Abstract
Protein syntheses mediated by cellular and viral internal ribosome entry sites (IRESs) are believed to have many features in common. Distinct mechanisms for ribosome recruitment and preinitiation complex assembly between the two processes have not been identified thus far. Here we show that the methylation status of rRNA differentially influenced the mechanism of 80S complex formation on IRES elements from the cellular sodium-coupled neutral amino acid transporter 2 (SNAT2) versus the hepatitis C virus mRNA. Translation initiation involves the assembly of the 48S preinitiation complex, followed by joining of the 60S ribosomal subunit and formation of the 80S complex. Abrogation of rRNA methylation did not affect the 48S complex but resulted in impairment of 80S complex assembly on the cellular, but not the viral, IRESs tested. Impairment of 80S complex assembly on the amino acid transporter SNAT2 IRES was rescued by purified 60S subunits containing fully methylated rRNA. We found that rRNA methylation did not affect the activity of any of the viral IRESs tested but affected the activity of numerous cellular IRESs. This work reveals a novel mechanism operating on a cohort of cellular IRESs that involves rRNA methylation for proper 80S complex assembly and efficient translation initiation.
INTRODUCTION
Initiation of translation on the majority of the cellular mRNAs is 5′ cap dependent. This process relies on the recruitment of the 43S initiation complex to the cap structure at the 5′ end of the mRNA promoted by cap-binding initiation factor eIF4F to form 48S initiation complex and subsequent scanning of the 48S complex across the 5′ untranslated region (UTR) in search of the initiator AUG codon. This major initiation pathway is widely believed to account for most cellular mRNA translation (19, 20). On the other hand, a minor cap-independent initiation pathway relying on the presence of internal ribosome entry sites (IRESs) has been described for a subset of viral mRNAs (for a recent review, see reference 15). Viral IRES-dependent translation has been shown to rely on both canonical and noncanonical interactions between IRES elements, canonical eukaryotic initiation factors (eIFs), IRES trans-acting factors (ITAFs; largely considered to act as noncanonical initiation factors), and 40S ribosomal subunits. As a rule, viral IRES-mediated translation does not require the cap-binding initiation factor eIF4E and in general has reduced requirements for other members of the eIF4F factor/complex (specifically, intact eIF4G) (15, 19). It has been further shown that certain cellular mRNAs may rely on similar translation initiation mechanism (21, 23, 24). Reports on the IRES elements in cellular mRNAs are emerging on a continuing basis; however, the mechanism(s) of their activation and function remains less well understood.
The best-studied viral IRES elements can be assigned (based on their structure and requirements for the canonical initiation factors) to four different types. Type 1 (e.g., poliovirus) and type 2 (e.g., encephalomyocarditis virus [EMCV]) IRESs require eIF4G and eIF4A but do not require eIF4E for their activity (10, 39, 41); type 3 (hepatitis C virus [HCV]) IRESs require eIF3 but not eIF4F (40, 47). Finally, type 4 IRESs (cricket paralysis virus [CrPV] and Taura shrimp virus [TSV]) can support initiation without the need for any canonical initiation factors at all (44, 59).
It has been shown that cellular IRES elements, similar to the viral counterparts in general, have reduced requirements for initiation factors eIF4E and eIF4G (for a review, see references 21, 23, and 24), thus resembling in this feature the type1 and type 2 viral IRES elements. However, certain cellular IRESs, such as those of p120 catenin and vascular endothelial growth factor (46), seem to have a strong requirement for eIF4G and also appear to be dependent on the activities of initiation factors eIF3 and eIF4A (c-myc and N-myc IRESs) (51). At the same time, a recently discovered cellular IRES element in c-Src mRNA was suggested to be able to bind 40S ribosomal subunits directly (2), a feature similar to that of type 3 and type 4 viral IRES elements. The majority of studies done so far have argued that cellular IRES-mediated translation has many features in common with viral IRES-mediated protein synthesis. To date, it remains unclear whether cellular IRES-mediated translation might have features distinct from those of viral IRES-mediated translation.
Here we show that protein synthesis mediated by a cohort of cellular IRES-containing mRNAs (unlike viral IRES-containing mRNAs) is critically dependent on the methylation of the rRNA. To our knowledge, this work for the first time provides insights into the mechanism leading to the reduced expression of a group of cellular IRESs under these conditions. In addition, we show that both viral IRES-mediated protein synthesis and cap-dependent translation remain refractory to the disruption of rRNA methylation.
rRNA methylation is a covalent modification of 2′-O-ribose, the biological role of which is unclear (11). It has been established that a C/D box small-nucleolar ribonucleoprotein (snoRNP) accomplishes 2′-O-methylation on 90S pre-rRNA and that fibrillarin, a component of the C/D box snoRNP, functions as a methyltransferase (38, 55, 58). The other common modification of rRNA is pseudouridylation (conversion of uridine to pseudouridine) mediated by the snoRNP complex of H/ACA guide snoRNA and the pseudouridine synthase dyskerin (37, 57). A mutation(s) in the DKC1 gene, encoding the pseudouridine synthase, reduces rRNA pseudouridylation and causes dyskeratosis congenita (DC), an X-linked disease characterized by premature aging and increased tumor susceptibility (32, 35). Reduced pseudouridylation in DC patients does not affect cap-dependent protein synthesis but results in impairment of IRES-mediated translation of p53 (5) and p27 mRNAs (6), thus contributing to the pathogenesis and development of cancer. Interestingly, inhibition of rRNA pseudouridylation was found to affect both cellular (p53 and p27) and viral (HCV and CrPV) IRES-mediated protein synthesis to similar extents (43).
We have already shown that, in contrast to the effects of reduced pseudouridylation, the inhibition of rRNA methylation results in an about 50% decrease in the expression of p27, p53, and sodium-dependent neutral amino acid transporter 2 (SNAT2) IRESs but not HCV and CrPV IRESs (8). To get further insights into the mechanism leading to the inhibition of cellular IRES-mediated translation under these conditions, we have used a combination of ex vivo and in vitro approaches.
We show here that both the polysomal abundance of the SNAT2 mRNA and system A amino acid transport activity are affected by the inhibition of rRNA methylation. We further show that the assembly of the 80S complexes (under conditions of reduced rRNA methylation) is significantly impaired on cellular (SNAT2), but not viral (HCV), IRES elements. At the same time, rRNA methylation did not affect 48S complex formation on both cellular and viral IRES elements, suggesting that the effects observed are due largely to the impairment of the subunit joining on the former mRNAs. We further show that this defect could be rescued by the addition of wild-type (containing fully methylated rRNA) 60S ribosomal subunits, suggesting that the defect involved changes in the interaction of the 48S preinitiation complex with the large ribosomal subunit. Finally, cap-dependent initiation was not affected under any of the conditions tested.
We have previously shown that the depletion of ribosomal protein L13a resulted in a reduction of rRNA methylation (8) accompanied by a decrease in cellular IRES-mediated protein synthesis. However, the nature of the link between L13a and rRNA methylation remained unclear. Here we show that L13a and fibrillarin associate and L13a may thus potentially regulate methyltransferase activity of the C/D box snoRNP complex. Overall, our results suggest that cellular IRES-mediated translation includes characteristic features distinct not only from cap-dependent translation but also from viral IRES-mediated protein synthesis. The dependence of cellular IRES-mediated protein synthesis on rRNA methylation reported here may provide a unique platform for the differentiation of cellular and viral IRES-mediated translation and for the discovery of new cellular IRES elements.
MATERIALS AND METHODS
Cells and culture conditions.
Human embryonic kidney 293T (HEK293T) and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, and 100 U/ml penicillin and streptomycin at 37°C and 5% CO2.
Cells were starved for amino acids by culturing in Krebs-Ringer bicarbonate buffer with 10% dialyzed FBS.
Plasmid construction.
The designs of the constructs m/CON, m/hpCON, m/hpSNAT2-1, and m/SNAT2-1/PA98 were previously published (14). The SNAT2 IRES sequence used in this study is the same sequence previously reported as SNAT2-1 (14). m/hpCrPV was constructed by replacing the SNAT2-1 IRES sequence in m/hpSNAT2-1 with the CrPV IRES sequence amplified from a CrPV IRES-containing construct (gift from Peter Sarnow, Stanford University, Stanford, CA). To obtain m/hpSNAT2-1/PA98, the same hairpin from m/hpCON was inserted between the transcription and translation start sites. The SNAT2-1 IRES sequence from this construct was replaced with the HCV IRES sequence to make the m/hpHCV/PA98 construct.
SNAT2-1 IRES-containing bicistronic reporter construct pRF-SNAT2-1 was previously reported (8). To generate the pRF-c-Src construct, the c-Src IRES was amplified by reverse transcription (RT)-PCR from HEK293T cell total RNA using primers based on the published sequence (2). The sequence was verified and replaced with the SNAT2-1 IRES sequence of the vector pRF-SNAT2-1. Similarly, pRF-EMCV was generated with the EMCV IRES sequence amplified from an EMCV IRES-containing construct (gift from Davide Ruggero, University of California, San Francisco, CA).
Translational status of IRES-containing cellular mRNAs.
