-1 is present both in the cytoplasm and in the nucleus. Immunoblot analysis was performed for hSMG-1 in nuclear and cytoplasmic extracts from primary normal foreskin fibroblasts (NFF). ATM was used as a control to demonstrate fractionation. (D) hSMG-1 does not localize to sites of DNA damage. DNA damage induced nuclear foci in response to 10 Gy of IR. NFF were fixed and stained with anti-γH2AX (red) and anti-hSMG-1 (Ab2, green) antibodies. Nuclei were detected with DAPI. (E) Detection of cytoplasmic granules in NFF in response to H2O2. The cells were incubated for 1 h after exposure, fixed, and stained with hSMG-1 (Ab2) antibody. (F) Immunofluorescent detection of hSMG-1 with three different antibodies in NFF. hSMG-1 antibodies (red) directed to different regions of hSMG-1 show cytoplasmic granule formation in response to heat.
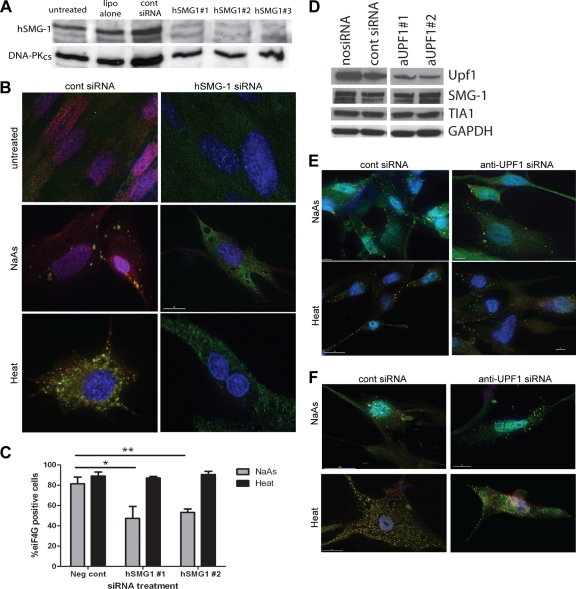
Fig. 5.
Effect of hSMG-1 or Upf1 disruption on SG formation. (A) Knockdown of hSMG-1 in NFF by three different siRNA oligonucleotides. hSMG-1 protein was detected by immunoblotting (Ab3), and the loading control was DNA-PKcs. (B) At 48 h after siRNA treatment, the NFF were treated with NaAs or heat for 1 h and then stained for hSMG-1 (Ab2, red) and eIF4G (green). Colocalization appears as yellow. Nuclei were detected with DAPI (blue). After siRNA treatment, the numbers of eIF4G SG-positive cells were reduced in NFF that had been treated with NaAs but not in heat-treated NFF. Control siRNA had no affect on SG formation in response to either agent. (C) Quantification of SG formation following treatment with two different anti-hSMG-1 or control siRNA. The percentage of NFF with eIF4G-positive granules was scored. The data represent the averages of three independent experiments, and error bars show the standard errors of the mean. Asterisks denote statistically significant differences in SG formation (*, P < 0.05; **, P < 0.01). (D) Knockdown of Upf1 expression in NFF using two different siRNA. Western blotting confirmed knockdown of Upf1 protein level but no affect on hSMG-1 or TIA-1 expression levels. GAPDH was probed for as a loading control. An irrelevant lane has been removed from the image, but all samples were run on the same gel. (E) Upf1 knockdown does not block stress granule formation. Approximately 64 h after anti-Upf1 siRNA treatment SG were induced with either NaAs or heat for 1 h. Fibroblasts were stained for TIA-1 (green) and Upf1 (red). SG formation could be seen in all treated samples. The data are representative of three independent experiments. (F) Cells were treated as for panel E but stained for hSMG-1 (green) and Upf1 (red). Nuclei were visualized with DAPI (blue). hSMG-1 was still recruited to SG following Upf1 knockdown. The data are representative of three independent experiments.
hSMG-1 localization to SG.
