Abstract
The S phase checkpoint response slows down replication in the presence of replication stress such that replication can resume normally once conditions are favorable. Both proper activation and deactivation of the checkpoint are crucial for genome stability. However, the mechanisms of checkpoint deactivation have been largely unknown. Here, we show that two highly conserved Saccharomyces cerevisiae ATP-dependent chromatin remodeling factors, Isw2 and Ino80, function to attenuate and deactivate S phase checkpoint activity. Genetic interactions revealed that these chromatin remodeling factors and the Rad53 phosphatases function in parallel in the DNA replication stress response. Following a transient replication stress, an isw2 nhp10 double mutant displays stronger and prolonged checkpoint activation without experiencing increased replication fork troubles. Isw2 and Ino80 are both enriched at stalled replication forks and physically and specifically interact with a single-stranded DNA binding protein, replication protein A (RPA). Based on these results, we propose that Isw2 and Ino80 are targeted to stalled replication forks via RPA and directly control the amplitude of S phase checkpoint activity and the subsequent deactivation process.
INTRODUCTION
The packaging of DNA into chromatin facilitates compaction of eukaryotic genomes into the cell nucleus. However, this compaction also presents challenges for DNA-dependent processes such as transcription, replication, and DNA repair by limiting access of DNA binding proteins to DNA. Thus, elucidating mechanisms of chromatin regulation is essential to understanding how these DNA-dependent processes are regulated. One mechanism that eukaryotes use to regulate chromatin structure operates through ATP-dependent chromatin remodeling complexes, which hydrolyze ATP to alter histone-DNA contacts by nucleosome sliding, histone exchange, and/or nucleosome or histone eviction. The in vivo roles of chromatin remodeling complexes have been best characterized in studies of transcription, but their functions in DNA repair, recombination, and replication have also been revealed.
ATP-dependent chromatin remodeling complexes are evolutionarily conserved throughout eukaryotes (14). We have been investigating functions of two Saccharomyces cerevisiae complexes, Isw2 and Ino80. Isw2 exists as either a two- or a four-subunit complex (20, 25, 45), while Ino80 consists of 15 subunits (21, 37, 38). Isw2 slides nucleosomes (13) toward nucleosome-free regions in the intergenic regions (49, 51), which causes repression of both coding (17, 35, 39) and noncoding (49, 51) RNA transcripts. Ino80 mutants show hypersensitivities to methyl methanesulfonate (MMS), a DNA alkylating agent, and to hydroxyurea (HU), an inhibitor of ribonucleotide reductase that causes depletion of deoxynucleoside triphosphate (dNTP) pools (37). Ino80 is enriched at DNA double-strand breaks (DSBs) and interacts with γH2AX to facilitate the DNA damage response (11, 27, 46, 47). More recently, Ino80 has been suggested to play a critical role in the replication stress response (12, 30, 31, 40), although a consensus has not been formed on which step(s) in the replication stress response requires Ino80.
We recently found that several Ino80 subunits (Nhp10, Ies2, Ies3, Ies5) function in parallel with Isw2 to facilitate growth in the presence of MMS (48). Double mutants with mutations in these two chromatin remodelers, such as the isw2 nhp10 mutant, display a prolonged S phase in the presence of MMS due to delayed replication in late-replicating regions. We further demonstrated that Isw2 and Ino80 facilitate replication fork progression rates and late origin firing in the presence of replication stress caused by MMS (48). This was the first finding of chromatin regulators having this role; however, the mechanisms by which Isw2 and Ino80 function in facilitating replication remained to be determined.
Replication stress, resulting from endogenous insults or exogenous treatment with replication inhibitors such as MMS or HU, triggers activation of the S phase checkpoint (15). Replication forks stall when they encounter obstacles, resulting in accumulation of single-stranded DNA (ssDNA). This leads to a high concentration of an ssDNA binding protein, replication protein A (RPA), which is thought to signal the recruitment of the other players in the highly conserved eukaryotic checkpoint cascade (56). The sensor 9-1-1 complex (Rad17, Ddc1, and Mec3), complex loader (Rad24), and central kinase Mec1 with its interacting partner Ddc2 are recruited to stalled replication forks (24, 26). Rad9 and Mrc1 are then also targeted to stalled forks to mediate the activation of the effector kinases Rad53 and Chk1 (1, 43). The activated checkpoint results in slowed replication, transcriptional induction of damage response genes, prevention of replication fork collapse, and suppression of late origin firing (15). The S phase checkpoint cascade allows cells to slow down replication to handle replication stress problems.
Due to the essential role of the S phase checkpoint in genome stability, activation mechanisms of the S phase checkpoint have been extensively studied. Indeed, premature or defective checkpoint activation leads to genome instability (28). In contrast, mechanisms of S phase checkpoint deactivation have not been well understood. The presence of an active mechanism to deactivate checkpoint activity was recently uncovered by the finding that Rad53 phosphatases play this role (44). The importance of proper deactivation of the S phase checkpoint was demonstrated by the fact that failure to deactivate the checkpoint can lead to defects in replication fork progression, resulting in incomplete DNA replication and thus lethality (44). Similarly, how the amplitude of S phase checkpoint activation is regulated remains unknown, despite the fact that checkpoint activation is a graded response (41). It has been established that checkpoint activity correlates with the presence of the activated, hyperphosphorylated form of the Rad53 kinase (32). Rad53 is phosphorylated by Mec1 during checkpoint activation (36, 42) and dephosphorylated by the Pph3 and Ptc2 phosphatases (29, 44). Aside from phosphatases, it remains to be discovered whether other players function in checkpoint deactivation.