The translational status of several endogenous cellular mRNAs containing IRESs, such as the SNAT2, CAT-1, and c-Myc mRNAs, was measured by assessing the ratio of polysomal to nonpolysomal mRNA fractions. Stress was induced by amino acid starvation (AAS) for IRES activation of SNAT2 and CAT-1 (14, 60) and by treatment with recombinant human TRAIL for activation of c-Myc IRES (52). Cytoplasmic extracts were prepared from control or stress-induced cells and resolved by centrifugation using 10 to 50% sucrose density gradients. One microgram of total RNA from the pooled nonpolysome and polysome fractions was reverse transcribed using random hexamer primers. PCR amplification was further carried out using SYBR green PCR master mix (Applied Biosystems, Foster City, CA). Primers were designed with the intent to produce 90- to 200-bp amplified product. The ratios of the CT values obtained from polysomal and nonpolysomal fractions were determined. The primer sequences used to quantify different mRNAs were as follows (F, forward; R, reverse): SNAT2(F), 5′-TGCCATGGCTAATACTGGAA-3′; SNAT2(R), 5′-TTGGCAGTCTTCAAAAGGAGA-3′; CAT-1(F), 5′-GTCTGTCTGTTCGCGATCCT-3′; CAT-1(R), 5′-AGAGGACAGCCTCGATCTTG-3′; c-Myc(F), 5′-TTTCGGGTAGTGGAAAACCA-3′; c-Myc(R), 5′-CACCGAGTCGTAGTCGAGGT-3′; GAPDH(F), 5′-AAGGTGAAGGTCGGAGTCAA-3′; GAPDH(R), 5′-AATGAAGGGGTCATTGATGG-3′.
In vitro transcription.
The SNAT2-1 IRES-containing, ApppG (unmethylated cap; Epicenter Biotechnologies, Madison, WI)-capped, poly(A)-tailed transcript was made in vitro from linearized m/hpSNAT2-1/PA98 using the T3 mMessage mMachine kit (Ambion, Austin, TX). The m7G-capped T7 gene 10 transcript was similarly transcribed from linearized pGEMEX-2 (Promega, Madison, WI). To generate radiolabeled RNA, the appropriate constructs were linearized, followed by in vitro transcription in the presence of [α-32P]UTP (Perkin-Elmer, Boston, MA). The full-length 32P-labeled RNA was gel purified before use in in vitro ribosome assembly reaction mixtures.
Preparation of translation-competent extracts and in vitro translation.
Translation-competent extracts from 293T cells were made by following previously published methods using HeLa cells (7, 33). In short, exponentially growing cells were harvested, washed thrice with cold phosphate-buffered saline, resuspended in lysis buffer (20 mM HEPES [pH 7.6], 10 mM K acetate, 0.5 mM MgCl2, 5 mM dithiothreitol [DTT], and proteinase inhibitor cocktail), and kept on ice for 15 min. Cells were homogenized by 15 strokes in a Dounce homogenizer, followed by centrifugation for 10 min at 13,000 × g at 4°C. The protein concentration of the supernatant was adjusted to 15 μg/μl (translation-competent extract), aliquoted, frozen in liquid nitrogen, and stored at −80°C until use. For in vitro translation, the in vitro-transcribed RNA and 50 μCi of [35S]methionine was added in 100 μg of the nuclease-treated extract in a 20-μl translation reaction mixture containing a 50 μM amino acid mixture (without methionine), 20 mM creatine phosphate, 80 ng/μl creatine kinase, 16 mM HEPES (pH 7.6), 0.8 mM ATP, 0.1 mM GTP, 0.1 mM spermidine, 100 mM K acetate, and 2 mM MgCl2. The newly synthesized and radiolabeled protein was resolved by SDS-PAGE, followed by autoradiography.
Elongation assay by measuring poly(U)-directed polyphenylalanine synthesis.
Translation-competent extracts from control, cycloleucine-treated, and L13a-depleted cells were prepared as described above. The elongation assay was performed as previously described (17, 36). In short, the extract was supplemented with 5 mM MgCl2 and 50 μM unlabeled amino acid without phenylalanine. Poly(U) was added at a final concentration of 2.2 μg/ml. The assay was performed with 5 μCi [3H]phenylalanine and 14 μl of lysate in a total volume of 20 μl for 1 h at 30°C. At the end of the incubation period, a 10-μl aliquot was withdrawn and subjected to trichloroacetic acid (TCA) precipitation with 1 ml of cold 25% TCA. The TCA precipitate was filtered through a Whatman filter, ethanol washed, dried, and counted in scintillation fluid.
Purification of 60S and 40S ribosomal subunits.
60S and 40S ribosomal subunits were isolated as described previously (39), with minor modifications. Cells were lysed by three rapid freeze-thaw cycles in cold breaking buffer (20 mM Tris [pH 7.6], 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mg/ml heparin), followed by centrifugation at 10,000 rpm for 20 min at 4°C. Ribosomes were pelleted by centrifugation in 2 M sucrose cushion at 43,000 rpm for 21 h at 4°C using a Beckman 70.1 Ti rotor and resuspended in buffer 1 (20 mM Tris-HCl [pH 7.6], 50 mM KCl, 4 mM MgCl2, 2 mM DTT, 0.25 M sucrose). KCl was added slowly to this suspension to a final concentration of 0.5 M, and incubation was continued on ice for 30 min, followed by centrifugation at 50,000 rpm for 5 h at 4°C in a Beckman TLA-110 rotor. The pellet was resuspended in buffer 2 (20 mM Tris [pH 7.5], 50 mM KCl, 4 mM MgCl2, 2 mM DTT) and incubated with 1 mM puromycin for 10 min at 4°C and then for 10 min at 37°C, followed by centrifugation in a 5 to 25% sucrose density gradient prepared in buffer 3 (20 mM Tris [pH 7.5], 0.5 M KCl, 4 mM MgCl2, 2 mM DTT) at 20,000 rpm for 18 h at 4°C using a Beckman SW32.1 Ti rotor. The 60S subunit peak fractions were combined and concentrated using Centricon (Millipore, Billerica, MA). For the 40S subunit, the corresponding peak fractions were used.
Sucrose density gradient analysis of complex formation during translation initiation.
Polysomes were analyzed from cytoplasmic extracts as described previously (8). In brief, cytosolic extracts were made from exponentially growing cells pretreated with 100 μg/ml cycloheximide (MP Biomedicals, Santa Ana, CA) for 15 min at 37°C before harvesting. Ten optical density units of cytoplasmic extracts were carefully layered over 10 to 50% linear sucrose density gradients in polysome buffer (PB; 10 mM HEPES [pH 7.5], 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 50 U recombinant RNasin [Promega, Madison, WI], and 0.1% NP-40 [Sigma, St. Louis, MO]) and centrifuged for 18 h at 17,000 rpm. The density gradients were unloaded by upward displacement using a programmable density gradient system with a UA-6 detector (Teledyne Isco, Lincoln, NE). In some cases, total RNA and total protein from fractions were precipitated with TRIzol or TCA, respectively, and used for RT-PCR or Western analysis.
To detect the 48S and 80S ribosomal complexes, in vitro-synthesized, 32P-labeled reporter RNAs (500,000 cpm) were incubated with translation-competent extract (20 mg/ml) in the presence of guanylyl-imidodiphosphate (GMP-PNP; 2 mM) or cycloheximide (0.5 mM), respectively. Initiation reactions were stopped by placement on ice and resolved by centrifugation on a 5 to 25% linear sucrose density gradient in PB. Fractions (1.2 ml) were collected, and radioactivity was determined by scintillation counting.
Dual-luciferase assays.
Cells were seeded into 6-well plates at a density of 2 × 105 cells/well. After 16 h, cells were transfected with bicistronic reporter vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) by following the vendor's protocol. Cell lysates were prepared after an additional 24 h, and dual-luciferase assays were performed using the DLR assay kit (Promega, Madison, WI). Ratios of firefly and Renilla luciferase-derived luminescence were recorded using a 20/20n luminometer (Turner BioSystems, Sunnyvale, CA). To perform the dual-luciferase assay under AAS conditions, transfected cells were starved for amino acids by culturing in AAS medium (Krebs-Ringer bicarbonate buffer with 10% dialyzed FBS) for 3 h before cell lysate preparation.
Amino acid transport analysis.
HeLa cells were seeded into a 24-well dish at a density of 5 × 104 cells/well. After 24 h, the cells were starved for amino acids for 3 h by culturing in AAS medium. The activity of transport system A was determined by measuring the uptake of [14C]methylaminoisobutyric acid (0.1 mM, 2 μCi/ml) for 1 min at 37°C in 0.25 ml of Krebs-Ringer bicarbonate buffer. The incubation was terminated with two rapid washes in 3 ml of ice-cold 300 mM urea. Ethanol extracts of cells were added to 0.6 ml of scintillation fluid and counted for radioactivity in a Wallac Microbeta Trilux counter (Perkin-Elmer, Boston, MA). Cell monolayers were then dissolved with 0.5% sodium deoxycholate in 1 N NaOH, and protein content was determined using a modified Lowry procedure (13).