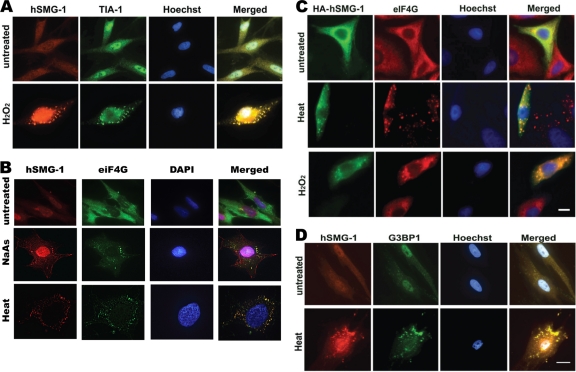
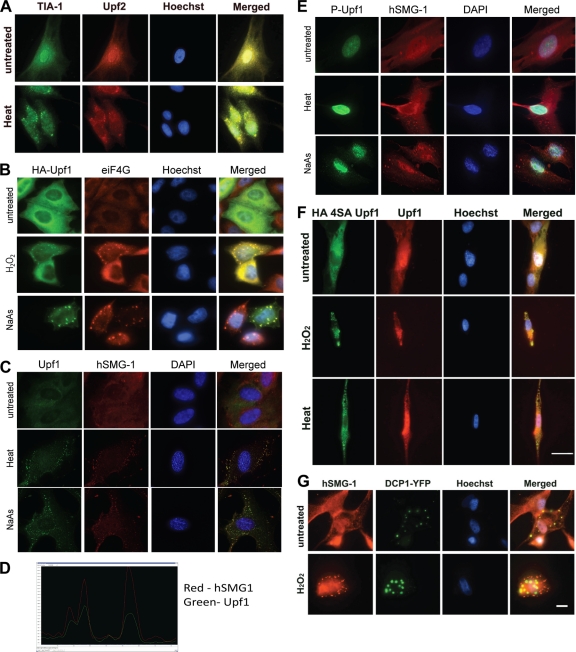
In fibroblasts, three major types of cytoplasmic granules have been described: vaults, processing bodies (P bodies) and SG. These structures are involved in regulation of translation, mRNA degradation and in stabilization and intracellular transport of mRNA (31–33, 49). Of these structures, only SG are significantly induced by stress-inducing agents (31). SG form in response to protein translation inhibition and contain the 48S complex and mRNA bound to T-cell internal antigen 1 (TIA-1) and many other proteins (19, 32). These granules contain proteins that either promote mRNA stability or lead to destabilization and degradation of specific mRNA (2). hSMG-1-positive granules, induced by treatment with H2O2, showed colocalization with TIA-1, indicating that these were indeed SG (Fig. 2A). SG were not detected with preimmune serum, copurified anti-GST antibody or in the absence of primary antibody (data not shown). A commonly used agent to induce SG is sodium arsenite (NaAs) (30). NaAs causes oxidative stress in cells, increases eIF2α phosphorylation, and causes translational arrest (2, 37). Exposure of cells to NaAs gave rise to SG, as determined by eIF4G and hSMG-1 staining (Fig. 2B). Heat was also investigated as an SG-inducing agent since it induces SG containing unique SG components (14, 29). Heat treatment induced hSMG-1-positive SG (Fig. 2B). SG induction in response to NaAs and heat occurred in at least 70% of cells (see Fig. 5C). Further confirmation of granule induction was carried out with overexpressed HA-tagged hSMG-1. Due to the difficulty in transfecting primary cell lines, transient transfection of HeLa cells with HA-hSMG-1 was performed. SG detected by anti-eIF4G antibody revealed colocalization with HA-hSMG-1 in SG after heat shock and H2O2 treatment (Fig. 2C). To confirm that the hSMG-1-positive structures were SG, we used another SG marker, G3BP1 (52). G3BP1 and hSMG-1 colocalized after treatment with heat (Fig. 2D). The number of SG per cell and the number of SG-positive cells varied (particularly with H2O2), and these granules showed a great deal of heterogeneity in size (data not shown), findings consistent with previous observations (27, 31).
Fig. 2.
hSMG-1 localizes to stress granules. (A) hSMG-1 (detected by Ab1, red) colocalizes with the SG marker TIA-1 (green) in response to H2O2. NFF were stained after H2O2 treatment (1.5 mM) for 1 h. (B) Colocalization of hSMG-1 (Ab2, red) with a second SG marker eIF4G (green) after treatment of NFF with heat (45°C) or 1 mM NaAs for 1 h. DAPI (blue) was used to stain nuclei. Colocalization (yellow) is indicated in a merged panel on the right. (C) An HA-tagged form of hSMG-1 localizes to SG. HeLa cells were transiently transfected with HA-hSMG-1 and incubated for 48 h in fresh medium prior to exposure to heat or H2O2, followed by staining with anti-HA or anti-eIF4G antibodies. (D) hSMG-1 (Ab1) (red) colocalizes with G3BP1 (green) in SG formed in NFF in response to heat. Hoechst was used to stain the nuclei.
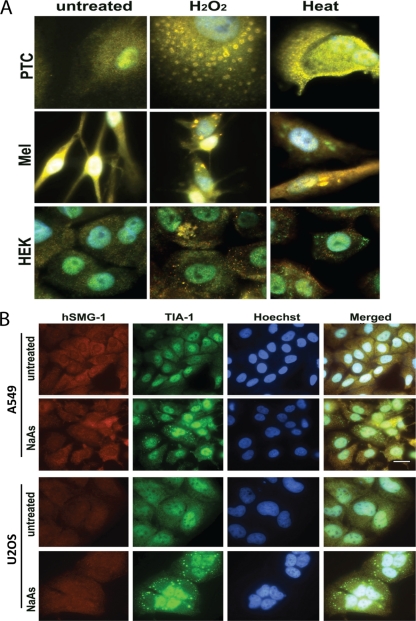
The universality of SG induction in other primary cells was also investigated. Figure 3A shows merged images of hSMG-1 (red) and TIA-1 (green) staining for SG induced in kidney proximal tubular cells, melanocytes, and human undifferentiated keratinocytes. With both H2O2 and heat treatment, hSMG-1 and TIA-1 colocalized for all of the primary cells. In contrast, the carcinoma cell lines U2OS and A549 were unable to form hSMG-1 staining granules but formed TIA-1-positive granules after treatment with NaAs (Fig. 3B).
Fig. 3.
hSMG-1 plays a central role in SG formation in primary cells but does not localize to SG in tumor cells. (A) Appearance of SG in primary cells in response to H2O2 or heat treatment. The images represent merged images between hSMG-1 (Ab1) (red) and TIA-1 (green). Nuclei were visualized with Hoechst stain. Abbreviations: PTC, human proximal tubular cells; MEL, primary melanocytes; HEK, undifferentiated keratinocytes. (B) A549 and U2OS cells were treated with NaAs for 1 h and stained for hSMG-1 (Ab1) (red) and TIA-1 (green). TIA-1-positive but hSMG-1-negative SG were formed in response to NaAs.