As noted above, we previously reported that the ATP-dependent chromatin remodeling factors Isw2 and Ino80 facilitate DNA replication in the presence of replication stress (48). We showed that Isw2 and Ino80 facilitate replication fork progression and activation of late-firing origins. However, the mechanisms by which these remodeling factors function have been unknown. In this article, we present our finding that Isw2 and Ino80 affect DNA replication through regulation of S phase checkpoint activity. We found that the isw2 nhp10 double mutant displays delayed S phase progression as well as stronger and prolonged Rad53 activation during the replication stress and recovery stages. Multiple lines of evidence clearly argue against the possibility that the stronger checkpoint activity of the isw2 nhp10 double mutant is due to increased replication fork troubles but rather that Isw2 and Nhp10 directly function in attenuating the checkpoint response. Furthermore, we discovered that Isw2 and Ino80 physically interact with RPA. Based on these results, we propose that Isw2 and Ino80 are targeted to stalled replication forks by RPA to directly attenuate the amplitude of the S phase checkpoint activity and facilitate checkpoint deactivation. Thus, our work uncovered a biological function not previously known to be associated with any ATP-dependent chromatin remodeling factors, as well as a novel mechanism to regulate S phase checkpoint activity.
MATERIALS AND METHODS
Yeast growth and cell synchronization.
Cells were grown at 30°C. Log-phase cells for arrest and release experiments were grown to an optical density at 600 nm (OD600) of 0.2 to 0.25. Arrest in G1 phase was accomplished by treatment with 5 μg ml−1 α-factor. Cells were released from α-factor arrest by filtration on 0.45-μm-pore-size nitrocellulose membranes, washed, and then suspended in a half volume of prewarmed media without α-factor. For experiments involving MMS or HU treatment of cells in liquid culture, MMS (Sigma) or HU (Sigma) was added to achieve a 0.02% (vol/vol) or a 200 mM final concentration, respectively. Cell synchrony was monitored by flow cytometry (48).
Coimmunoprecipitation (co-IP).
Cells (in 200 ml of yeast extract-peptone-dextrose [YPD]) were washed once each with water supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and with buffer H 0.15 M KCl (25 mM HEPES KOH [pH 7.6], 2 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 150 mM KCl, 0.02% NP-40) supplemented with 1× protease inhibitors (1 mM PMSF, 2 μM pepstatin, 0.6 μM leupeptin, 2 mM benzamidine, 2 μg/ml chymostatin A). To prepare cell lysates, pellet was resuspended in 1 ml of buffer H 0.15 M KCl supplemented with 1× protease inhibitors and subjected to bead beating, followed by high-speed centrifugation. FLAG immunoprecipitation (IP) was done using 4 μg of monoclonal anti-FLAG M2 antibody (Sigma) cross-linked to 6 μl of protein G Dynabeads (Invitrogen) per IP. Each IP consisted of 200 μl of cell lysate and anti-FLAG M2 beads, bound for 90 min at 4°C. IP products were washed 3 times with 0.5 ml buffer H 0.15 M KCl supplemented with 1× protease inhibitors. Western analysis was performed with monoclonal anti-FLAG M2 antibody (Sigma), 9E10 monoclonal primary antibody (anti-c-myc; Covance), and IRDye 800CW goat anti-mouse secondary antibody (LiCor Biosciences), followed by scanning using an Odyssey scanner (LiCor Biosciences).
Rad53 ISA and Western blot analyses.
For in situ autophosphorylation (ISA), cells were grown in YPD as described above, and 25 ml was harvested at each time point. Protein samples from trichloroacetic acid (TCA)-treated cells were prepared as described previously (33), except they were processed using an 8% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Anti-Rad53 (EL7E1) and antitubulin were used in Western blot analyses.
Spot tests.
Serial dilutions (5-fold) of saturated culture were spotted on YPD agar plates with or without drugs. Plates containing drugs were used within 24 h of preparation. For gradient plate assays, stationary-phase cells were spotted 24 times across a YPD agar plate using a Biomek 2000 workstation (Beckman Coulter). Concentration gradients of MMS were set up as previously described (50). The indicated drug concentrations signify the concentration of the bottom agar.
Hydroxyurea survival assay.
Cells were arrested in G1 phase with α-factor and released into S phase in prewarmed YPD media containing 200 mM hydroxyurea. At the indicated time points, equal aliquots of diluted cells were plated in duplicate on YPD plates. Plates were grown at 30°C for 2 days (for wild- type [WT] and isw2 nhp10 cells) and 3 days (for ino80 cells) until surviving colonies were large enough for counting. Survival, assessed by the average number of colonies, was calculated as a percentage of untreated colony numbers.
Fluorescence microscopy.
Cells (1.5 ml) were harvested, pelleted, and resuspended in 100 μl of paraformaldehyde at room temperature for 15 min. Cells were pelleted, washed once in 1 ml of 0.1 M potassium phosphate–1.2 M sorbitol buffer, and resuspended in 100 μl of the same buffer. Cells were sonicated prior to microscopy. A total of 200 cells were scored twice for each biological replicate.
RESULTS
Genetic interactions with Rad53 phosphatases suggest that Isw2 and Ino80 function in the S phase checkpoint.
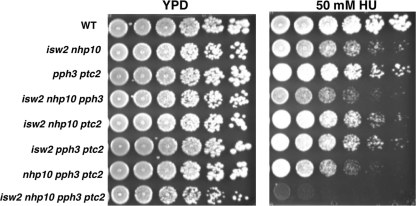
The first clue that Isw2 and Ino80 may function in the S phase checkpoint came from our observation that the isw2 nhp10 mutant has a phenotype similar to that of the pph3 ptc2 mutant. Nhp10 is a subunit of the Ino80 complex. Pph3 and Ptc2 are Rad53 phosphatases that function to deactivate Rad53, the effector kinase of the S phase checkpoint (44). Both isw2 nhp10 and pph3 ptc2 mutants display slow growth on the replication inhibitor MMS and defects in replication fork progression rates (44, 48). To determine functional relationships among Isw2, Nhp10, Pph3, and Ptc2, we examined the growth phenotypes of a quadruple isw2 nhp10 pph3 ptc2 mutant. We found that the quadruple mutant was additively HU sensitive (Fig. 1). Addition of either an isw2 or nhp10 mutation to the pph3 and ptc2 phosphatase mutations also resulted in increased HU sensitivity. Interestingly, the quadruple mutant displayed slower growth on normal YPD media. The striking growth defect of the quadruple mutant on HU demonstrated that Isw2 and Ino80 function in a pathway that is independent from and parallel to that of the Rad53 phosphatases. In addition, this suggested that Isw2 and Ino80 might have functions similar to those of the Rad53 phosphatases in the replication stress response. Thus, we decided to explore whether Isw2 and Ino80 also function to deactivate the S phase checkpoint.