Metabolic labeling of cells to examine rRNA methylation.
rRNA methylation was determined by [3H]methylmethionine labeling of the de novo-synthesized rRNA as previously described (8). In short, 1 × 106 HEK293T cells were preincubated in 1 ml of methionine-free DMEM for 30 min, followed by pulse-labeling with 50 μCi of [3H]methylmethionine (Perkin-Elmer, Boston, MA) for 1 h. To test rRNA methylation in L13a-depleted or cycloleucine-treated cells, cells were subjected to treatment with short hairpin RNA (shRNA) against L13a or 2 mg/ml cycloleucine, followed by pulse-labeling. Depletion of L13a was confirmed by immunoblot analysis using anti-L13a antibody. To monitor total RNA synthesis in parallel, the same number of cells were pulse-labeled with 50 μCi of [3H]uridine for 1 h, followed by extraction of the total RNAs. At the end of the pulse period, cells were washed and total RNAs were extracted, resolved through a 1% agarose gel, and transferred to Hybond N+ membrane. Dried membranes were treated with Amplify (Amersham, Arlington Heights, IL) and exposed to film (Kodak, Rochester, NY).
Determination of methylation efficiency at different rRNA sites.
Site-specific methylation ratios were determined under conditions in which rRNA methylation was inhibited by either cycloleucine treatment or L13a depletion. Several known sites for rRNA methylation, such as the nucleotides at positions 1612, 1858, 3810, and 4198 in the 28S rRNA and 37, 1489, 1713, and 1803 in the 18S rRNA, respectively (4, 31), were selected for methylation ratio quantification by the previously published method (4, 30). In short, RT reactions using primers 3′ to the methylation sites were carried out with either a low (10 μM) or a high (1 mM) deoxynucleoside triphosphate (dNTP) concentration. Quantitative PCRs (qPCRs) were performed using primer pairs corresponding to the 5′ and 3′ regions of the methylation sites. ΔCT values were obtained after normalizing the target CT values with the CT value of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The methylation ratio of each site was determined from the ratio of the ΔCT values obtained using high and low dNTP concentrations.
Immunofluorescence studies.
293T cells were transfected with plasmids expressing L13a-hemagglutinin (HA) fusion constructs. At 24 h posttransfection, cells were fixed and then incubated with a mouse monoclonal antibody against the HA tag (Molecular Probes, Eugene, OR) and a rabbit polyclonal antibody against fibrillarin (Abcam, Cambridge, MA). Cells were then incubated with appropriate color-conjugated secondary antibodies (Invitrogen, Carlsbad, CA). Standard 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) staining was also performed. Cells were then imaged in a Nikon TE-2000-E inverted epifluorescence microscope. z stacks were obtained, and a deconvolution algorithm was used to process the image stacks.
RESULTS
Translation and polysomal abundance of the SNAT2 IRES-containing reporter mRNA decrease upon L13a depletion and/or inhibition of rRNA methylation.
Using bicistronic reporter constructs, we have previously shown that the activity of several cellular (p53, p27, SNAT2), but not viral (HCV, CrPV), IRESs substantially decreased under conditions of inhibition of rRNA methylation (8). To get further insights into the mechanism leading to this decrease, we have performed here a detailed study using the SNAT2 IRES as a model (14). We chose the SNAT2 IRES because it was previously shown to be active in the absence of stress (14). Expression of reporter constructs containing the 5′ UTR of the SNAT2 mRNA was unaffected by the inclusion of a stable hairpin in front of the IRES (14). We first investigated the polysomal association of the SNAT2 IRES-containing reporter mRNA under normal conditions and under the conditions of inhibition of rRNA methylation and compared it to that of the CrPV IRES-containing mRNA. Inhibition of rRNA methylation was induced by depleting cells of L13a or treating them with cycloleucine (a methylation inhibitor) as described before (8). Monocistronic reporter constructs with a stable hairpin structure in the 5′ UTRs of reporter mRNAs were used (Fig. 1A). SNAT2 IRES polysomal association was measured using the m/hpSNAT2 construct described previously (14). The construct m/hpCrPV (Fig. 1A) was made by replacing the SNAT2 IRES sequence in m/hpSNAT2 with the CrPV IRES (54). The m/CON and m/hpCON constructs (Fig. 1A) were used as reference standards. 293T cells were transfected with these four constructs. For all constructs, transfections were carried out using cells stably expressing shRNA against L13a (i.e., L13a-depleted cells). Cells expressing an irrelevant shRNA were used as controls. Inhibition of rRNA methylation and L13a depletion in cycloleucine-treated and/or L13a-depleted cells were confirmed by metabolic labeling of RNA with [3H]methylmethionine and immunoblotting with anti-L13a specific antibody, respectively, as previously described (8) (Fig. 1B). Consistent with our previous data, pulse-labeling with [3H]uridine showed an unaltered rate of general rRNA synthesis when rRNA methylation is inhibited (Fig. 1B). Lysates equivalent to 10 optical density units of RNA isolated from the transfected cells were resolved by sucrose density gradient centrifugation. Polysomal fractions were collected, and the presence of reporter luciferase (luc) mRNA in the fractions was detected by RT-PCR using luc-specific primers. RT-PCR analyses revealed that under L13a-depleted conditions, the SNAT2 IRES-containing reporter luc mRNA shifted from the heavy polyribosome fractions to the lighter ones, compared to the cells transfected with the same construct and control shRNA (Fig. 1C). The polyribosomal distribution of the m/CON, m/hpCON, and m/hpCrPV reporter constructs was also unaffected under the L13a-depleted conditions (Fig. 1C). Endogenous β-actin mRNA from the same fractions was also detected and used as a reference for loading (Fig. 1D). All of the polysomal profiles obtained from the transfected cell lysates were almost identical (data not shown). The polysomal profile obtained from the m/hpSNAT2-transfected and L13a-depleted cells is shown as an example (Fig. 1C). This observation corroborated our previous findings showing that inhibition of rRNA methylation affects the activity of the SNAT2 IRES (8) and further suggested that the translation of the SNAT2 IRES-containing mRNA has most probably been affected at the initiation step of mRNA translation.
Fig. 1.
Inhibition of rRNA methylation reduces polyribosomal association and translation of SNAT2 IRES-containing monocistronic RNA. (A) Diagram of the monocistronic reporter harboring IRES elements and hairpin RNA upstream of the luc reporter. (B) Status of rRNA methylation and L13a depletion in 293T cells. L13a was depleted from 293T cells by expressing shRNA against L13a. The extent of depletion was confirmed by immunoblot analysis using an anti-L13a antibody (lower panel). The same blot was probed for actin as a loading control. To determine the level of rRNA methylation in L13a-depleted and cycloleucine-treated cells, pulse-labeling with [3H]methylmethionine was performed. rRNA synthesis was examined by pulse-labeling with [3H]uridine. Total RNA was resolved by 1% denaturing agarose gel electrophoresis and transfer to nylon membrane, followed by autoradiography (upper panel). (C) Reduced polyribosomal association of SNAT2 IRES-containing reporter RNA in L13a-depleted cells. SNAT2 and CrPV IRES element-containing reporter RNA constructs were transfected into cells expressing shRNA against L13a or a control shRNA. Cell lysates were made from transfected cells, and ribosomal fractions were resolved by centrifugation using a 10 to 50% sucrose density gradient. Continuous UV monitoring of A254 detected the 40S, 60S, 80S, and polyribosome peaks. All of the polyribosomal profiles obtained from cells transfected with the different constructs were almost identical, and the profile obtained from m/hpSNAT2-transfected and L13a-depleted cells is shown at the top as a representative example. The association of the luciferase reporter RNA with polyribosomes was determined by RT-PCR analysis of the total RNA isolated from each fraction using luciferase-specific primers. (D) The association of the β-actin mRNAs with polyribosomes was determined by RT-PCR analysis of the total RNA isolated from the same fractions as those described for panel C using β-actin-specific primers. (E) Translation-competent extracts from rRNA methylation-deficient cells failed to support the in vitro translation of SNAT2 IRES-containing reporter RNA. Translation-competent extracts were prepared from untreated, cycloleucine-treated, or L13a-depleted 293T cells as indicated; 200 ng of SNAT2 IRES-containing reporter mRNA (as shown at the top) was subjected to in vitro translation using this extract in the presence of [35S]methionine. m7G-capped T7 gene 10 RNA (100 ng) was cotranslated in the same reaction as a control.
The SNAT2 IRES-dependent translation was further reconstituted in translation-competent extracts from 293T cells using a chimeric reporter SNAT2-IRES-luc mRNA construct carrying an unmethylated cap (ApppG) at the 5′ end, followed by a stable hairpin structure and the SNAT2 IRES element (a diagram of the chimeric mRNA is shown in Fig. 1E). The unmethylated cap and the hairpin structure together block cap- and scanning-dependent translation. The SNAT2 IRES-driven translation of the reporter luc and the cap-driven translation of T7 phage gene 10 were efficient in competent extracts from untreated 293T cells. In contrast, SNAT2 IRES-driven translation, but not cap-driven translation, was substantially reduced when translation-competent extracts from either L13a-depleted or cycloleucine-treated cells were used (Fig. 1E).