Since hSMG-1 has an established role in NMD, we investigated the possibility that its localization to SG was connected with NMD and/or mRNA decay. Recruitment of hSMG-1 in this process is generally mediated by Upf2 and Upf3b stably associated with the EJC (24). Therefore, we determined whether Upf2 was localized to SG. Upf2 localized with TIA-1 to SG in response to heat (Fig. 4A). Another key member of this mRNA surveillance complex, Upf1, was also present in SG induced by H2O2, heat, and NaAs, as shown by colocalization with eIF4G or hSMG-1 (Fig. 4B and C). This colocalization was quantified, yielding a PCC of 0.7832, where a PCC value between 0.5 and 1 is considered a positive association. A line profile of fluorescence in the red and green channels also confirmed the overlapping signals (Fig. 4D). A key step in the targeting of NMD is phosphorylation of Upf1 at four specific sites by hSMG-1 (S1073, S1078, S1096, and S1116), which appears to be critical for recognition of the PTC (24). A Upf1 phospho-specific antibody recognizing pS1078 and p1096 failed to detect phosphorylation in SG, suggesting that Upf1 phosphorylation is not required for SG formation or that it occurs at different sites (Fig. 4E). The localization of a form of Upf1 (HA4SAUpf1), nonphosphorylatable at all four sites, confirms the findings obtained with the P-Upf1 antibody (Fig. 4F). These observations are in keeping with a report by Gardner (16), which showed that, in response to hypoxia and arsenic, Upf1 localized to SG and NMD was inhibited (16). However, hSMG-1 mediated NMD-dependent phosphorylation at the remaining 23 predicted PIKK phosphorylation sites (NetPhosK [http://www.cbs.dtu.dk]) was not investigated and cannot be ruled out. To determine whether hSMG-1 could be regulating another form of mRNA decay, we determined whether hSMG-1 localized to P bodies. We used a construct encoding the P-body-specific marker decapping protein 1 (DCP1), which is required for mRNA decapping and degradation (49). No significant colocalization between hSMG-1- and DCP-1-positive granules was observed after treatment of fibroblasts with H2O2, providing further confirmatory evidence that hSMG-1 was localized specifically to SG under these conditions (Fig. 4G).
Fig. 4.
Localization of NMD proteins to SG but lack of evidence for a role for hSMG-1 in mRNA decay. (A) Upf2 colocalizes with TIA-1 in SG. NFF were fixed with or without heat treatment and stained for TIA-1 (green) and Upf2 (red). Colocalization (yellow) following heat treatment is shown in the merged panel on the right. (B) Upf1 also localizes to SG. HeLa cells were transiently transfected with HA-Upf1. After 48 h, the cells were treated with H2O2 and NaAs and then stained with anti-HA (green) and anti-eIF4G (red) antibodies. (C) Endogenous Upf1 localizes to hSMG-1. NFF were treated with NaAs or heat and after 1 h stained for Upf1 (green) and hSMG-1 (Ab2, red). (D) Upf1 colocalizes with hSMG-1. The line profile shows overlapping fluorescence signals for Upf1 (green) and hSMG-1 (red). Image analysis was performed using the Softworx computer program. Line profiles were determined by drawing a line through the cytoplasm of SG positive fibroblasts. (E) Upf1 is not phosphorylated in heat- and NaAs-induced SG. NFF were treated with NaAs or heat and, after 1 h, stained with phospho-specific antibodies recognizing sites phosphorylated on Upf1 during NMD. P-Upf1 (green) was not detected in hSMG-1 (Ab2, red) positive SG but could be clearly seen in the nucleus. (F) Localization of a nonphosphorylatable form of Upf1, HA4SAUpf1, to SG. NFF were transiently transfected with HA4SAUpf1 and incubated for 48 h in fresh medium prior to exposure to H2O2 and heat, followed by staining with anti-HA and anti-Upf1 antibodies. (G) hSMG-1 (Ab1) does not localize to P bodies. NFF were transiently transfected with the P-body-specific marker DCP1-YFP and incubated for 48 h prior to treatment with H2O2 for 1 h and followed by the detection of YFP and staining with antibodies against hSMG-1.
hSMG-1 is required for NaAs-induced SG formation or maintenance.
To further establish the importance of hSMG-1 in SG formation, three different siRNA duplexes were used to reduce cellular levels of hSMG-1. In these experiments we focused on heat and NaAs as inducing agents because they reproducibly induced SG in a high percentage of cells. Figure 5A shows that all siRNA sequences significantly reduced but did not ablate hSMG-1 protein expression. After siRNA treatment of fibroblasts, SG formation was decreased after NaAs treatment but not after exposure to heat (Fig. 5B). The percentage of cells containing eIF4G-positive SG was the same after heat treatment but was significantly reduced in NaAs-treated cells (Fig. 5C). siRNA knockdown of hSMG-1 also appeared to decrease SG formation in response to H2O2 (data not shown), although the very low and variable percentage of cells forming SG in response to H2O2 meant these data were not statistically significant. These data point to an important role for hSMG-1 in the formation or stability of a subgroup of SG, although more complete inhibition of SG formation may occur if total hSMG-1 knockdown was achieved. This experiment also demonstrates that heat-induced SG formation is hSMG-1 independent. To further examine the role NMD might play in SG regulation, we knocked down Upf1 expression with siRNA (Fig. 5D). As determined by Western blotting, the protein level of Upf1 was decreased by two independent siRNA sequences. The Upf1 siRNA treatment did not affect the protein levels of either hSMG-1 or TIA-1 (Fig. 5D). siRNA treatment in this experiment did not completely knock down Upf1 expression; however, if the amount of siRNA used or the length of incubation was increased, fibroblast cell viability was severely compromised (data not shown). Therefore, we used these conditions to examine the role of Upf1 in SG formation. After siRNA knockdown of Upf1, the fibroblasts were treated with either NaAs or heat for 1 h to induce SG formation. The cells were then immunostained for Upf1 and TIA-1, and SG formation was quantified. Knockdown of Upf1 showed a small decrease in SG formation in response to both NaAs and heat, although this was not statistically significant (data not shown). However, in many cells treated with anti-Upf1 siRNA, sufficient Upf1 protein was present to see it recruited to SG (Fig. 5E). Decreased SG formation after Upf1 knockdown was not due to inhibition of hSMG-1 recruitment to SG since hSMG-1 clearly localized in SG even after Upf1 depletion (Fig. 5F). Therefore, whether Upf1 plays a direct role in SG formation or whether depletion of Upf1 generally suppresses cellular responses is currently unclear.