Fig. 1.
Isw2 and Ino80 function in parallel to the Rad53 phosphatases Pph3 and Ptc2. The results of a spot assay performed as a genetic-interaction test are shown. Strains were grown to saturation, and then 5-fold serial dilutions were plated on YPD plates with or without 50 mM HU. The relevant genotypes of the strains are shown on the left. Plates were grown for 2 days at 30°C.
Isw2 and Ino80 attenuate S phase checkpoint activity and facilitate checkpoint deactivation.
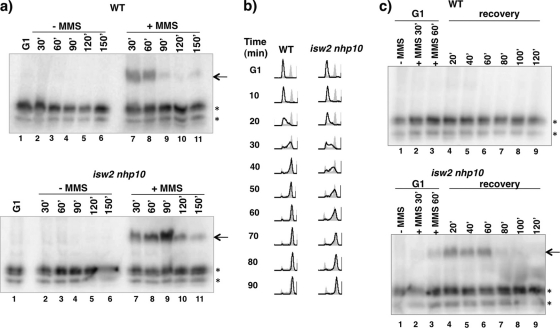
To test whether Isw2 and Nhp10 are involved in S phase checkpoint regulation, we first analyzed checkpoint activity by a Rad53 in situ autophosphorylation (ISA) assay. Only checkpoint-activated Rad53 exhibits in situ autophosphorylation activity in vitro that is apparent upon incubation with radiolabeled ATP (33). When WT and isw2 nhp10 cells were released from G1 phase into S phase in the constitutive presence of MMS, both cells displayed Rad53 ISA activity (Fig. 2 a). However, isw2 nhp10 cells exhibited noticeably stronger and prolonged Rad53 ISA activity, showing that Isw2 and Nhp10 negatively affect S phase checkpoint activity.
Fig. 2.
Isw2 and Ino80 attenuate S phase checkpoint activity and facilitate efficient checkpoint deactivation during and after MMS treatment. (a) Rad53 ISA assay during MMS treatment. WT and isw2 nhp10 cells were arrested in G1 and released into YPD in the absence or presence of 0.02% (vol/vol) MMS. Cells were harvested during G1 arrest and every 30 min during release until 150 min postrelease. The arrow shows the Rad53 ISA band, and asterisks designate other kinases with ISA activity that served as a loading control. (b) Flow cytometry analysis after transient MMS treatment. WT and isw2 nhp10 cells were arrested in G1 by α-factor treatment. α-Factor was readded for 75 min, and 0.02% (vol/vol) MMS was added 15 min later for a total of 60 min of MMS treatment during G1. Cells were released into S phase in YPD and collected at the indicated time points after release. DNA content was analyzed by flow cytometry (black lines). Gray profiles correspond to the results obtained with reference asynchronous cells collected before G1 arrest. (c) Rad53 ISA assay during recovery from MMS treatment. Cells were harvested before MMS was added, at two time points during MMS treatment in G1, and every 20 min during S phase recovery. The arrow shows the Rad53 ISA band, and asterisks designate other kinases with ISA activity that served as a loading control.
We next tested whether Isw2 and Nhp10 affect S phase checkpoint activity during recovery from MMS treatment. To this end, cells were arrested in G1 phase and treated with MMS during the last hour of arrest. Then, MMS was washed out, and cells were allowed to recover and proceed through S phase. Flow cytometry analysis revealed a 20- to 30-min delay in S phase progression in isw2 nhp10 cells compared to wild-type (WT) results (Fig. 2b). Considering that WT cells complete the S phase in about 45 to 50 min (Fig. 2b), the 20- to 30-min delay seen with isw2 nhp10 cells reveals that isw2 nhp10 cells exhibit a marked delay in recovering from a transient MMS exposure. Budding index analysis revealed that only 5 min of the delay can be accounted for by slower kinetics of isw2 nhp10 cell release from G1 arrest (data not shown).
When WT cells were transiently treated with MMS in G1 phase, no Rad53 ISA activity was detected in the subsequent S phase, revealing that the checkpoint was deactivated by the first S phase time point (see lane 4 on Fig. 2c, top). In contrast, Rad53 ISA activity was detected within 20 min, 40 min, and 60 min during the recovery of isw2 nhp10 cells (see lanes 4, 5, and 6 on Fig. 2c, bottom). Thus, isw2 nhp10 cells display stronger activation of the S phase checkpoint and slower deactivation of the checkpoint that correlate with their delay in S phase progression. Since genetic evidence strongly suggests that DNA damage repair is not impaired in the isw2 nhp10 mutant (see reference 48 and below), these results imply that Isw2 and Ino80 affect S phase checkpoint activity after DNA damage repair.
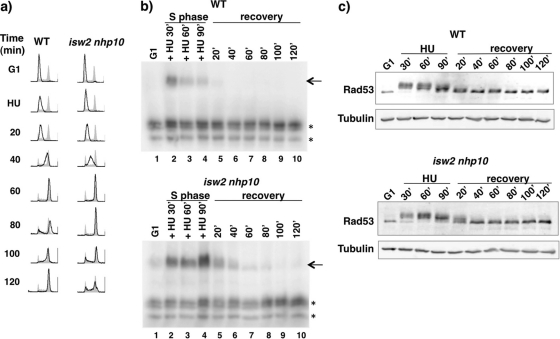
The experiments using MMS described above were complicated by the fact that replication forks stall in S phase while DNA damage is being repaired, raising the possibility that differences in the rate of DNA repair can affect how much replication forks stall. To independently monitor S phase checkpoint activity of isw2 nhp10 cells in a more synchronous manner, we transiently treated cells with HU. Cells were arrested in G1 phase and released into HU for 90 min. HU was then washed out, and cells were allowed to recover and proceed through the S phase. Flow cytometry analysis revealed a noticeable delay in S phase progression of isw2 nhp10 cells compared to WT results (Fig. 3 a). We performed the Rad53 ISA assay to monitor both Rad53 activation (in the presence of HU) and deactivation (during recovery). Both WT and isw2 nhp10 cells displayed Rad53 activity during the 90 min of HU treatment, with noticeably stronger Rad53 activity in isw2 nhp10 cells (compare lanes 2 to 4 in Fig. 3b). During the recovery, only isw2 nhp10 cells showed persistent Rad53 activity within 20 min and 40 min (lanes 5 and 6 in Fig. 3b). Rad53 protein levels were equivalent for WT and isw2 nhp10 cells and thus cannot explain the difference in Rad53 activity (Fig. 3c). Figure 3c also shows that the slow-migrating phosphorylated form of Rad53 shifts to the nonphosphorylated form more slowly in isw2 nhp10 cells, consistent with the Rad53 ISA results (Fig. 3b). These results collectively establish that Isw2 and Nhp10 attenuate S phase checkpoint activation and facilitate checkpoint deactivation.