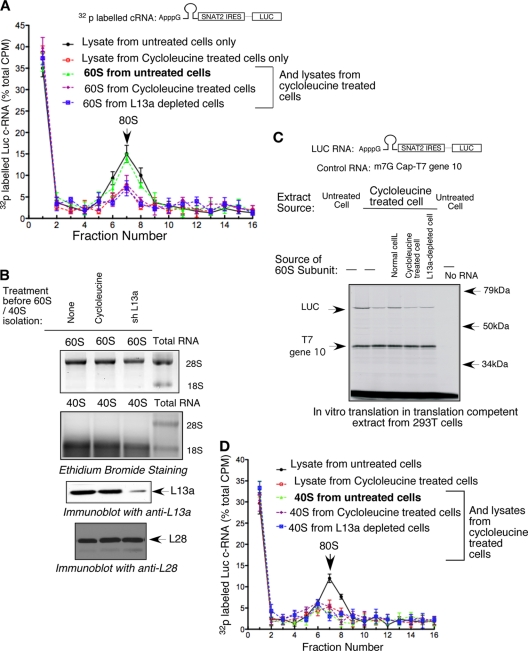
The SNAT2 IRES failed to support efficient assembly of the 80S initiation complex but allowed formation of the 48S complex under conditions of inhibition of rRNA methylation.
To determine the step at which the initiation of SNAT2 IRES-driven translation was inhibited under the conditions of rRNA methylation, we examined the assembly of the 48S and 80S ribosomal complexes using translation-competent extracts from 293T cells. A chimeric reporter mRNA containing a nonmethylated ApppG cap, a strong hairpin structure in the 5′ UTR, and a SNAT2 IRES upstream of the luc reporter described above (Fig. 1D) was further used to monitor the assembly of the 48S and 80S complexes on the SNAT2 IRES. m7G-capped and ApppG-capped luc RNAs were used, respectively, as positive and negative controls to verify the ability of ApppG to block cap-dependent initiation. Diagrams of the chimeric RNAs are shown at the top of Fig. 2A. The assembly of 48S and 80S ribosomal complexes on the SNAT2 IRES was conducted in the presence of the nonhydrolyzable GTP analog GMP-PNP and cycloheximide, respectively (Fig. 2A). To determine whether the assembly of either 48S or 80S complexes on the SNAT2 IRES was sensitive to the inhibition of rRNA methylation, we reconstituted the ribosome assembly using cell extracts from L13a-depleted (Fig. 2B) or cycloleucine-treated (Fig. 2C) cells. Since fibrillarin is the essential component of the C/D box snoRNP-containing rRNA methyltransferase complex (38, 58), we also reconstituted the ribosome assembly using fibrillarin-depleted cells (Fig. 2B). The extent of fibrillarin depletion was tested by immunoblot analysis (Fig. 3). Substantial inhibition of 80S complex formation was observed in L13a-depleted, fibrillarin-depleted (Fig. 2B), and cycloleucine-treated (Fig. 2C) extracts compared to that in control cell extracts made from control shRNA-expressing or untreated cells. In contrast, 48S complex assembly was unaffected (Fig. 2B and C). To further verify that the inhibition of 80S ribosomal complex formation is specific for the SNAT2 IRES, we performed the assembly reaction using m7G-capped (Fig. 2D) or HCV IRES-containing luc reporter constructs (Fig. 2E and F) and found that 80S ribosomal complex formation was completely unaffected by the lack of rRNA methylation caused by cycloleucine or by L13a depletion. These results show that 48S preinitiation complex formation on the SNAT2 IRES is independent of rRNA methylation; however, rRNA methylation is required for efficient 80S complex formation.
Fig. 2.
Inhibition of rRNA methylation significantly inhibits 80S but not 48S initiation complex formation on SNAT2 IRES-containing monocistronic RNA. (A) Assembly of 48S and 80S initiation complexes using synthetic SNAT2 IRES-containing monocistronic RNA. A diagram of the constructs is shown at the top. 32P-labeled mRNAs were synthesized from these constructs. Ribosomal complex formation on these mRNAs (500,000 cpm) was reconstituted in translation-competent extract from 293T cells in the presence of 2 mM GMP-PNP (to detect 48S) or 0.5 mM cycloheximide (to detect 80S) and resolved by sucrose density gradient centrifugation. (B and C) Inhibition of rRNA methylation by L13a depletion, fibrillarin depletion, and cycloleucine treatment causes inhibition of the 80S but not the 48S ribosomal complex on SNAT2 IRES-containing reporter RNA. Translation initiation reaction mixtures were reconstituted using the translation-competent extracts made from either L13a- or fibrillarin-depleted (B) or cycloleucine-treated (C) 293T cells in the presence of 2 mM GMP-PNP or 0.5 mM cycloheximide (CHX) and 32P-labeled SNAT2 IRES-containing reporter RNA. The reaction product was subjected to sucrose density gradient centrifugation, and radioactivity from each fraction was determined. The extent of L13a depletion and inhibition of rRNA methylation was confirmed by the same procedure used for the results shown in Fig. 1C. (D) The 48S and 80S initiation complex formed on m7G-capped reporter RNA is insensitive to the inhibition of rRNA methylation. Translation initiation reaction mixtures were reconstituted using translation-competent extracts from cycloleucine-treated cells and m7G-capped reporter luciferase RNA. The 48S and 80S initiation complexes were identified as described for panels A to C. (E and F) The 48S and 80S initiation complexes formed on HCV IRES-containing reporter RNA are insensitive to the inhibition of rRNA methylation caused by either L13a depletion or cycloleucine treatment. A diagram of the construct used is shown in each panel. The 48S and 80S initiation complexes were identified as described for panels A to C.
Fig. 3.

RNA interference-mediated knockdown of fibrillarin. 293T cells were transfected using synthetic siRNA against human fibrillarin (Dharmacon). The knockdown of fibrillarin was confirmed by immunoblot analysis of the transfected lysate using antibody against human fibrillarin (Abcam, Cambridge, United Kingdom).
To test whether lack of rRNA methylation may also inhibit elongation, we measured the poly(U)-dependent synthesis of polyphenylalanine at a high Mg2+ concentration in the presence of [3H]phenylalanine (17, 18, 36). In this experiment, we used translation-competent extracts from cycloleucine-treated or L13a-depleted cells. Results presented in Fig. 4 show that rates of chain elongation remained unaffected under the conditions of inhibition of rRNA methylation by either means.
Fig. 4.
Inhibition of rRNA methylation or L13a depletion failed to inhibit elongation on poly(U) RNA. Cell lysates were made from cycloleucine-treated or L13a-depleted cells. Elongation was carried out with these extracts using poly(U) RNA, and the incorporation of [3H]phenylalanine into polyphenylalanine was measured (upper panel). The inhibition of rRNA methylation and L13a depletion in the cells used to prepare lysates were tested by pulse-labeling with [3H]methylmethionine, pulse-labeling with [3H]uridine, and immunoblot analysis with anti-L13a antibody. The top panel shows that the elongation stage of protein synthesis is insensitive to inhibition of rRNA methylation. The bottom panel shows the status of rRNA methylation and L13a depletion of the cells used in the elongation experiments whose results are shown at the top.
Impaired 80S assembly on the SNAT2 IRES caused by inhibition of rRNA methylation can be restored by purified wild-type 60S but not 40S ribosomal subunits.
The inhibition of 80S, but not 48S, complex formation on the SNAT2 IRES under conditions of abrogation of rRNA methylation suggests that the defect is related to the subunit joining step and is most likely associated with some abnormalities in the 60S ribosomal subunit, bearing undermethylated rRNA. Therefore, we investigated whether the defect in 80S initiation complex formation on the SNAT2 IRES can be corrected by replenishing the assembly reaction mixture with purified 60S ribosomal subunits isolated from wild-type/untreated cells. 80S complex formation was reconstituted as described above, by using the 32P-labeled chimeric mRNA containing the SNAT2 IRES fused to the luc reporter (diagrammed at the top of Fig. 5A) and cell extracts from cycloleucine-treated (and untreated control) cells and in the presence or absence of undermethylated or wild-type 60S ribosomal subunits.
Fig. 5.
