Abstract
Swine are an important source of proteins worldwide but are subject to frequent viral outbreaks and numerous infections capable of infecting humans. Modern farming conditions may also increase viral transmission and potential zoonotic spread. We describe here the metagenomics-derived virome in the feces of 24 healthy and 12 diarrheic piglets on a high-density farm. An average of 4.2 different mammalian viruses were shed by healthy piglets, reflecting a high level of asymptomatic infections. Diarrheic pigs shed an average of 5.4 different mammalian viruses. Ninety-nine percent of the viral sequences were related to the RNA virus families Picornaviridae, Astroviridae, Coronaviridae, and Caliciviridae, while 1% were related to the small DNA virus families Circoviridae, and Parvoviridae. Porcine RNA viruses identified, in order of decreasing number of sequence reads, consisted of kobuviruses, astroviruses, enteroviruses, sapoviruses, sapeloviruses, coronaviruses, bocaviruses, and teschoviruses. The near-full genomes of multiple novel species of porcine astroviruses and bocaviruses were generated and phylogenetically analyzed. Multiple small circular DNA genomes encoding replicase proteins plus two highly divergent members of the Picornavirales order were also characterized. The possible origin of these viral genomes from pig-infecting protozoans and nematodes, based on closest sequence similarities, is discussed. In summary, an unbiased survey of viruses in the feces of intensely farmed animals revealed frequent coinfections with a highly diverse set of viruses providing favorable conditions for viral recombination. Viral surveys of animals can readily document the circulation of known and new viruses, facilitating the detection of emerging viruses and prospective evaluation of their pathogenic and zoonotic potentials.
INTRODUCTION
The need to monitor viruses in both human and animal species to better understand emerging infectious diseases has recently received attention under a “one-health” concept (23, 30, 36, 46, 73, 97). Swine are the natural reservoir of a large variety of viruses capable of causing human diseases, including hepatitis E virus (74), Nipah virus (75), and pandemic H1N1 influenza virus (43). The potential zoonotic transmission of swine noroviruses, sapelovirus, and rotaviruses has also been discussed (6, 71). Because of the nearly ubiquitous use of pigs as farm animals and their frequent involvement in viral zoonoses, we selected feces from healthy and diarrheic piglets from a high-density U.S. farm for an unbiased metagenomics analysis of their fecal virome.
Porcine diarrhea can have an important impact on the swine industry, where cases can remain without identified viral or bacterial etiology. For humans in the United States, >40% of cases of diarrhea remain unexplained after extensive testing for all known diarrheic pathogens (85). Worldwide, diarrhea is one of the leading infectious causes of childhood death (http://www.who.int/topics/diarrhea/en/) (19, 49, 107). The recent introduction of human rotavirus vaccines has had a major impact on reducing diarrhea-caused morbidity (28).
Using a viral metagenomics approach, recently enhanced by high-throughput sequencing technologies, several studies have shown a previously unrecognized level of viral genetic diversity and identified novel viral species in animals, plants, other host species, and diverse environmental sources (4, 13, 14, 24, 26, 27, 38, 39, 102, 109). We describe here the virome in feces of healthy and diarrheic piglets from a single farm. A very high level of enteric infection with both known and previously uncharacterized viral species was found, reflecting a high degree of viral transmission in these intensely farmed animals.
MATERIALS AND METHODS
Stool specimens.
Stool samples were collected from a commercial farm with 1,000 sows producing piglets in North Carolina. Feces samples were collected from 12 piglets with diarrhea between the ages of 23 and 30 days and from 24 healthy pigs of age 19 to 30 days. Fecal content was obtained directly from the distal colon and rectum of euthanized animals and frozen at −80°C.
Viral nucleic acid purification.
Stool samples were resuspended in 10 volumes of phosphate-buffered saline (PBS) and vigorously vortexed for 5 min. Three hundred microliters of supernatant was collected after centrifugation (5 min, 15,000 × g) and filtered through a 0.45-μm filter (Millipore) to remove eukaryotic and bacterial cell-sized particles. The filtrates enriched in viral particles were treated with a mixture of DNases (Turbo DNase from Ambion, Baseline-ZERO from Epicentre, and benzonase from Novagen) and RNase (Fermentas) to digest unprotected nucleic acid at 37°C for 90 min (20). Viral nucleic acids were then extracted using the QIAamp viral RNA extraction kit (Qiagen).