Fig. 3.
Isw2 and Ino80 attenuate S phase checkpoint activity and facilitate efficient checkpoint deactivation during and after HU treatment. (a) Flow cytometry analysis during and after transient HU treatment. WT and isw2 nhp10 cells were arrested in G1 and released into YPD containing 200 mM HU for 90 min. Then, cells were released into the S phase in YPD and collected at the indicated time points after release. Flow cytometry was performed as described for Fig. 2b. (b) Rad53 ISA assay during and after HU treatment. Cells were harvested during G1 arrest, every 30 min during HU treatment, and every 20 min during S phase recovery. The arrows show the Rad53 ISA band, and asterisks designate other kinases with ISA activity that served as a loading control. (c) Rad53 Western blot analysis during and after HU treatment. Western blot analysis of cells grown under transient HU conditions was performed as described for panel b. The blot was probed with anti-Rad53 and antitubulin antibodies.
Evidence against increased replication fork problems in isw2 nhp10 cells.
There are two possible causes for stronger and prolonged checkpoint activation of isw2 nhp10 cells. First, Isw2 and Nhp10 may function in replication fork protection or in repair of MMS-induced DNA damage. If this were the case, isw2 nhp10 cells should experience increased replication fork problems, which cause stronger and prolonged checkpoint activation. The second possibility is that Isw2 and Nhp10 may attenuate S phase checkpoint activity and facilitate checkpoint deactivation; thus, isw2 nhp10 cells would have stronger and more constitutive checkpoint activity, leading to slower S phase progression. We favor the latter possibility because of numerous independent lines of evidence against fork protection and DNA damage repair problems in isw2 nhp10 cells (full details in Discussion).
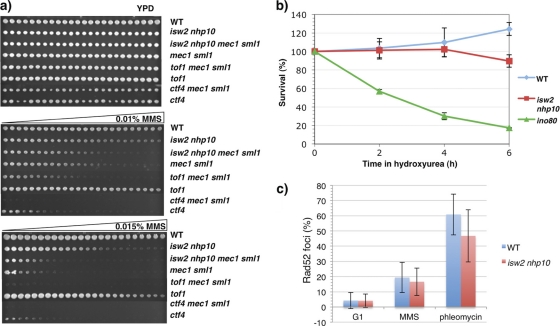
To further rule out the possibility that Isw2 and Nhp10 function in replication fork protection, we largely abrogated the S phase checkpoint through deletion of MEC1, the central kinase of the checkpoint. MEC1 must be deleted in an sml1 deletion mutant background for the mutant to remain viable, as mec1 mutation results in the depletion of intracellular nucleotide levels that can be relieved by deletion of Sml1, an inhibitor of ribonucleotide reductase (55). Abrogating the checkpoint in a strain with replication fork problems is expected to strongly decrease viability; thus, the isw2 nhp10 mec1 sml1 quadruple mutant would be predicted to display an additive MMS sensitivity if Isw2 and Nhp10 aided fork protection. On the other hand, if Isw2 and Nhp10 function in the S phase checkpoint, the quadruple mutant would be expected to show MMS sensitivity identical to that of either the isw2 nhp10 or mec1 sml1 mutant (whichever mutant has the higher MMS sensitivity). Examination of growth on MMS gradient plates revealed that the isw2 nhp10 mec1 sml1 quadruple mutant showed MMS sensitivity identical to that of the mec1 sml1 mutant (Fig. 4 a). To confirm that the MMS sensitivity of mec1 sml1 mutant cells could be exacerbated by adding mutations that increase replication fork problems, we created ctf4 mec1 sml1 and tof1 mec1 sml1 triple mutants. Ctf4 is required for coupling of DNA polymerase alpha and the MCM helicase (16), and Tof1 is required for stabilization of replication forks (3, 22). As shown in Fig. 4a, both ctf4 mec1 sml1 and tof1 mec1 sml1 mutants exhibited strongly additive MMS sensitivity, demonstrating that the mec1 mutation has a devastating effect on MMS sensitivity when mutant cells have increased replication fork problems. Therefore, the epistatic relationship of the isw2 nhp10 and mec1 sml1 double mutations strongly argues against the possibility that Isw2 and Nhp10 function in replication fork protection. In addition, the fact that the isw2 nhp10 mutation does not increase MMS sensitivity in the mec1 background supports our model that Isw2 and Nhp10 function in the S phase checkpoint pathway.
Fig. 4.
Evidence against increased replication fork troubles in isw2 nhp10 cells. (a) Spot assay for genetic-interaction test. Strains were grown to saturation and pinned on YPD plates with or without a gradient of 0.01% (vol/vol) or 0.015% (vol/vol) MMS. Plates were grown for 2 days at 30°C. (b) Viability assay following HU treatment. Percentages of surviving colonies at each time point determined relative to untreated cells are shown. Average percent survival and standard deviation (error bars) values, representing the results of three experimental replicates for WT and ino80 cells and 4 experimental replicates for isw2 nhp10 cells, are shown. (c) Quantitation of Rad52 foci after drug treatment. Cells were arrested in G1 and released into either 0.02% MMS or 6 μg/ml phleomycin for 60 min. Cells were fixed with paraformaldehyde and viewed by fluorescence microscopy. The averages and standard deviations (error bars) of the percentages of Rad52-YFP foci were determined by counting 200 cells twice each from two independent experiments.