Role of hSMG-1 kinase activity in SG formation.
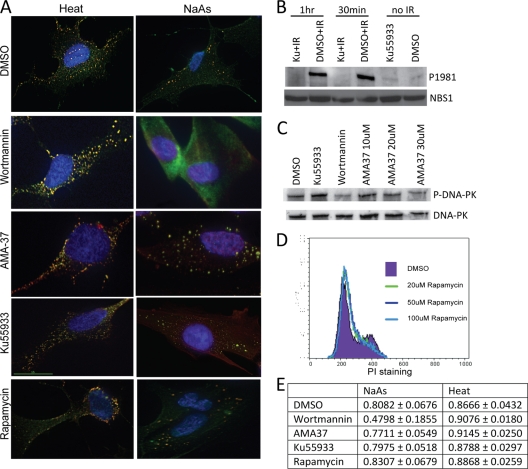
hSMG-1 may be required for SG formation for either of two reasons: (i) hSMG-1 kinase activity may be required for formation or stability of SG or, (ii) alternatively, the hSMG-1 protein may physically be required. In common with other members of the PIKK family, hSMG-1 phosphorylates substrates at (S/T)Q motifs in response to DNA damage (7, 59). Since there is no specific hSMG-1 inhibitor, we investigated the role of hSMG-1 kinase activity in SG formation using the PIKK inhibitor wortmannin (48). Wortmannin exposure for 2 h prior to stress treatment blocked SG formation after treatment with NaAs but did not inhibit SG formation induced by heat treatment (Fig. 6A). This further supported the hSMG-1-independent formation of heat-induced SG previously observed (Fig. 5B and C). Wortmannin appeared to also inhibit SG formation in the small number of cells that responded to H2O2 (data not shown). To investigate whether inhibition of other PIKK family members may have resulted in the observed decrease in SG formation, specific inhibitors of the related PIKK family members were used. ATM (Ku55933) and DNA-PK (AMA-37) inhibitors did not significantly inhibit SG formation in response to either NaAs or heat treatment (Fig. 6A). We also checked for the involvement of another member of the PIKK family mammalian target of rapamycin (mTOR) that controls cell growth and survival (51). Inhibition of mTOR by rapamycin also failed to interfere with heat- or NaAs-induced SG (Fig. 6A). All inhibitors were shown to be active under these conditions, as evidenced by the inhibition of radiation-induced ATM autophosphorylation measured by S1981 phosphorylation, inhibition of DNA-PKcs autophosphorylation on S2056, and rapamycin inhibition of mTOR, which blocked the movement of cells from G1 into S phase (Fig. 6B to D). Quantification of the immunofluorescent images showed far less colocalization after wortmannin treatment than after treatment with any other inhibitor (a PCC of 0.8082 for DMSO versus a PCC of 0.4798 for wortmannin) (Fig. 6E). These results suggest that PIKK activity is essential for SG formation in response to NaAs but not to heat.
Fig. 6.
Role for PIKK kinase activity in SG formation. (A) Wortmannin inhibits SG formation in response to NaAs treatment but not heat treatment. Cells were NaAs or heat treated after 2 h of incubation with the indicated inhibitor. NFF were then stained with anti-hSMG-1 (Ab2, red) and anti-eIF4G (green) antibodies, with the colocalization appearing as yellow. Nuclei were detected with DAPI (blue). Inhibition of ATM (Ku55933), DNA-PKcs (AMA37), or mTOR (rapamycin) did not interfere with formation of SG in response to either treatment. The data are representative of two to four independent experiments. (B) Effect of ATM-specific inhibitor on ATM autophosphorylation at S1981. NFF were incubated with Ku55933 for 2 h prior to 10 Gy of IR and then incubated for 30 min or 1 h prior to the preparation of cell extracts. Immunoblotting was carried out with a phospho-specific antibody for ATM S1981. Nbs1 protein was used as a loading control. (C) Effect of different inhibitors on DNA-PK autophosphorylation at S2056. NFF were incubated with either Ku55933 (ATM inhibitor), wortmannin (general PIKK inhibitor), or AMA37 (DNA-PKcs inhibitor) for 2 h prior to exposure to 10 Gy of IR. Extracts were prepared and immunoblotted either with the phospho-specific antibody (P2056-DNA-PK) or an anti-DNA-PKcs antibody. Total DNA-PKcs detection was used as a loading control. (D) Rapamycin prevents passage of NFF from G1 to S phase. Cells were exposed to concentrations of rapamycin from 20 to 100 μM prior to analysis by flow cytometry using propidium iodide (PI) staining. (E) PCCs for cells treated with PIKK inhibitors prior to SG formation. These coefficients were determined using Softworx software for a defined region of the cytoplasm of fibroblasts. The data show the averages of at least five measurements from different cells from at least two independent experiments and the standard deviations of the measurements.