Library construction and pyrosequencing.
Viral nucleic acid libraries containing both DNA and RNA viral sequences were constructed by random RT-PCR amplification as previously described (102, 103). One hundred picomoles of a random primer containing a 20-base fixed sequence at the 5′ end followed by a randomized octamer at the 3′ end (8N) was used in a reverse transcription reaction with SuperScript III reverse transcriptase (Invitrogen). A single round of DNA synthesis was then performed using Klenow fragment polymerase (New England BioLabs), followed by PCR amplification of nucleic acids using primers consisting of only the 20-base fixed portion of the random primer.
A total of 36 different random primers (containing distinct 20-base fixed sequences) were used for the 36 porcine fecal samples (Table 1). The DNA products of these random reverse transcription-PCR (RT-PCR) amplifications were pooled in equimolar amounts, and fragments of the appropriate size were purified from a 2% agarose gel for 454 pyrosequencing using the FLX titanium kit.
Table 1.
Viruses detected in porcine fecal samples
| Health status of pig | Age of pig (days) | Sample no. | No. of reads | Known viruses detected (no. of reads) | Divergent viruses detected (no. of reads) | No. of mammalian viruses |
|---|---|---|---|---|---|---|
| Diarrhea | 23 | 21 | 17,429 | Coronavirus (465), kobuvirus (1,470), enterovirus (9,649), teschovirus (4), sapelovirus (6) | Circovirus-like virus (1,404) | 5 |
| 22 | 17,600 | Coronavirus (2), enterovirus (41), teschovirus (5), kobuvirus (7) | Astrovirus (10,473), bocavirus (1,204), circovirus-like virus (3) | 6 | ||
| 23 | 20,081 | Coronavirus (216), kobuvirus (85), enterovirus (13,965), sapelovirus (1,995) | Astrovirus (2,253), bocavirus (37) | 6 | ||
| 24 | 8,899 | Coronavirus (421), kobuvirus (33), enterovirus (6,166), sapelovirus (42) | Astrovirus (776) | 5 | ||
| 25 | 10,731 | Coronavirus (210), kobuvirus (764), sapovirus (1,884), enterovirus (1,701), sapelovirus (13) | Astrovirus (4,378), circovirus-like virus (30) | 6 | ||
| 26 | 14,836 | Coronavirus (18), kobuvirus (548), enterovirus (85), | Astrovirus (8,625) | 4 | ||
| 30 | 51 | 23,868 | Coronavirus (367), kobuvirus (846), enterovirus (734), sapovirus (232), sapelovirus (218) | Astrovirus (13,990), bocavirus (8), circovirus-like virus (25) | 7 | |
| 52 | 13,035 | Coronavirus (18), kobuvirus (15), sapovirus (3,874), enterovirus (1,157), sapelovirus (8) | Astrovirus (11,414), posavirus 1 and 2 | 6 | ||
| 53 | 7,546 | Coronavirus (375), kobuvirus (580), enterovirus (760), sapovirus (1,819), teschovirus (26) | Bocavirus (3), circovirus-like virus (44) | 6 | ||
| 54 | 10,596 | Coronavirus (90), kobuvirus (257), enterovirus (1,139), sapovirus (1,488), sapelovirus (40) | Astrovirus (1,849) | 6 | ||
| 55 | 19,498 | Coronavirus (11), kobuvirus (20), enterovirus (6), sapovirus (3,369) | Astrovirus (11,703), bocavirus (3) | 6 | ||
| 56 | 5,291 | Sapelovirus (3) | Astrovirus (8) | 2 | ||
| Healthy | 19 | 1 | 23,786 | Kobuvirus (1,047) | None | 1 |
| 2 | 31,994 | Kobuvirus (25,648), sapovirus (94) | Astrovirus (17) | 3 | ||
| 3 | 17,332 | Kobuvirus (9,445) | None | 1 | ||
| 4 | 23,577 | Kobuvirus (17,734) | None | 1 | ||
| 5 | 16,109 | Kobuvirus (83), teschovirus (80) | None | 2 | ||
| 6 | 16,608 | Kobuvirus (6,006) | None | 1 | ||