We next examined the viability of cells after they have been subjected to prolonged exposure to HU. Cells with fork protection defects are expected to display significantly decreased viability during prolonged HU exposure, because collapsed forks cannot complete replication. We arrested cells in G1 phase and released them into media containing HU. The viability of WT cells was similar to that of isw2 nhp10 cells after 2 and 4 h of HU exposure (Fig. 4b). isw2 nhp10 cells exhibited slightly decreased viability (∼90% survival) only after 6 h in HU. After this extensive time in HU, the stronger checkpoint activation and delayed checkpoint deactivation defects of isw2 nhp10 cells may have contributed to the slightly reduced viability. In contrast, an ino80 null mutant (in a strain S288C background, since the ino80 null mutation is lethal in the strain W303 background used in this study) displayed a significant loss of viability after all time points in HU (Fig. 4b), as previously reported (30, 40).
We further examined formation of Rad52-YFP foci in isw2 nhp10 cells. Rad52 functions in homologous recombination and is targeted to the sites of DNA breaks, such as collapsed replication forks or DNA double-strand breaks (23). Treatment of cells with phleomycin, a double-strand break-inducing agent, induced Rad52 foci similarly in WT and isw2 nhp10 cells (Fig. 4c), which is consistent with our earlier conclusion that Isw2 and Nhp10 do not have a detectable role in DNA double-strand-break repair (48). Compared to untreated cell results, MMS treatment induced only modest, similar increases in numbers of Rad52 foci in both WT and isw2 nhp10 cells (Fig. 4c). This result strongly supports our conclusion that the isw2 nhp10 mutant does not have increased replication fork troubles in the presence of replication stress. Collectively, our results argue strongly against fork protection or DNA damage repair problems in isw2 nhp10 cells and support the idea that Isw2 and Nhp10 function to control the amplitude of activation and the subsequent S phase checkpoint deactivation process.
Isw2 and Ino80 physically interact with RPA.
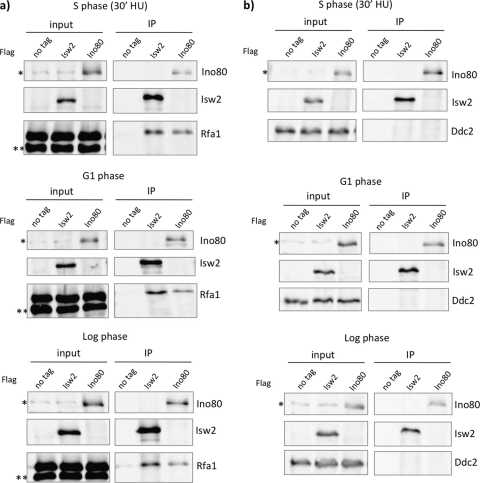
Stalled replication forks generate excess ssDNA that is bound by RPA, and accumulation of RPA signals recruitment of the S phase checkpoint proteins to stalled forks (56). It has been previously reported that both Isw2 and Ino80 are also enriched at stalled replication forks (30, 40, 48). We predict that Isw2 and Ino80 may be also be enriched at stalled forks resulting from mutations in the replicative helicase. We next asked how Isw2 and Ino80 are targeted to stalled replication forks. A high-throughput mass spectrometry analysis recently found that several Isw2 and Ino80 subunits physically associate with RPA in the presence of MMS-induced replication stress (6). To further analyze these interactions using a more stringent approach, we immunoprecipitated Isw2 and Ino80 to examine whether RPA coimmunoprecipitated in the presence of HU. Western blot analysis revealed that both FLAG-tagged Isw2 and Ino80 coimmunoprecipitated with Myc-tagged Rfa1, the largest subunit of RPA (Fig. 5 a). To test whether these interactions are dependent on replication stress, we harvested cells grown during G1 phase and log phase. Isw2 and Ino80 both coimmunoprecipitated with Rfa1 under these conditions, although slightly more Rfa1 appeared to coimmunoprecipitate with Isw2 and Ino80 during the S phase in the presence of HU (Fig. 5a). The physical interaction of Isw2 and Ino80 with Rfa1 during G1 phase suggests that their interaction is independent of replication stress and also does not have to occur at stalled replication forks. Given that stoichiometric amounts of Rfa2 and Rfa3 copurify with Rfa1 from yeast cells in the presence or absence of HU (data not shown), we believe that both Isw2 and Ino80 interact with the RPA complex in both G1 phase and S phase in the presence of HU.
Fig. 5.
Isw2 and Ino80 physically interact with RPA. (a) Co-IP experiment using Isw2 and Ino80 with Rfa1. Strains were harvested during S phase (30 min after release in the presence of 200 mM HU), G1 arrest, or log phase. FLAG immunoprecipitations were performed using untagged strains and Isw2-3xFLAG and Ino80-3xFLAG strains in which Rfa1 was 13× Myc tagged. Nonspecific, cross-reacting bands are marked by asterisks on the Western blot. (b) Co-IP experiment using Isw2 and Ino80 with Ddc2. Cells were harvested under same conditions as those described for panel a. FLAG immunoprecipitations were performed using untagged strains and Isw2-3xFLAG and Ino80-3xFLAG strains in which Ddc2 was 13× Myc tagged. A nonspecific cross-reacting band is marked by an asterisk on the Western blot.
To further confirm that Isw2 and Ino80 can physically interact with Rfa1 independently of the presence of chromatin, we tested whether Ddc2 coimmunoprecipitated. Ddc2 is the interaction partner of Mec1 and is targeted to stalled replication forks in an RPA-dependent fashion (5). We found that Ddc2 did not coimmunoprecipitate with Isw2 or Ino80 during the S phase in the presence of HU or in G1 phase or log phase (Fig. 5b). We therefore conclude that Isw2 and Ino80 specifically interact with RPA in a DNA-independent fashion, although we cannot completely exclude the possibility that the interactions take place on DNA. The physical interaction of Isw2 and Ino80 with Rfa1 is consistent with the remodeling complexes having roles in the S phase checkpoint and suggests the possibility that Isw2 and Ino80 are targeted to stalled replication forks via physical interactions with RPA.