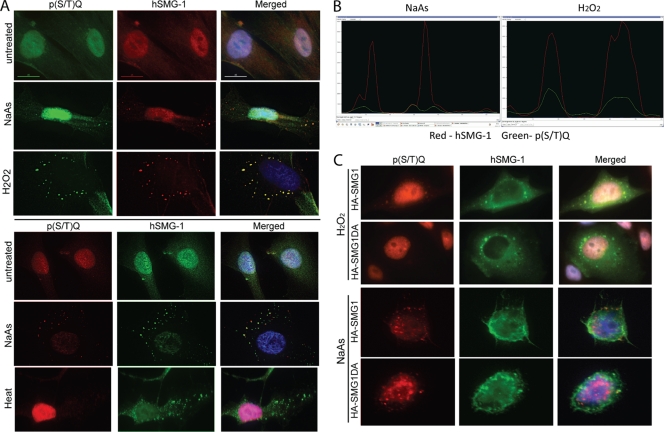
We looked for the presence of potential PIKK substrates in SG by staining with a phospho-specific antibody against p(S/T)Q sites. In response to NaAs and heat treatment, speckles of p(S/T)Q staining could be observed in the cytoplasm of cells (Fig. 7A). These sites were not completely colocalized with hSMG-1 but were often overlapping or associated with hSMG-1-positive granules (PCC of 0.4497; see also the left panel of Fig. 7B). In contrast, SG formed in response to heat were strongly hSMG-1 positive, but no phosphorylated (S/T)Q sites were detected (Fig. 7A, lower panel). In response to H2O2, a much stronger phosphorylated (S/T)Q signal, more completely colocalizing with hSMG-1, was detected (PCC of 0.6988) (Fig. 7A, right panel of Fig. 7B). These data imply that PIKK activity may be important in the regulation or stability of NaAs- and H2O2-induced SG but not heat-induced SG.
Fig. 7.
Detection of phosphorylated proteins in SG and importance of protein kinase activity of hSMG-1. (A) NaAs and H2O2 treatment of NFF induced the appearance of proteins phosphorylated at (S/T)Q sites, and these sites partially colocalize in SG with hSMG-1. Heat induced SG formation (hSMG-1 positive) without detectable phosphorylation at (S/T)Q sites. Staining in the upper panel used an older batch of rabbit anti-P(S/T)Q antibody: P(S/T)Q (green), hSMG-1 (Ab2, red), and DAPI (blue). In the lower panel, a newer batch of rabbit anti-P(S/T)Q antibody was used: P(S/T)Q (red), hSMG-1 (Ab2, green), and DAPI (blue). (B) P(S/T)Q signals associate with hSMG-1 after SG induction. The left panel shows a line profile indicating regions of overlapping fluorescence signals for P(S/T)Q (green) and hSMG-1 (red) following NaAs treatment. The right-hand panel shows greater overlap of the P(S/T)Q (green) signal with hSMG-1(red) after H2O2 treatment. Image analysis was performed using the Softworx computer program. Line profiles were determined by drawing a line through the cytoplasm of SG-positive fibroblasts. (C) Overexpression of kinase-deficient hSMG-1 does not inhibit SG formation. HeLa cells were transfected with either HA-hSMG-1 or HA-hSMG-1-DA (kinase deficient). The cells were treated with 1 mM H2O2 or NaAs for 1 h and then stained with anti-HA (green) and anti-P(S/T)Q (red) antibodies. In cells transfected with HA-hSMG-1-DA, no P(S/T)Q sites were observed in response to H2O2 treatment.
To further investigate the specific requirement of hSMG-1 kinase activity in SG formation in response to NaAs and H2O2, a kinase-deficient mutant of hSMG-1 (HA-hSMG-1-DA) was transiently transfected into HeLa cells. Kinase-dead hSMG-1 localized as efficiently as wild-type hSMG-1 to SG induced by H2O2 or NaAs (Fig. 7C). In SG containing wild-type HA-SMG-1, phosphorylated (S/T)Q signal was observed in response to both H2O2 and NaAs (Fig. 7C). In SG containing the kinase-deficient HA-hSMG-1-DA, phosphorylated (S/T)Q sites were undetectable in response to H2O2 treatment (Fig. 7C), suggesting that hSMG-1 is responsible for the observed phosphorylation. However, phosphorylated (S/T)Q sites were still present in NaAs-induced SG containing the kinase-dead hSMG-1, indicating that another member of the PIKK family is capable of phosphorylation of these sites. Collectively, the experiments described here show that hSMG-1 is recruited to SG and its presence, but not kinase activity, is essential for NaAs- and H2O2-induced, but not heat-induced, SG formation.
DISCUSSION
hSMG-1 has been shown to play several distinct roles in the cellular response to stress, including its involvement in NMD (59), in the protection of cells against TNF-α- or granzyme B-induced apoptosis (41, 46), and in activating the G1/S checkpoint (18). More recently Chen et al. (10) found that hSMG-1 inhibited HIF-1α transactivation activity in part by suppressing MAP kinase ERK1 in hypoxia. These results suggested that hypoxic conditions were efficient in activating hSMG-1 to restrain hypoxia-induced malignancy. Furthermore, mutations of the hSMG-1 kinase domain have been observed in breast cancer, lung adenocarcinoma, and kidney and stomach cancer (11, 53, 57; http://www.sanger.ac.uk/genetics/CGP/cosmic). In short, SMG-1 has known roles in NMD and genome maintenance and is implicated in regulation of oxidative stress, apoptosis and hypoxia responses. We have identified here a novel role for hSMG-1 in the formation of SG.