The defect in 80S initiation complex assembly on SNAT2 IRES-containing reporter RNA resulting from an rRNA methylation defect can be corrected by using the purified 60S ribosomal subunit. (A) The 80S ribosomal complex was reconstituted using 32P-labeled SNAT2 IRES-containing reporter RNA and translation-competent extract from either untreated or cycloleucine-treated cells as described in the legend to Fig. 2. A 30-fold molar excess of the purified 60S ribosomal subunit isolated from untreated, cycloleucine-treated, or L13a-depleted cells was added to the assembly reaction mixture. The extent of inhibition of rRNA methylation was confirmed by the same procedure used for the results shown in Fig. 1B. (B) Purified 60S and 40S ribosomal subunits isolated from untreated, cycloleucine-treated, or L13a-depleted cells. The 28S and 18S rRNAs and the level of L13a were shown by ethidium bromide staining and immunoblotting with anti-L13a antibody. As a control, the same blot was subjected to immunoblot analysis to detect L28, which was not affected. (C) The purified 60S subunit rescues the translation of SNAT2 IRES-containing reporter RNA in translation-competent extracts from rRNA methylation-deficient cells. Translation-competent extracts were prepared from untreated, cycloleucine-treated, and L13a-depleted 293T cells; 200 ng of SNAT2 IRES-containing reporter mRNA (shown at the top) was subjected to in vitro translation using this extract in the presence of [35S]methionine. The m7G-capped T7 gene 10 RNA (100 ng) was cotranslated in the same reaction as a control. The purified 60S subunit isolated either from normal cells or from cycloleucine-treated or L13a-depleted cells was added to the in vitro translation reaction mixture before the addition of [35S]methionine. (D) The defect in 80S initiation complex assembly on SNAT2 IRES-containing reporter RNA caused by inhibition of rRNA methylation cannot be corrected by complementation with the purified 40S ribosomal subunit. The 80S ribosomal complex was reconstituted using 32P-labeled SNAT2 IRES-containing reporter RNA and translation-competent extract from either untreated or cycloleucine-treated cells as described in the legend to Fig. 2. A 30-fold molar excess of the purified 40S ribosomal subunit isolated from untreated, cycloleucine-treated, or L13a-depleted cells was added to the assembly reaction mixture.
As expected, substantial inhibition of 80S complex formation was observed when translation-competent extracts from cycloleucine-treated cells were used in the assembly reaction mixture (Fig. 5A). Interestingly, replenishing this assembly reaction mixture with purified 60S ribosomal subunits from untreated cells resulted in substantial recovery of 80S complex formation (Fig. 5A). At the same time, the recovery of 80S complex formation was not observed when 60S ribosomal subunits (Fig. 5B) isolated from cycloleucine-treated or L13a-depleted cells were used (Fig. 5A). Consistent with this result, 60S ribosomal subunits from normal cells (Fig. 5B) were able to rescue the translation of the luc reporter driven by the SNAT2 IRES using a translation-competent extract from cells where rRNA methylation was inhibited (Fig. 5C). On the other hand, 60S subunits from cycloleucine-treated or L13a-depleted cells (Fig. 5B) were ineffective (Fig. 5C). Since rRNA methylation also occurs on the 40S ribosomal subunit, we also tested the possibility that the fully methylated 40S ribosomal subunit from normal cells (Fig. 5B) could rescue 80S complex formation. The results in Fig. 5D demonstrate that this is not the case.
Cycloleucine treatment and/or L13a depletion affects global rather than site-specific rRNA methylation.
Next, we tested whether impairment of 80S complex formation on the SNAT2 IRES results from global or perhaps site-specific inhibition of rRNA methylation. To address this question, we compared the methylation efficiencies of several previously identified rRNA methylation sites, e.g., at nucleotide positions 1612, 1858, 3810, and 4198 in the 28S rRNA and 37, 1489, 1713, and 1803 in the 18S rRNA, respectively (4, 31). The extent of RNA methylation was measured by an RT-based assay (4, 30). This method relies on the inability of reverse transcriptase to read through the methylation site in the presence of a limiting dNTP concentration (4, 30). Results presented in Fig. 6 show that all of the methylation sites tested were equally affected by the inhibition of rRNA methylation caused by cycloleucine treatment or L13a depletion. Taken together, these results indicate that the defect in 80S complex assembly on the SNAT2 IRES is due to global rather than site-specific inhibition of rRNA methylation.
Fig. 6.
Inhibition of rRNA methylation by cycloleucine treatment and L13a depletion is not site specific. Site-specific methylation ratios at several known methylation site on 28S or 18S rRNA from cycloleucine-treated or L13a-depleted cells were determined. An RT-based method using limiting and nonlimiting dNTP concentrations, followed by qPCR, was used (see Materials and Methods for details).
Disruption of rRNA methylation abrogates the AAS-induced activation of SNAT2 IRES activity.
The SNAT2 IRES was previously shown to be active during nutritional stress caused by AAS (14). Here we determined whether the activity of the SNAT2 IRES during AAS of 293T cells requires rRNA methylation. To measure SNAT2 IRES activity under these conditions, we used the bicistronic RLuc-FLuc reporter construct (Fig. 7A). rRNA methylation was disrupted in 293T cells by cycloleucine treatment or L13a depletion and confirmed by labeling with [3H]methylmethionine (data not shown). Cells were transfected with the bicistronic construct and either left untreated or amino acid starved with or without disruption of rRNA methylation. We measured the change in SNAT2 IRES activity by determining the ratio of firefly and Renilla luciferase activities. Results showed upregulation of SNAT2 IRES activity under conditions of AAS, as previously reported (14), and substantial inhibition of SNAT2 IRES activity in cells with cycloleucine treatment or L13a depletion. However, AAS did not lead to rescue of the inhibition of SNAT2 IRES activity mediated by abrogation of rRNA methylation (Fig. 7A). AAS causes a decrease in cell volume (14), and SNAT2-mediated transport of neutral amino acids is essential for regulatory volume recovery and cell survival (14). IRES-mediated enhanced synthesis of SNAT2 protein under AAS conditions contributes to the induction of system A transport activity (14). Therefore, we tested whether inhibition of rRNA methylation could compromise system A transport activity by directly measuring the uptake of the system A substrate [14C]methylaminoisobutyric acid. Our results show that inhibition of rRNA methylation by cycloleucine leads to inhibition of the AAS-mediated induction of system A transport activity (Fig. 7B).
Fig. 7.
AAS fails to rescue the SNAT2 IRES and system A transport activity from inhibition caused by the disruption of rRNA methylation. (A) Cycloleucine treatment and L13a depletion cause an inhibition of SNAT2 IRES activity that cannot be rescued by AAS. A diagram of the bicistronic construct harboring the SNAT2 IRES element between R-Luc and F-Luc is shown at the top. The bicistronic construct was transfected into untreated, amino acid-starved, or L13a-depleted cells and into cells that were treated simultaneously with AAS medium and cycloleucine or L13a depleted. IRES activities were measured by measuring the ratio of F-Luc to R-Luc. The results of each experiment are averages of triplicates, and the error bars were drawn on the basis of three independent experiments. (B) AAS failed to rescue the system A transport activity from inhibition caused by the disruption of rRNA methylation (left panel). Untreated or cycloleucine-treated HeLa cells were initially grown in normal medium, and a 1-min influx of [14C]methylaminoisobutryic acid was measured after the cells were incubated for 3 h in AAS medium. The ratio of incorporation of [3H]methylmethionine and [3H]uridine remained unchanged under AAS conditions (right panel). 293T cells were subjected to either AAS or simultaneous AAS and cycloleucine treatment and labeled with [3H]methylmethionine and [3H]uridine separately. Cells were lysed, TCA precipitated, filtered through a Whatman GF/C filter, and counted in a scintillation counter. (C) L13a expression under AAS and cycloleucine treatment or L13a depletion. WT, wild type.
AAS does not affect the rRNA methylation status of the cell.
Previously, we showed that inhibition of a number of cellular IRES activities, including that of SNAT2 IRES, was not due to L13a depletion per se but rather was a direct consequence of decreased rRNA methylation (8). Results from these studies clearly show the requirement of rRNA methylation for SNAT2 IRES activation and upregulation of IRES activation by AAS. It remained possible, however, that AAS could enhance rRNA methylation either directly or through modulation of the elevated expression of L13a. Therefore, we measured the levels of rRNA methylation and L13a expression under conditions of AAS. Results presented in Fig. 7B (right panel) and C show no change (enhancement) in rRNA methylation or L13a expression levels under conditions of AAS.
rRNA methylation is required for the efficient translation of endogenous SNAT2, Cat-1, and c-Myc mRNAs.
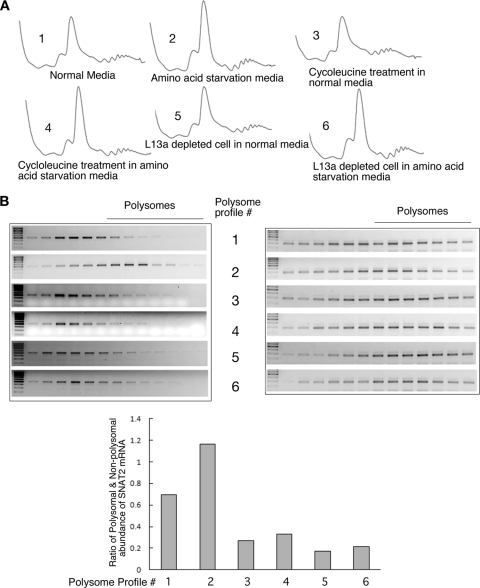
In all of the experiments described above, the requirement for rRNA methylation in the activation of the SNAT2 IRES was demonstrated by the use of monocistronic SNAT2 IRES-containing reporter RNA (Fig. 1, 2, and 5) and/or the bicistronic reporter constructs (Fig. 7). To check whether rRNA methylation indeed affects the translation of endogenous SNAT2 mRNA, we further looked at the polysomal association of the endogenous SNAT2 mRNA. Cytoplasmic extracts from cells grown under different conditions, such as AAS (to activate SNAT2 IRES activity) or cycloleucine treatment/L13a depletion (to inhibit rRNA methylation), were used in the subsequent set of experiments. Figure 8A shows the polysome profiles obtained under each condition by sucrose density gradient centrifugation analysis. Ratios of polysomal to nonpolysomal SNAT2 mRNA abundance were measured semiquantitatively by determining band intensities using the ImageJ64 program (NIH). As expected, increased polysomal association of endogenous SNAT2 mRNA under AAS conditions was observed (Fig. 8B, profile 2). In contrast, the ratio of the polysomal versus nonpolysomal SNAT2 mRNA abundance was reduced when cycloleucine-treated or L13a-depleted cells were used (Fig. 8B, lower panel). Measurement of this ratio by qPCR analysis also showed reduced polyribosomal abundance of the SNAT2 mRNA under cycloleucine-treated and L13a-depleted conditions (Fig. 9). Together, these results show that efficient translation of endogenous SNAT2 mRNA requires rRNA methylation.