| 23 | 11 | 12,641 | Kobuvirus (243), coronavirus (603), enterovirus (10,417), teschovirus (7) | Astrovirus (10) | 5 | |
| 12 | 13,065 | Kobuvirus (19), coronavirus (578), enterovirus (8,718), sapovirus (789), sapelovirus (7) | Astrovirus (738) | 6 | ||
| 13 | 22,141 | Kobuvirus (12,727), coronavirus (3), enterovirus (14), sapelovirus (423) | Astrovirus (3,705) | 5 | ||
| 14 | 25,995 | Kobuvirus (12,343), coronavirus (66), enterovirus (237), sapelovirus (895) | Astrovirus (6,512) | 5 | ||
| 15 | 13,100 | Kobuvirus (1,123), coronavirus (15), enterovirus (3,248), teschovirus (7), sapelovirus (4,245) | Astrovirus (2,847) | 6 | ||
| 16 | 18,622 | Kobuvirus (6,574), coronavirus (106), enterovirus (7,267), sapovirus (75) | None | 4 | ||
| 31 | 18,886 | Kobuvirus (3,435), coronavirus (228), enterovirus (12,044), teschovirus (6), sapelovirus(9) | Astrovirus (1,504), circovirus-like virus (18) | 6 | ||
| 32 | 15,414 | Kobuvirus (10,538), enterovirus (155), teschovirus (3) | Astrovirus (369) | 4 | ||
| 33 | 18,564 | Kobuvirus (3,343), coronavirus (2), enterovirus (630), teschovirus (14), sapelovirus (534) | Astrovirus (9,673) | 6 | ||
| 34 | 13,363 | Kobuvirus (870), coronavirus (5), enterovirus (291), sapelovirus (110), teschovirus (2) | Astrovirus (7,445) | 6 | ||
| 35 | 11,280 | Kobuvirus (5), coronavirus (10), enterovirus (24), sapelovirus (27) | Astrovirus (8,062) | 4 | ||
| 36 | 18,484 | Kobuvirus (15,598), coronavirus (13), enterovirus (105), teschovirus (25) | None | 4 | ||
| 30 | 41 | 7,549 | Enterovirus (42), sapovirus (8), sapelovirus (2) | Astrovirus (788), bocavirus (4), circovirus-like virus (41) | 5 | |
| 42 | 7,482 | Enterovirus (43), sapovirus (1,598), sapelovirus (355) | Astrovirus (2,021), bocavirus (8) | 5 | ||
| 43 | 18,597 | Enterovirus (24), sapovirus (4), | Astrovirus (14,295), bocavirus (41) | 4 | ||
| 44 | 13,778 | Sapovirus (7,368), teschovirus (2), enterovirus (41) | Astrovirus (4,887) | 4 | ||
| 45 | 13,846 | Coronavirus (5), kobuvirus (198), enterovirus (394), sapovirus (9,445) | Astrovirus (8), posavirus 2 | 5 | ||
| 46 | 12,551 | Coronavirus (160), kobuvirus (52), enterovirus (528), sapovirus (733), sapelovirus (100) | Astrovirus (1,265), posavirus 2 | 6 |
Data assembly and processing.
A total of 652,000 pyrosequence reads (average length, 235 bases) were initially generated. Sequence reads were assigned to 36 data bins based on the different 20-base fixed-region sequences of the primers used. Each read was then trimmed of the fixed primer sequences plus eight additional nucleotides (encoded by the 8N part of the random primers), leaving 570,000 sequences longer than 100 bp for further analyses. Trimmed sequences within each group were assembled into contigs using MIRA software, with a criterion of ≥95% identity over at least 35 bp. The assembled contigs and singlet sequences were then compared to GenBank using BLASTx. Using BLASTx search, sequences with E values of ≤10−5 were classified as likely originating from a eukaryotic virus, bacterium, phage, eukaryote, other, or unknown based on the taxonomic origin of the sequence with the best E value.
Viral genome sequencing.
PCR primers were designed based on the viral sequences with sequence matches to known virus. PCR to bridge sequence gaps, inverse PCR, and 3′ and 5′ rapid amplification of cDNA ends (RACE) were used to complete viral genomes.