SG are related to other mRNA-protein complexes, including P bodies, germ cell granules, and neuronal RNA granules (8), all of which contain a variety of RNA-binding proteins, translation factors, and RNA decay machinery that can change depending on exposure to cellular stress (31). However, P bodies and SG are the only types present in fibroblasts. Typically, SG contain the 48S preinitiation complex composed of the small ribosomal subunit and mRNA, together with a number of translation initiation factors, poly(A)-binding protein, proteins that regulate mRNA translation, and proteins involved in cell signaling pathways (2, 44). SG are commonly described as structures that sequester RNA during times of cellular stress either to promote degradation, to stabilize mRNA for rapid translation once the stress has abated, or to promote translation of specific mRNA during stress (2). Nascent mRNA transcripts can also be exported from the nucleus and targeted directly to SG (8). This finding shows that while SG form in response to translational arrest, not all transcripts in SG are associated with stalled ribosomal complexes. More recently, alternative functions for SG have been described. SG have been shown to harbor proteins involved in the regulation of apoptosis such as TRAF2, RACK, and FAST (5, 34, 36). These data suggest that SG act to limit apoptosis, while cells adapt to stress, since inhibition of SG formation during stress can lead to decreased cell survival (8). Furthermore, SG have been implicated in cellular responses to viral infection (8), although the role SG play here is unclear since many viruses disrupt or induce SG formation to benefit their own life cycle. Previous work has shown that in mammalian cells NMD factors, including Upf1 and NMD mRNA targets, can traffic through P bodies (12) and localize to SG (16) when NMD is inhibited by specific inhibitors or in response to hypoxia.
We showed here that hSMG-1 is recruited to SG in response to a range of cellular stresses and that knockdown of hSMG-1 strongly reduced SG formation in response to NaAs but not to heat. Although use of the PIKK inhibitor wortmannin prevented SG formation in response to NaAs, overexpression of a kinase-dead version of hSMG-1 did not. These data suggest that the role of hSMG-1 in stress responses is likely to be dependent on the type of cellular stress encountered. There are potentially three facets to the involvement of hSMG-1 in SG: (i) the mechanism of hSMG-1 recruitment to SG, (ii) the requirement for hSMG-1 as a protein facilitating SG formation following certain stresses, and (iii) the role of PIKK, including hSMG-1, kinase activity in either SG formation, function, or disassembly.
Mechanism of hSMG-1 recruitment to SG.
Stress-induced signaling leading to phosphorylation of eIF2α and SG formation can be initiated by at least four different kinases (GCN2, PERK, HRI, and PKR) (22, 54). These kinases function in conjunction with other signaling pathways to coordinate the cellular response to a specific stress. Some of these pathways, converging on eIF2α, may have additional parallel effects facilitating recruitment of hSMG-1 to SG. The recruitment of hSMG-1 to SG in response to all stresses tested may be linked to inhibition of NMD. During the response to hypoxia Upf1 localized to SG and under the same conditions NMD was inhibited, although a causative link between these phenomena was not established (16). Furthermore, a very recent study showed that a variety of cellular stresses (including the SG-inducing agent NaAs) resulted in the phosphorylation of eIF2α and inhibition of NMD (55). Although in neither study was a causative link between SG formation and NMD inhibition established. In addition, a hyperphosphorylated form of Upf1 accumulated in P bodies in response to chemical inhibition of NMD, which blocks NMD at a step following Upf1 phosphorylation (12). In the present study, we observed the recruitment of hSMG-1, Upf1, and Upf2 to SG, but Upf1 was not phosphorylated at known NMD sites under these conditions, indicating that active NMD was not occurring within SG. This result is supported by the recent finding (55) that NMD was inactivated after treatment with the SG-inducing agent NaAs used here. Since Upf1 detected in P bodies after NMD inhibition was hyperphosphorylated, the unphosphorylated form we detected in SG may have an NMD independent role, or it may represent a different form of NMD inhibition more similar to that observed during hypoxia (12, 16). It is likely that under all of the stresses examined here, NMD is inhibited (55). Therefore, the recruitment of hSMG-1 and Upf1 to SG, without being essential for SG formation, may provide the basis of a mechanism for NMD inhibition in response to the phosphorylation of eIF2α (55). Alternatively, hSMG-1 may be required to traffic or process specific transcripts in SG under stress conditions such as heat shock, where it is not essential for SG formation.
Requirement of hSMG-1 as a protein facilitating SG formation following certain stresses.
hSMG-1 clearly plays a role in the formation or stability of SG after treatment with NaAs or H2O2 but not heat treatment. Knockdown of hSMG-1 with siRNA reduced the formation of SG in response to these agents, but a kinase-dead version of hSMG-1 also strongly localized to SG. This construct has previously been shown to act in a dominant-negative manner (24). Therefore, the data suggest that the presence of the hSMG-1 protein, but not its kinase activity, is required for SG formation in response to some stresses. This role for hSMG-1 has not been described before. Using an RNA-mediated interference-based screen, Ohn et al. (45) identified 100 human genes required for SG assembly. hSMG-1 was not identified in that screen, which may be due to differences in the cell lines used or that knockdown of hSMG-1 induces apoptosis (7), removing these cells from further analysis. To determine whether the requirement for hSMG-1 in SG formation was related to NMD, we attempted to define the role of the hSMG-1 substrate Upf1 in response to NaAs using siRNA knockdown. However, substantial Upf1 depletion decreased cell viability. This is not surprising since Upf1 has previously been shown to be required for embryonic development (40) and another NMD component Upf2 was shown to be essential for the viability of hematopoietic stem cells (56). Consequently, the decrease in SG formation observed with Upf1 knockdown can be interpreted in two ways: (i) Upf1 is required for SG formation in response to both NaAs and heat, but sufficient Upf1 depletion could not be achieved in order to see a dramatic effect, or (ii) Upf1 is not required for SG formation, and the small decrease observed here may be due to decreased viability or functionality of the cells. At this stage, it is still unclear what role Upf1 plays in SG formation. Combined, these data suggest that hSMG-1 is required for SG formation in response to NaAs in a manner that may be related to its role in NMD, but its retention in SG is independent of its kinase activity. Known roles for hSMG-1 are associated with its kinase activity. An understanding of this novel role for hSMG-1 will require further biochemical analysis of the protein and its stress induced interaction partners and/or isolation of different classes of SG to see what is unique about SG where hSMG-1 is required for their formation. Currently, isolation of SG in this manner is not possible. The differential requirement of hSMG-1 for SG formation may reflect the ability of cells to survive after insult by different stresses or reflect different mechanisms of translation or NMD inhibition induced (8).