The ATPase activity of Isw2 is required for its function in the S phase checkpoint.
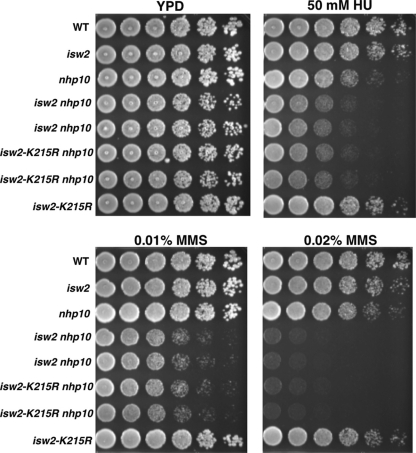
We asked whether the ATPase activity of Isw2 was required for its function(s) in the S phase checkpoint. We could not address whether the ATPase activity of Ino80 was required, because the catalytically inactive mutant is lethal in the W303 strain background used in this study. We found that a catalytically inactive Isw2 mutant, carrying isw2-K215R in combination with nhp10, displayed MMS and HU sensitivity identical to that of isw2 nhp10 cells (Fig. 6). The isw2-K215R mutation does not affect the subunit composition of the Isw2 complex. Therefore, this result demonstrates that the ATPase activity of Isw2 is essential for its function in the S phase checkpoint. The Isw2 complex exists in two forms, as either a two (Itc1, Isw2)- or a four (Itc1, Isw2, Dls1, Dpb4)-subunit complex (20, 25). Deletion of DLS1, together with deletion of NHP10, resulted in elevated sensitivity to MMS and HU, although the sensitivity was slightly weaker than that of the isw2 nhp10 mutant (data not shown). This result demonstrates that the four-subunit form of Isw2 plays the major, but not an exclusive, role in regulating S phase checkpoint activity.
Fig. 6.
The ATPase activity of Isw2 is required for its function in response to replication stress. Strains were grown to saturation, and then 5-fold serial dilutions were plated on YPD plates with or without 50 mM HU, 0.01% (vol/vol) MMS, or 0.02% (vol/vol) MMS. Plates were grown for 2 days at 30°C. The relevant genotypes are shown on the left. Two independent isolates each of isw2 nhp10 and isw2-K215R nhp10 strains are shown.
DISCUSSION
In this article, we describe a previously unknown role for ATP-dependent chromatin remodeling complexes in the S phase checkpoint. To our knowledge, this is the first report of any chromatin remodeling factors functioning to attenuate the amplitude of S phase checkpoint activation and facilitate checkpoint deactivation. Our conclusion that stronger activation of S phase checkpoint leads to slower replication fork progression is consistent with a previous report on Rad53 phosphatase mutants (44). Our genetic analysis suggested that ISW2 and NHP10 function in parallel to the Rad53 phosphatases and in the same pathway as MEC1. The striking sensitivity to HU of the quadruple isw2 nhp10 pph3 ptc2 mutant demonstrates that the Rad53 Pph3 and Ptc2 phosphatases and Isw2 and Ino80 chromatin remodeling factors are major players in S phase checkpoint control. Although proper deactivation of the S phase checkpoint is vital for cells, the only known regulators of this process are the Rad53 phosphatases (18). Our work identified a previously unknown mechanism regulating the amplitude of activation as well as facilitating deactivation of the S phase checkpoint. Cells in S phase are in a vulnerable state, especially during replication stress and recovery stages. During these times, we imagine that Isw2 and Ino80 ensure that cells can progress through S phase without extensive delay. Isw2 and Ino80 may also help recycle or distribute checkpoint proteins to the rest of the chromosomes to achieve proper replication of the entire genome.
One alternative possibility to explain our findings is that Isw2 and Nhp10 function in replication fork protection or DNA damage repair. If this were the case, stronger checkpoint activation and delayed checkpoint deactivation would be the results of the increased replication fork problems in isw2 nhp10 cells. However, we have established numerous independent lines of evidence arguing against that possibility. First, isw2 nhp10 in combination with mec1 sml1 shows epistatic MMS sensitivity instead of increased MMS sensitivity. Second, WT and isw2 nhp10 cells display similar survival rates after prolonged HU exposure, in contrast to the significantly decreased survival of ino80 null cells. Furthermore, isw2 nhp10 and WT cells exhibit similar numbers of Rad52 foci in the presence of MMS. Additional evidence against increased fork problems in isw2 nhp10 cells includes the following (48): (i) WT and isw2 nhp10 cells display identical levels of viability after treatment with a wide range of MMS concentrations; (ii) transcript array analysis reveals no indication of increased DNA damage in isw2 nhp10 cells in the presence of MMS; and (iii) isw2 and nhp10 mutations cause additive MMS sensitivity with all known DNA repair pathways, thus demonstrating that they do not function in these pathways. These results collectively and strongly argue against fork protection defects in isw2 nhp10 cells. Furthermore, the fact that the isw2 nhp10 mutation does not increase MMS sensitivity in the mec1 background demonstrates that ISW2 and NHP10 function within the MEC1 pathway. The two major functions of the central kinase Mec1 in the S phase checkpoint, replication fork protection and activation of Rad53, are separable (8, 9). Our results strongly suggest that Isw2 and Nhp10 function in the Rad53 activation, but not the replication fork protection, role of Mec1.
Recent studies suggested that Ino80 is required for the DNA replication stress response. These studies were done using an ino80 null mutant, which is extremely slow growing in the S288C background. ino80 is hypersensitive to replication inhibitors, and one report showed that Ino80 stabilizes a stalled replisome and thus is required for efficient fork progression (30). Another report demonstrated that ino80 cells displayed decreased DNA synthesis after transient HU treatment and suggested that Ino80 functions to aid in fork restarting (40). Recently, yet another report showed that, in the presence of MMS, Ino80 functions in DNA damage tolerance pathways and not fork progression or stability (12). Collectively, these reports suggest the possibility that the Ino80 complex has multiple functions in DNA damage and the replication stress response. Alternatively, it is possible that Ino80 has one major function and that an ino80 null mutation causes severe trouble in the replication stress response that can lead to pleotropic phenotypes, depending on the assay systems.