Fig. 8.
The increase in the polyribosomal abundance of endogenous SNAT2 mRNA under AAS conditions is abrogated by cycloleucine treatment and L13a depletion. (A) Polyribosomal profiles obtained from untreated, amino acid-deprived, cycloleucine-treated, and L13a-depleted cells. Cell lysates were made from cycloleucine-treated or L13a-depleted cells, followed by AAS. Polysomes were resolved by centrifugation through 10 to 40% sucrose density gradients and collecting the fractions through continuous monitoring of UV absorption. (B) Polyribosomal abundance of endogenous SNAT2 mRNA. Total RNA was isolated from each fraction, and SNAT2 mRNA was identified by RT-PCR analysis using primers specific for human SNAT2 mRNA (left). The final value was normalized to the ratio of polysomal and nonpolysomal abundance of β-actin (right). Semiquantitative measurements of the polysome-to-nonpolysome ratio of SNAT2 mRNA (bottom) were performed by the ImageJ64 program (NIH).
Fig. 9.
Ratio of abundances of polysomal and nonpolysomal fractions of IRES-containing cellular mRNAs, e.g., SNAT2, CAT-1, and c-Myc, shows translational inhibition by cycloleucine treatment and L13a depletion. (A) Resolution of polysomal and nonpolysomal fractions by sucrose density gradient centrifugation. The fractions designated polysomal and nonpolysomal are shown. (B) Cells were subjected to AAS with or without cycloleucine treatment or L13a depletion. Polysomal and nonpolysomal fractions were made by sucrose density gradient centrifugation. Total RNA was isolated from polysomal and nonpolysomal fractions. The ratio of the polysomal and nonpolysomal fraction abundances of GAPDH, SNAT2, and CAT-1 mRNAs was determined by ΔCT values obtained by qPCR using specific primers. (C) Cells were subjected to treatment with TRAIL with or without cycloleucine treatment and L13a depletion. The abundances of the polysomal and nonpolysomal fractions of the GAPDH and c-Myc mRNAs were determined as described for panel B.
In addition to the SNAT2 IRES, we tested two other cellular IRES-containing mRNAs for their polyribosomal abundance under conditions of inhibition of rRNA methylation. IRES-mediated translation of cellular CAT-1 (60) and c-Myc mRNA (9, 52, 53) was upregulated by amino acid deprivation and by TRAIL-induced apoptosis, respectively. To investigate the efficiency of CAT-1 and c-Myc IRES translation under these conditions, we isolated polysomal and nonpolysomal fractions of mRNAs from amino acid-starved and TRAIL-treated cells and estimated mRNA abundance by qPCR. qPCR analysis of these mRNA pools showed significant increases in the polysomal abundance of CAT-1 and c-Myc mRNAs upon AAS and TRAIL treatment, respectively. However, the polysomal abundance of GAPDH mRNA was reduced under these conditions, as might be expected, showing the inhibition of cap-dependent translation. Interestingly, similarly to SNAT2, IRES-mediated expression of both CAT-1 and c-Myc was reduced under the conditions of inhibition of rRNA methylation, suggesting a common mechanism for all cellular IRES-containing mRNAs (Fig. 9).
c-Src, but not EMCV, IRES is sensitive to inhibition of rRNA methylation.
Although the mechanism of IRES-mediated translation remains not well understood, there are several examples of viral and cellular IRESs that have very different requirements for canonical initiation factors. In our experiments, we used the CrPV and HCV IRES elements, which have minimal requirements for canonical initiation factors (for example, they do not require eIF4E, eIF4G, and eIF4A, the subunits of the eIF4F complex) and are capable of direct binding to the 40S ribosomal subunits. The canonical factor requirements for the SNAT2 IRES (as well as the p53 and p27 IRESs used in our previous studies) are not known yet. It thus remained possible that the differences in activation of the viral and cellular IRESs reported here are due to the differences in their requirements for canonical initiation factors. To test this possibility, we measured the activities (under conditions of inhibition of rRNA methylation) of the two additional IRES elements found in c-Src and the EMCV mRNAs, respectively. The c-Src IRES was suggested to be able to bind the 40S ribosomal subunits directly (2) and thus is believed to have reduced requirements for canonical initiation factors, a feature reminiscent of the HCV-like and CrPV-like IRESs (40). On the other hand, the EMCV IRES is well known not to bind the 40S ribosomal subunits directly and to require most of the canonical initiation factors, with the exception of eIF4E, to initiate translation (22). Using FLuc-Rluc bicistronic reporter constructs, we tested the sensitivity of c-Src and EMCV IRES-dependent translation to the inhibition of rRNA methylation induced by either cycloleucine treatment or L13a depletion. Our results clearly indicate that IRES-mediated translation driven by the c-Src IRES, but not the EMCV IRES, element was sensitive to the inhibition of rRNA methylation (Fig. 10). This result also shows that sensitivity to rRNA methylation may not be related to the requirement for canonical initiation factors.
Fig. 10.
c-Src, but not EMCV, IRES activity is inhibited by disruption of rRNA methylation. A diagram of the bicistronic constructs harboring the c-Src or EMCV IRES element between R-Luc and F-Luc is shown at the top. Untreated, L13a-depleted, or cycloleucine-treated 293T cells were transfected with these constructs. IRES activities were measured by calculating the ratios of F-Luc to R-Luc. The results of the experiments are averages of triplicates, and the error bars were drawn on the basis of three independent experiments (middle). The extent of rRNA methylation and synthesis in these cells was examined by pulse-labeling with [3H]methylmethionine and [3H]uridine (bottom).
L13a associates with the methyltransferase protein fibrillarin, a component of the C/D box snoRNP.
Results from our previous (8) and present studies demonstrate the essential role of L13a in rRNA methylation. It is also known that fibrillarin, a component of the C/D box snoRNP, is responsible for RNA methylation activity in eukaryotic cells (38, 55, 58). We therefore hypothesized that L13a may be part of the fibrillarin-containing methyltransferase holoenzyme complex and thus may be directly and/or indirectly required for its activity. We therefore decided to check whether L13a and fibrillarin may colocalize. To test this, we transfected 293T cells with plasmids expressing an L13a-HA fusion construct and determined the localization of HA-tagged L13a and endogenous fibrillarin by microscopy using a monoclonal mouse antibody against the HA tag and a polyclonal rabbit antibody against human fibrillarin. HA-tagged L13a and fibrillarin were stained green and red, respectively, using appropriate secondary antibodies conjugated with Alexa Fluor 488 and Alexa Fluor 594. The nucleus was stained with DAPI. The results presented in Fig. 11 show the colocalization of L13a and fibrillarin within the nucleus, as evidenced by the merging of the green staining of HA-tagged L13a and the red staining of fibrillarin inside the DAPI-stained nucleus. In the lower panel, an adjacent L13a-untransfected cell revealing only staining for fibrillarin is shown for comparison (Fig. 11). Control untransfected cells or cells probed with conjugated secondary antibodies alone without the primary antibodies showed no color, thus confirming the specificity of the antibodies.
Fig. 11.
Colocalization and association of L13a with fibrillarin. Immunofluorescence studies showing the colocalization of L13a and fibrillarin. 293T cells were transfected with plasmids expressing L13a-HA fusion constructs. At 24 h posttransfection, cells were fixed and probed with a monoclonal antibody against the HA tag (Molecular Probes) and a polyclonal rabbit antibody against fibrillarin (Abcam). Cells were then incubated with appropriate color-conjugated secondary antibodies (Invitrogen, Carlsbad, CA). Nuclei were stained with DAPI.