Role of PIKK activity in SG formation, function, or disassembly.
The final aspect of hSMG-1 function in SG is the potential role of PIKK activity in the cellular response to stress. Phosphorylation of PIKK target (S/T)Q sites was detected in or associated with SG in response to NaAs and H2O2 treatment. Wortmannin inhibition blocked formation of SG in response to these stresses, indicating that PIKK activity is essential for SG formation under these conditions. Interestingly, overexpression of a kinase-dead version of hSMG-1 in HeLa cells did not prevent SG formation and, in fact, the kinase-dead form localized to SG with an efficiency similar to that of wild-type hSMG-1 in response to both stimuli, although this may be due to the ability of hSMG-1 to dimerize (42). To further complicate the picture, H2O2-induced phosphorylation of (S/T)Q sites was inhibited by kinase-dead hSMG-1, but NaAs-induced phosphorylation was not. These data suggest either that hSMG-1 kinase activity is not involved in SG formation or that hSMG-1 kinase activity can be compensated for by another PIKK family member in this situation. However, hSMG-1 kinase activity may be involved in SG regulation in response to H2O2 since hSMG-1-dependent phosphorylation sites were observed in these SG. Other PIKK family members have been implicated in SG formation. In the RNAi screen discussed above, the PIKK family member, DNA-PKcs was detected, indicating that its kinase activity may also be important (45). However, in the presence of a specific DNA-PKcs inhibitor we did not observe interference with SG formation in response to either NaAs or heat treatment. We also failed to prevent SG formation using the ATM-specific inhibitor KU55933 (21) or rapamycin, an inhibitor of mTOR (51), despite wortmannin strongly inhibiting SG formation. At the concentration used wortmannin inhibition of activity of proteins other that members of the PIKK family should be minimal. Potentially, multiple PIKK family members may be involved in an overlapping or redundant manner in SG regulation, as they are in the nuclear DNA damage response. If so, inhibition of more than one would be required to see a reduction in SG formation in response to NaAs or H2O2.
Interestingly, PIKK family members have also been implicated in NMD-independent RNA degradation involving Upf1. Histone mRNA stability may also be controlled by DNA-PK-mediated phosphorylation of Upf1 (43), and ATR may also phosphorylate Upf1 during histone mRNA degradation (25, 26). hSMG-1 involvement in this process has not been investigated. How this process may relate to a role for PIKK in SG regulation will require further investigation.
Overall, we show that hSMG-1 is recruited to SG in response to heat, NaAs, and H2O2 treatment. Our data suggest that the physical presence of the hSMG-1 protein is required for formation of a subset of SG independently of its protein kinase activity and that protein phosphorylation by PIKKs, including hSMG-1, may be involved in the regulation and/or turnover of SG in response to specific stresses.
ACKNOWLEDGMENTS
We thank Paul Anderson and Nancy Kedersha (Harvard Medical School) for the DCP1-eYFP construct, L. E. Maquat (University of Rochester Medical Centre) for the Upf1 and Upf2 antibodies and constructs, and Dianne Watters (Griffith University) for the rapamycin. We thank Glen Boyle (Queensland Institute of Medical Research) for the primary fibroblasts, Nicholas Saunders (University of Queensland) for the primary keratinocytes, C. Percy (University of Queensland) for the human kidney proximal tubular cells, Aine Farrell for her tissue culture expertise, and the Lavin laboratory for stimulating discussions. We also thank Tracey Laing for typing the manuscript.
We thank the Australian Research Council for funding and the National Health and Medical Research Council of Australia for a Peter Doherty Fellowship to T.L.R.
Footnotes
Published ahead of print on 12 September 2011.