Either way, the phenotypes of the isw2 nhp10 cells described in this article are different from those of the ino80 null mutant. We are confident that the phenotypes caused by the nhp10 mutation reflect Ino80 complex functions, as NHP10 and multiple other Ino80 subunit genes exhibit identical genetic interactions with ISW2 (48). In addition, purification of Nhp10 yields only Ino80 complex and no other proteins, showing that Nhp10 is present exclusively in the Ino80 complex in vivo (27). It is therefore likely that the nhp10 mutation affects specific subsets of Ino80 function, which enabled us to uncover its functions that parallel those of Isw2.
It is likely that ISWI subfamily members and Ino80 play similar roles in metazoan cells. Mammalian ISWI complexes have been implicated in promoting DNA replication in heterochromatin regions (10) and are recruited to replication foci through interaction with the DNA clamp PCNA (34). Human Ino80 was also recently suggested to facilitate DNA replication by binding to chromatin at PCNA foci (19). Depletion of either mammalian ISWI or human Ino80 leads to a delay in S phase progression similar to the phenotype of S. cerevisiae isw2 nhp10 cells. While mammalian ISWI and Ino80 complexes have not been reported to have roles in the control of S phase checkpoint activity, their roles in facilitating replication suggest possible evolutionary functional conservation.
A distantly related member of the SWI2/SNF2 ATPase superfamily in mammalian cells, the ssDNA annealing helicase SMARCAL1/HARP, was found to directly bind RPA at sites of replication stress (4, 7, 52, 54). One similarity between SMARCAL1, Isw2, and Ino80 is that they all physically interact with RPA in the presence and absence of DNA damage or replication stress. However, SMARCAL1 has been suggested to protect stalled replication forks, unlike Isw2 and Nhp10. In addition, SMARCAL1 does not appear to exhibit chromatin remodeling activity, while the annealing helicase activity of ISWI and Ino80 complexes has not been detected (53). These results show that RPA functions as a key player in the DNA replication stress response by providing a platform to recruit a large number of enzymes with different biochemical activities that are required for DNA transactions at stalled replication forks.
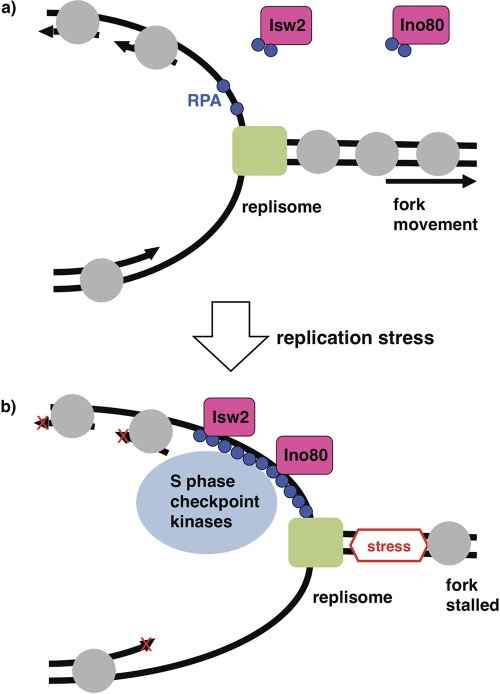
The enrichment of Isw2 and Ino80 at stalled forks and their physical interactions with RPA suggest that these chromatin remodeling factors function directly in the S phase checkpoint (Fig. 7). How do Isw2 and Ino80 control the amplitude of S phase checkpoint activation and facilitate deactivation? One interesting possibility is that they function by physically removing checkpoint proteins from stalled replication forks. While chromatin remodeling enzymes act on histones, some enzymes in the same SWI2/SNF2 ATPase superfamily can function on nonhistone proteins. For example, Mot1 modulates transcription by removing TATA binding protein from DNA (2). Isw2 and/or Ino80 might act similarly to remove S phase checkpoint proteins from DNA. Consistent with this possibility, the ATPase activity of Isw2 is required for its function. One possible explanation is that Isw2 and/or Ino80 physically remove RPA bound to ssDNA at stalled forks. We tested this possibility by an RPA band shift in vitro as well as by kinetic analyses of RPA levels at stalled forks in WT and isw2 nhp10 cells in vivo, but we did not see any evidence that Isw2 or Ino80 affects RPA binding to ssDNA (data not shown). Alternatively, Isw2 and/or Ino80 may function by changing the chromatin structure around stalled replication forks, which could affect the levels of checkpoint proteins that accumulate. Although Ino80 has been shown to function by replacing histone variant Htz1 with canonical histone H2A (31), a htz1 mutation did not rescue the sensitivity of isw2 nhp10 mutant to replication inhibitors, suggesting that Isw2 and Ino80 attenuate S phase checkpoint activity independently of the presence of Htz1 (data not shown). Elucidating the molecular mechanisms by which Isw2 and Ino80 control S phase checkpoint activity would be a significant next step.
Fig. 7.
A model for functions of Isw2 and Ino80 at stalled replication forks. (a) In the absence of replication stress, the replisome (green rectangle) moves smoothly. Isw2 and Ino80 physically interact with RPA (blue circles) independently of replication stress and DNA. Gray circles denote nucleosomes. (b) When a replication fork encounters replication stress, the fork stalls. RPA then accumulates and targets Isw2, Ino80, and checkpoint proteins. Isw2 and Ino80 at stalled replication forks attenuate the S phase checkpoint response and facilitate the subsequent deactivation process.
ACKNOWLEDGMENTS
We thank Linda Breeden (courtesy of Marco Foiani) for EL7E1 antibody and Doug Koshland for antitubulin antibody. We are grateful to Oscar Aparicio and Dave Toczyski for helpful discussion and suggestions and to Huilin Zhou for his initial discovery of RPA interactions with Isw2 and Ino80. We thank Tsukiyama laboratory members, Sue Biggins, and Harmit Malik for comments on the manuscript.