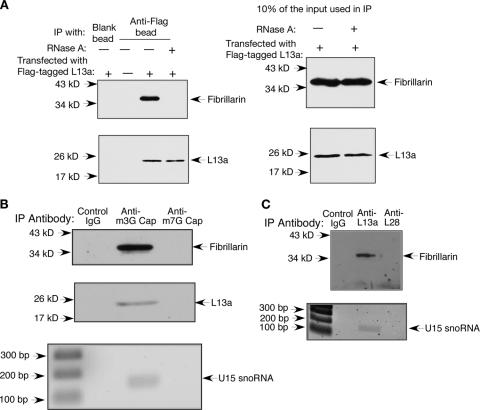
The colocalization of L13a and fibrillarin suggests that these two proteins may indeed interact. We further decided to check, by the use of coimmunoprecipitation, whether this is indeed the case. 293T cells were transfected with Flag-L13a, and immunoprecipitation was done using anti-Flag antibody-conjugated agarose beads. Coimmunoprecipitation of fibrillarin with L13a was further confirmed by immunoblot analysis using anti-fibrillarin antibody (Fig. 12A). Fibrillarin is a C/D box snoRNP component (38, 55, 58), and it remained possible that interaction between fibrillarin and L13a is not direct and is RNA dependent, in case snoRNA functions as an anchor between these two proteins. To test whether the interaction between fibrillarin and L13a is RNA dependent, the transfected cell lysate was digested with RNase A before immunoprecipitation. Results show complete abrogation of the interaction between fibrillarin and L13a after RNase A treatment (Fig. 12A). To directly test for the presence of snoRNA in the same complex as L13a and fibrillarin, we immunoprecipitated snoRNA using an antibody against the m3G cap. This strategy is based on the fact that many box C/D snoRNAs are hypermethylated using the C/D box itself as a cis-acting element for hypermethylation, resulting in the conversion of the m7G cap to an m3G cap (49). In addition, antibodies against the m3G cap have been successfully used to isolate C/D box snoRNPs from mammalian cell extracts (28). We thus performed immunoprecipitation from 293T cell nuclear lysate using an anti-m3G cap antibody (Synaptic Systems, Goettingen, Germany). The presence of fibrillarin and L13a was detected in the anti-m3G cap immunoprecipitate by immunoblot analysis (Fig. 12B). RNA was extracted from an aliquot of the same immunoprecipitate, and the presence of a C/D box snoRNA U15 was detected by random-primer RT and PCR amplification using a primer specific for human U15 snoRNA (third panel of Fig. 12B). The identity of the amplified product was confirmed by sequencing. However, fibrillarin, L13a, and U15 snoRNA were not detected when an anti-m7G cap antibody was used. Since many C/D box snoRNAs, such as U3, U8, U13, U14, and U16, are m3G capped and also associate with fibrillarin (48, 56), we could not rule out the possibility of their presence in the anti-m3G cap immunoprecipitate containing fibrillarin and L13a. To further test the association of U15 snoRNA with L13a and fibrillarin, immunoprecipitation was performed using an anti-L13a antibody. The results presented in Fig. 12C show the presence of fibrillarin and U15 snoRNA in the anti-L13a immunoprecipitate. However, no amplification product was observed in RT-PCR using specific primers for H/ACA box snoRNA, e.g., U17 and U19 and another C/D box snoRNA, U3 (data not shown).
Fig. 12.
L13a associates with the methyltransferase protein fibrillarin and a C/D box snoRNA, U15. (A) Association of L13a with fibrillarin is RNA dependent. 293T cells were transfected with plasmids expressing L13a-Flag fusion constructs. At 24 h posttransfection, cell lysates were made and incubated with or without RNase A. Immunoprecipitation (IP) was carried out with anti-Flag antibody-coated beads (or blank beads as a control). The fibrillarin present in the immunoprecipitate was determined by immunoblotting with an antifibrillarin antibody (Abcam) (top left and right). The blot was reprobed with an anti-Flag antibody to confirm immunoprecipitation (bottom left and right). (B) Coimmunoprecipitation of L13a, fibrillarin, and U15 snoRNA using an antibody against the m3G cap. Immunoprecipitation was carried out using an antibody against the m3G cap (Synaptic Systems, Goettingen, Germany). The immunoprecipitate was resolved by SDS-PAGE, followed by immunoblot analysis with an anti-fibrillarin antibody (Abcam, Cambridge, United Kingdom) (top) and an anti-L13a antibody (middle). RNA was extracted from an aliquot of the immunoprecipitate, followed by random-primer RT and PCR analysis with a human U15 snoRNA (GenBank accession number AF281313)-specific primer pair (bottom). (C) Association of endogenous L13a with fibrillarin and U15 snoRNA. Immunoprecipitation was carried out using an antibody against L13a. Fibrillarin (top) and U15 snoRNA (bottom) were detected in the immunoprecipitate as described for panel B.
Taken together, these results suggest that L13a and fibrillarin colocalize in the same compartment within the nucleus and at least one C/D box snoRNA, such as U15, may serve as an anchor between the two. This result is also consistent with the RNase sensitivity of their association. To conclude, these results support the role of L13a as a component of the fibrillarin- and C/D box snoRNA-containing methyltransferase holoenzyme complex.
DISCUSSION
Our studies reveal a novel feature of cellular IRES-mediated translation, i.e., its dependence on the extent of rRNA methylation. We thus believe that our findings may provide a novel platform allowing the differentiation of cellular IRES-mediated translation from both cap-dependent and viral IRES-mediated translation. All of the cellular IRESs tested in the present study and our previous studies, including p27, p53, SNAT2, CAT-1, c-Myc, and c-Src, appeared to be sensitive to rRNA methylation. However, cap-dependent translation and the translation driven by various viral IRES elements, including EMCV, HCV, and CrPV, remained refractory to rRNA methylation. This implies that cellular IRES-mediated translation may have some features distinct from those of viral IRES-mediated translation.
It should be noted that, until now, most of the data in the literature have supported the view that the mechanisms of viral and cellular IRES-mediated translation would have many features in common. Both mechanisms are thought to have reduced requirements for canonical initiation factors and rely on canonical and noncanonical interactions between IRESs, eIFs, and 40S ribosomal subunits (for a review, see references 15 and 19). Many viral and cellular IRESs are also thought to require conformational flexibility induced by eIFs and ITAFs for efficient 40S binding (for a review, see reference 12). Further, both viral and cellular IRESs were found to rely on the same set of ITAFs for their function. For example, polypyrimidine tract-binding protein serves as an ITAF for the HCV IRES (3), as well as for the Apaf1 (34) and Bag-1 cellular IRESs (42). La autoantigen is an ITAF for the HCV IRES (1) and also for the cellular BIP (29) and XIAP (16) IRESs (for a review, see references 23 and 24). However, to our knowledge, there was no example demonstrating that the modes of activation of cellular and viral IRES elements are substantially different. Our present work strongly argues in favor of such a differentiation.
Our results provide evidence that inhibition of rRNA methylation leads to substantial inhibition of 80S ribosomal complex formation on the cellular (SNAT2) but not the viral (HCV) IRES element. It should be noted that, at the same time, abrogation of rRNA methylation does not inhibit 48S complex formation on either viral or cellular IRESs. Further, our results show that the observed defect in 80S ribosomal complex formation can be rescued by purified 60S but not 40S ribosomal subunits isolated from normal cells. These results clearly show that the observed defect in 80S complex formation is confined largely to the 60S ribosomal subunit.
However, the question of why the deficiency in rRNA methylation in the 60S ribosomal subunit results in inhibition of 80S complex formation on the SNAT2 IRES but not the viral IRESs and/or capped mRNAs remains open. We speculate that the sensitivity of cellular IRES-mediated translation to the inhibition of rRNA methylation may originate from combined effects of rRNA methylation on the conformation of the 48S complex formed on the cellular IRESs and the conformation of the 60S ribosomal subunit that preclude further efficient subunit joining. In other words, the conformation of the RNA-bound 40S ribosomal subunit may differ for cellular IRES, viral IRES and cap-dependent translation without influencing 48S assembly. However, those 48S complexes may show significant differences in tolerance for fully methylated and undermethylated 60S ribosomal subunit joining (Fig. 13). Alternatively, subunit joining on a cellular IRES may require an additional, as-yet-unidentified factor that fails to interact with methylation-deficient ribosomes. Experimental validation of this hypothesis remains challenging because of the absence of a minimal reconstitution system for ribosome binding to the SNAT2 IRES (our model for cellular IRES-mediated expression used for ribosome assembly experiments). Establishment of such a system may also require the identification of the additional ITAFs necessary for SNAT2 ribosome binding. Future experiments involving such an in vitro reconstitution system with minimal components are necessary to get further insights into the mechanism under investigation. Toeprinting and/or RNase footprinting experiments, in particular, may shed light on the efficiency of subunit joining and possible associated conformational changes within the ribosomal subunits.
Fig. 13.
The conformation of the 48S complex formed on the SNAT2 IRES creates a difference in tolerance for normal and undermethylated 60S ribosomal subunits. The conformations of the 48S initiation complexes formed in cap-dependent, SNAT2 IRES-dependent, and HCV IRES-dependent translations may be different. For the SNAT2 IRES, the conformation of the 48S subunit may not tolerate the joining of the undermethylated 60S subunit. However, the 48S complex formed in cap-dependent and HCV IRES-dependent translations may not discriminate between the normal 60S subunit and the undermethylated 60S subunit and allow its joining.
It is also important to mention that, in spite of the many reports supporting the existence of cellular IRES-mediated translation (50), skeptical views questioning the presence of IRES elements in cellular mRNAs have been presented (45). It was argued that most cellular translation is cap dependent (25, 26). Our results clearly demonstrate that cellular IRES-mediated translation has certain features distinct from the cap-dependent initiation mechanism.