REFERENCES
- 1. Andegeko Y., et al. 2001. Nuclear retention of ATM at sites of DNA double-strand breaks. J. Biol. Chem. 276:38224–38230 [DOI] [PubMed] [Google Scholar]
- 2. Anderson P., Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson P., Kedersha N. 2009. Stress granules. Curr. Biol. 19:R397–R398 [DOI] [PubMed] [Google Scholar]
- 4. Arias-Palomo E., et al. 2011. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 25:153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10:1324–1332 [DOI] [PubMed] [Google Scholar]
- 6. Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318:798–801 [DOI] [PubMed] [Google Scholar]
- 7. Brumbaugh K. M., et al. 2004. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell 14:585–598 [DOI] [PubMed] [Google Scholar]
- 8. Buchan J. R., Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chawla R., Azzalin C. M. 2008. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet. Genome Res. 122:194–201 [DOI] [PubMed] [Google Scholar]
- 10. Chen R. Q., et al. 2009. Kinome siRNA screen identifies SMG-1 as a negative regulator of hypoxia-inducible factor-1α in hypoxia. J. Biol. Chem. 284:16752–16758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ding L., et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durand S., et al. 2007. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol. 178:1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eulalio A., Behm-Ansmant I., Izaurralde E. 2007. P bodies: at the crossroads of posttranscriptional pathways. Nat. Rev. Mol. Cell. Biol. 8:9–22 [DOI] [PubMed] [Google Scholar]
- 14. Farny N. G., Kedersha N. L., Silver P. A. 2009. Metazoan stress granule assembly is mediated by P-eIF2α-dependent and -independent mechanisms. RNA 15:1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frangioni J. V., Neel B. G. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179–187 [DOI] [PubMed] [Google Scholar]
- 16. Gardner L. B. 2008. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 28:3729–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gatei M., et al. 2001. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3-related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites: in vivo assessment using phospho-specific antibodies. J. Biol. Chem. 276:17276–17280 [DOI] [PubMed] [Google Scholar]
- 18. Gehen S. C., Staversky R. J., Bambara R. A., Keng P. C., O'Reilly M. A. 2008. hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene 27:4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gewandter J. S., Bambara R. A., O'Reilly M. A. 2011. The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle 10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gharahdaghi F., Weinberg C. R., Meagher D. A., Imai B. S., Mische S. M. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20:601–605 [DOI] [PubMed] [Google Scholar]
- 21. Hickson I., et al. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159 [DOI] [PubMed] [Google Scholar]
- 22. Holcik M., Sonenberg N. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell. Biol. 6:318–327 [DOI] [PubMed] [Google Scholar]
- 23. Isken O., Maquat L. E. 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9:699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kashima I., et al. 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaygun H., Marzluff W. F. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12:794–800 [DOI] [PubMed] [Google Scholar]
- 26. Kaygun H., Marzluff W. F. 2005. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 25:6879–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kedersha N., Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30(Pt. 6):963–969 [DOI] [PubMed] [Google Scholar]
- 28. Kedersha N., Anderson P. 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90:155–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J., Anderson P. 2002. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13:195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kedersha N., et al. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kedersha N., et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kedersha N. L., Rome L. H. 1986. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J. Cell Biol. 103:699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim W. J., Back S. H., Kim V., Ryu I., Jang S. K. 2005. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 25:2450–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kozlov S., Gueven N., Keating K., Ramsay J., Lavin M. F. 2003. ATP activates ataxia-telangiectasia mutated (ATM) in vitro: importance of autophosphorylation. J. Biol. Chem. 278:9309–9317 [DOI] [PubMed] [Google Scholar]
- 36. Li W., Simarro M., Kedersha N., Anderson P. 2004. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol. Cell. Biol. 24:10718–10732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu S. X., et al. 2005. Mitochondrial damage mediates genotoxicity of arsenic in mammalian cells. Cancer Res. 65:3236–3242 [DOI] [PubMed] [Google Scholar]
- 38. Masse I., et al. 2008. A novel role for the SMG-1 kinase in life span and oxidative stress resistance in Caenorhabditis elegans. PLoS One 3:e3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McIlwain D. R., et al. 2010. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. U. S. A. 107:12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Medghalchi S. M., et al. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10:99–105 [DOI] [PubMed] [Google Scholar]
- 41. Meslin F., et al. 2011. hSMG-1 is a granzyme B-associated stress-responsive protein kinase. J. Mol. Med. 89:411–421 [DOI] [PubMed] [Google Scholar]
- 42. Morita T., et al. 2007. Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J. Biol. Chem. 282:7799–7808 [DOI] [PubMed] [Google Scholar]
- 43. Muller B., Blackburn J., Feijoo C., Zhao X., Smythe C. 2007. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J. Cell Biol. 179:1385–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nadezhdina E. S., Lomakin A. J., Shpilman A. A., Chudinova E. M., Ivanov P. A. 2010. Microtubules govern stress granule mobility and dynamics. Biochim. Biophys. Acta 1803:361–371 [DOI] [PubMed] [Google Scholar]
- 45. Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oliveira V., et al. 2008. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J. Biol. Chem. 283:13174–13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roberts T. L., Dunn J. A., Sweet M. J., Hume D. A., Stacey K. J. 2011. The immunostimulatory activity of phosphorothioate CpG oligonucleotides is affected by distal sequence changes. Mol. Immunol. 48:1027–1034 [DOI] [PubMed] [Google Scholar]
- 48. Sarkaria J. N., et al. 1998. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 58:4375–4382 [PubMed] [Google Scholar]
- 49. Sheth U., Parker R. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 51. Shor B., Gibbons J. J., Abraham R. T., Yu K. 2009. Targeting mTOR globally in cancer: thinking beyond rapamycin. Cell Cycle 8:3831–3837 [DOI] [PubMed] [Google Scholar]
- 52. Solomon S., et al. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stephens P., et al. 2005. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat. Genet. 37:590–592 [DOI] [PubMed] [Google Scholar]
- 54. Thomas M. G., Loschi M., Desbats M. A., Boccaccio G. L. 2011. RNA granules: the good, the bad, and the ugly. Cell Signal. 23:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang D., et al. 2011. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis Mol. Cell. Biol. 31:3670–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weischenfeldt J., et al. 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22:1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wong M. P., et al. 2003. Chromosomal aberrations of primary lung adenocarcinomas in nonsmokers. Cancer 97:1263–1270 [DOI] [PubMed] [Google Scholar]
- 58. Yamashita A., et al. 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 23:1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15:2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]