This work was supported by Chromosome Metabolism and Cancer Training grant NIH T32 CA09657 to T. J. Au and grant RO1GM058465 to T. Tsukiyama.
Footnotes
Published ahead of print on 19 September 2011.
REFERENCES
- 1. Alcasabas A. A., et al. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958–965 [DOI] [PubMed] [Google Scholar]
- 2. Auble D. T., et al. 1994. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920–1934 [DOI] [PubMed] [Google Scholar]
- 3. Bando M., et al. 2009. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J. Biol. Chem. 284:34355–34365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bansbach C. E., Betous R., Lovejoy C. A., Glick G. G., Cortez D. 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 23:2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barlow J. H., Lisby M., Rothstein R. 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell 30:73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen S. H., Albuquerque C. P., Liang J., Suhandynata R. T., Zhou H. 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 285:12803–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciccia A., et al. 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23:2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cobb J. A., Bjergbaek L., Shimada K., Frei C., Gasser S. M. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cobb J. A., et al. 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 19:3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins N., et al. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627–632 [DOI] [PubMed] [Google Scholar]
- 11. Downs J. A., et al. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979–990 [DOI] [PubMed] [Google Scholar]
- 12. Falbo K. B., et al. 2009. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat. Struct. Mol. Biol. 16:1167–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fazzio T. G., Tsukiyama T. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333–1340 [DOI] [PubMed] [Google Scholar]
- 14. Flaus A., Martin D. M., Barton G. J., Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34:2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedel A. M., Pike B. L., Gasser S. M. 2009. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 21:237–244 [DOI] [PubMed] [Google Scholar]
- 16. Gambus A., et al. 2009. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 28:2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433 [DOI] [PubMed] [Google Scholar]
- 18. Heideker J., Lis E. T., Romesberg F. E. 2007. Phosphatases, DNA damage checkpoints and checkpoint deactivation. Cell Cycle 6:3058–3064 [DOI] [PubMed] [Google Scholar]
- 19. Hur S. K., et al. 2010. Roles of human INO80 chromatin remodeling enzyme in DNA replication and chromosome segregation suppress genome instability. Cell. Mol. Life Sci. 67:2283–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iida T., Araki H. 2004. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jónsson Z. O., Jha S., Wohlschlegel J. A., Dutta A. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16:465–477 [DOI] [PubMed] [Google Scholar]
- 22. Katou Y., et al. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078–1083 [DOI] [PubMed] [Google Scholar]
- 23. Lisby M., Rothstein R., Mortensen U. H. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. U. S. A. 98:8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Majka J., Niedziela-Majka A., Burgers P. M. 2006. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell 24:891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McConnell A. D., Gelbart M. E., Tsukiyama T. 2004. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 24:2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melo J. A., Cohen J., Toczyski D. P. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morrison A. J., et al. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775 [DOI] [PubMed] [Google Scholar]
- 28. Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617–656 [DOI] [PubMed] [Google Scholar]
- 29. O'Neill B. M., et al. 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. U. S. A. 104:9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papamichos-Chronakis M., Peterson C. L. 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 15:338–345 [DOI] [PubMed] [Google Scholar]
- 31. Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L. 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144:200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pellicioli A., Lee S. E., Lucca C., Foiani M., Haber J. E. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293–300 [DOI] [PubMed] [Google Scholar]
- 33. Pellicioli A., et al. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poot R. A., et al. 2004. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat. Cell Biol. 6:1236–1244 [DOI] [PubMed] [Google Scholar]
- 35. Ruiz C., Escribano V., Morgado E., Molina M., Mazon M. J. 2003. Cell-type-dependent repression of yeast a-specific genes requires Itc1p, a subunit of the Isw2p-Itc1p chromatin remodelling complex. Microbiology 149:341–351 [DOI] [PubMed] [Google Scholar]
- 36. Sanchez Y., et al. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357–360 [DOI] [PubMed] [Google Scholar]
- 37. Shen X., Mizuguchi G., Hamiche A., Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541–544 [DOI] [PubMed] [Google Scholar]
- 38. Shen X., Ranallo R., Choi E., Wu C. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12:147–155 [DOI] [PubMed] [Google Scholar]
- 39. Sherriff J. A., Kent N. A., Mellor J. 2007. The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol. Cell. Biol. 27:2848–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimada K., et al. 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18:566–575 [DOI] [PubMed] [Google Scholar]
- 41. Shimada K., Pasero P., Gasser S. M. 2002. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16:3236–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun Z., Fay D. S., Marini F., Foiani M., Stern D. F. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395–406 [DOI] [PubMed] [Google Scholar]
- 43. Sun Z., Hsiao J., Fay D. S., Stern D. F. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272–274 [DOI] [PubMed] [Google Scholar]
- 44. Szyjka S. J., et al. 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 22:1906–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsukiyama T., Palmer J., Landel C. C., Shiloach J., Wu C. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438:379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Attikum H., Fritsch O., Hohn B., Gasser S. M. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788 [DOI] [PubMed] [Google Scholar]
- 48. Vincent J. A., Kwong T. J., Tsukiyama T. 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 15:477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Whitehouse I., Rando O. J., Delrow J., Tsukiyama T. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031–1035 [DOI] [PubMed] [Google Scholar]
- 50. Xiao W., Chow B. L., Rathgeber L. 1996. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr. Genet. 30:461–468 [DOI] [PubMed] [Google Scholar]
- 51. Yadon A. N., et al. 2010. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol. Cell. Biol. 30:5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuan J., Ghosal G., Chen J. 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev. 23:2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yusufzai T., Kadonaga J. T. 2008. HARP is an ATP-driven annealing helicase. Science 322:748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yusufzai T., Kong X., Yokomori K., Kadonaga J. T. 2009. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 23:2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao X., Muller E. G., Rothstein R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329–340 [DOI] [PubMed] [Google Scholar]
- 56. Zou L., Elledge S. J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548 [DOI] [PubMed] [Google Scholar]