In addition to elucidating the role of rRNA methylation in cellular IRES-mediated protein synthesis here, we have provided insights into the possible role of L13a in the mechanism of rRNA methylation. We previously showed that depletion of L13a in mammalian cells causes significant inhibition of rRNA methylation (8). However, the link between L13a depletion and inhibition of rRNA methylation remained undefined. A very recent study has revealed the structural basis for site-specific ribose methylation by box C/D RNP in the archaeon Sulfolobus solfataricus (27). Ribosomal protein L7Ae was shown to be one of the key players in the formation of the active C/D RNP complex containing fibrillarin. The possibility that L13a has a similar role in the formation of the active C/D RNP complex in eukaryotes cannot be excluded. To gain insights into the L13a-dependent mechanism of rRNA methylation, our study employed a combination of in situ staining and immunoprecipitation. The data revealed an RNA-dependent association of L13a and fibrillarin and the presence of C/D box snoRNA U15 in the RNP complex containing L13a and fibrillarin. It has been previously shown that fibrillarin is an active subunit of the C/D box snoRNP complex and is the principal component of the methyltransferase activity responsible for rRNA methylation (38, 58). Our present data suggest that L13a might be a critical regulator of the methyltransferase activity of the C/D box snoRNP complex.
It remains an open question, however, whether any physiological conditions that might affect SNAT2 IRES activity (such as AAS) might also regulate rRNA methylation. Our results indicate that AAS does not affect rRNA methylation (Fig. 7B, right panel). It remains possible that AAS-induced IRES activity relies on the activation/induction of an as-yet-unknown and rate-limiting ITAF(s) upstream of rRNA methylation. This idea is also consistent with our findings showing the inability of AAS to activate the IRES when rRNA methylation is inhibited. On the other hand, it remains possible that viruses evolved from an rRNA methylation-sensitive IRES activity to an insensitive one in order to protect viral translation from any antiviral response of the cell that may target rRNA methylation. Future experiments are required to answer these and other questions.
ACKNOWLEDGMENTS
This work was supported by NIH HL79164 and American Heart Association (AHA) G.I.A0855555D to B.M., NIH RO1-DK53307 and DK60596 to M.H., AI 059267 to S.B., AHA S.D.G0730120N to A.A.K., an AHA postdoctoral fellowship to S.C., an AHA predoctoral fellowship to P.D., and a predoctoral training fellowship to the Center for Lung Biology (NIH T32HL076125 to J.A.).
We sincerely thank Francesca Gaccioli for providing some critical reagents.
Footnotes
Published ahead of print on 19 September 2011.
REFERENCES
- 1. Ali N., Pruijn G. J., Kenan D. J., Keene J. D., Siddiqui A. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 275:27531–27540 [DOI] [PubMed] [Google Scholar]
- 2. Allam H., Ali N. 2010. Initiation factor eIF2-independent mode of c-Src mRNA translation occurs via an internal ribosome entry site. J. Biol. Chem. 285:5713–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anwar A., Ali N., Tanveer R., Siddiqui A. 2000. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 275:34231–34235 [DOI] [PubMed] [Google Scholar]
- 4. Belin S., et al. 2009. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One 4:e7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bellodi C., Kopmar N., Ruggero D. 2010. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 29:1865–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellodi C., et al. 2010. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 70:6026–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergamini G., Preiss T., Hentze M. W. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaudhuri S., et al. 2007. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 13:2224–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cobbold L. C., et al. 2010. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene 29:2884–2891 [DOI] [PubMed] [Google Scholar]
- 10. de Breyne S., Yu Y., Unbehaun A., Pestova T. V., Hellen C. U. 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U. S. A. 106:9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Decatur W. A., Fournier M. J. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278:695–698 [DOI] [PubMed] [Google Scholar]
- 12. Filbin M. E., Kieft J. S. 2009. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 19:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franchi-Gazzola R., et al. 2001. Amino acid depletion activates TonEBP and sodium-coupled inositol transport. Am. J. Physiol. Cell Physiol. 280:C1465–C1474 [DOI] [PubMed] [Google Scholar]
- 14. Gaccioli F., et al. 2006. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2alpha phosphorylation and cap-independent translation. J. Biol. Chem. 281:17929–17940 [DOI] [PubMed] [Google Scholar]
- 15. Hellen C. U. 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta 1789:558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holcik M., Korneluk R. G. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hussey G. S., et al. 2011. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell 41:419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ilan J. 1978. Translation of maternal messenger ribonucleoprotein particles from sea urchin in a cell-free system from unfertilized eggs and product analysis. Dev. Biol. 66:375–385 [DOI] [PubMed] [Google Scholar]
- 19. Jackson R. J., Hellen C. U., Pestova T. V. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johannes G., Carter M. S., Eisen M. B., Brown P. O., Sarnow P. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. U. S. A. 96:13118–13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. King H. A., Cobbold L. C., Willis A. E. 2010. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 38:1581–1586 [DOI] [PubMed] [Google Scholar]
- 22. Kolupaeva V. G., Lomakin I. B., Pestova T. V., Hellen C. U. 2003. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 23:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komar A. A., Hatzoglou M. 2011. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 10:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Komar A. A., Hatzoglou M. 2005. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 280:23425–23428 [DOI] [PubMed] [Google Scholar]
- 25. Kozak M. 2003. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene 318:1–23 [DOI] [PubMed] [Google Scholar]
- 26. Kozak M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13–37 [DOI] [PubMed] [Google Scholar]
- 27. Lin J., et al. 2011. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 469:559–563 [DOI] [PubMed] [Google Scholar]
- 28. Lübben B., Marshallsay C., Rottmann N., Luhrmann R. 1993. Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 21:5377–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macejak D. G., Sarnow P. 1991. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature 353:90–94 [DOI] [PubMed] [Google Scholar]
- 30. Maden B. E. 2001. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 25:374–382 [DOI] [PubMed] [Google Scholar]
- 31. Maden B. E. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39:241–303 [DOI] [PubMed] [Google Scholar]
- 32. Meier U. T. 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Michel Y. M., Poncet D., Piron M., Kean K. M., Borman A. M. 2000. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275:32268–32276 [DOI] [PubMed] [Google Scholar]
- 34. Mitchell S. A., Spriggs K. A., Coldwell M. J., Jackson R. J., Willis A. E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11:757–771 [DOI] [PubMed] [Google Scholar]
- 35. Mochizuki Y., He J., Kulkarni S., Bessler M., Mason P. J. 2004. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl. Acad. Sci. U. S. A. 101:10756–10761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monnier A., et al. 2001. Evidence for regulation of protein synthesis at the elongation step by CDK1/cyclin B phosphorylation. Nucleic Acids Res. 29:1453–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ni J., Tien A. L., Fournier M. J. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565–573 [DOI] [PubMed] [Google Scholar]
- 38. Omer A. D., Ziesche S., Ebhardt H., Dennis P. P. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U. S. A. 99:5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pestova T. V., Hellen C. U., Shatsky I. N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pestova T. V., Shatsky I. N., Hellen C. U. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pickering B. M., Mitchell S. A., Spriggs K. A., Stoneley M., Willis A. E. 2004. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 24:5595–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ruggero D., et al. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299:259–262 [DOI] [PubMed] [Google Scholar]
- 44. Schüler M., et al. 2006. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 13:1092–1096 [DOI] [PubMed] [Google Scholar]
- 45. Shatsky I. N., Dmitriev S. E., Terenin I. M., Andreev D. E. 2010. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cells 30:285–293 [DOI] [PubMed] [Google Scholar]
- 46. Silvera D., et al. 2009. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell Biol. 11:903–908 [DOI] [PubMed] [Google Scholar]
- 47. Siridechadilok B., Fraser C. S., Hall R. J., Doudna J. A., Nogales E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310:1513–1515 [DOI] [PubMed] [Google Scholar]
- 48. Smith C. M., Steitz J. A. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669–672 [DOI] [PubMed] [Google Scholar]
- 49. Speckmann W. A., Terns R. M., Terns M. P. 2000. The box C/D motif directs snoRNA 5′-cap hypermethylation. Nucleic Acids Res. 28:4467–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spriggs K. A., Bushell M., Willis A. E. 2010. Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40:228–237 [DOI] [PubMed] [Google Scholar]
- 51. Spriggs K. A., et al. 2009. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol. Cell. Biol. 29:1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stoneley M., et al. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stoneley M., et al. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28:687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thompson S. R., Gulyas K. D., Sarnow P. 2001. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. U. S. A. 98:12972–12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tran E. J., Zhang X., Maxwell E. S. 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 22:3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tycowski K. T., Shu M. D., Steitz J. A. 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 7:1176–1190 [DOI] [PubMed] [Google Scholar]
- 57. Wang C., Query C. C., Meier U. T. 2002. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 22:8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang H., Boisvert D., Kim K. K., Kim R., Kim S. H. 2000. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 19:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilson J. E., Pestova T. V., Hellen C. U., Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511–520 [DOI] [PubMed] [Google Scholar]
- 60. Yaman I., et al. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113:519–531 [DOI] [PubMed] [Google Scholar]