Phylogenetic analysis.
Sequences characterized in the current study and sequences representing the currently known diversity of astroviruses, bocaviruses, circoviruses and circovirus-like viruses, and picornavirus-like viruses in clades 1 and 6 (61) and astroviruses were aligned using the program MUSCLE. The latter data set was supplemented with additional porcine-derived virus-like posavirus sequences and homologous sequences from the RNA-dependent RNA polymerase (RdRp) of recently described cDNA recovered from the nematode Ascaris suum (see Table S2 in reference 61). The RdRp alignment used is provided in Fig. S1 in the supplemental material. Abbreviations of viruses correspond to those used in reference 61. Translated amino acid sequences were aligned using the program MUSCLE (33) with default settings. Maximum-likelihood phylogenetic trees of each alignment were generated using the program GARLI (114) using the amino acid model of Whelan and Goldman (106). One hundred bootstrap replications of the data were run to determine robustness of branching, with values indicated at each branching point when they were greater than 70%. For each data set, the maximum clade credibility tree was plotted and annotated with bootstrap values generated by the program CONSENSE in the PHYLIP 3.65 package using the tree output from GARLI. The GenBank accession numbers of the reference sequences used in the phylogenetic analysis are shown in the trees.
Nucleotide sequence accession numbers.
The genomes of known and new viruses described in detail here were deposited in GenBank under the following accession numbers: PAstV2-43, JF713710; PAstV5-33, JF713711; PAstV2-51, JF713712; PAstV4-35, JF713713; PBoV3-22, JF713714; PBoV3-23, JF713715; po-circo-like virus 21, JF713716; po-circo-like virus 22, JF713717; po-circo-like virus 41, JF713718; po-circo-like virus 51, JF713719; posavirus 1, JF713720; and posavirus 2, JF713721. All pyrosequence reads were deposited under short-read archive accession number SRA030664.
RESULTS
Overview of sequence data.
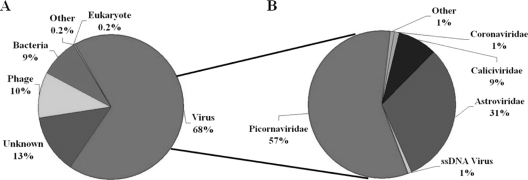
A total of 24 feces samples from healthy piglets and 12 feces samples from diarrheic piglets were collected on a high-density farm. Viral particles and their nucleic acids were enriched by filtration and nuclease treatment prior to nucleic acid extraction, random RT-PCR-based amplification, and pyrosequencing to generate over 600,000 sequence reads (102, 103). Sequence contigs were generated using reads from each of the 36 samples and classified based on best BLASTx expectation (E) scores. Summaries of the taxonomic classifications are shown in Fig. 1 A. Thirteen percent of all the sequence reads had no significant similarity to any sequences in GenBank, lower than the percentages of unclassified sequences in previous viral metagenomic studies of human and bat feces (65, 103). The most abundant fraction of viral sequences showed matches to mammalian viruses (Fig. 1B).
Fig. 1.
Sequence classification of porcine data sequences based on BLASTx (E value of ≤10−5). (A) Percentages of sequences with similarity to those of eukaryotes, bacteria, phages, and eukaryotic viruses and to unclassifiable sequences. (B) Percentages of most abundant eukaryotic viral sequence reads classified by viral families.
Virome of porcine stools.
Ninety-eight percent of sequence reads with best BLASTx matches to mammalian viruses were to RNA viruses from the families Picornaviridae, Astroviridae, Coronaviridae, and Caliciviridae (Fig. 1B). One percent of these sequences were related to the single-stranded DNA (ssDNA) viruses in the Circoviridae and Parvoviridae families. Virtual translations of these viral sequences showed a wide range of protein similarity to known viruses, indicating the presence of previously known and unknown viral species.
Previously characterized viruses identified here consisted of kobuviruses, enteroviruses, sapoviruses, sapeloviruses, coronaviruses, and teschoviruses (Table 1). Astroviruses and bocaviruses showing substantial sequence divergence from known porcine viruses as well as highly divergent Rep-encoding circular DNA and RdRp-encoding RNA genomes were also identified and their genomes characterized (Table 1). The most highly represented mammalian viruses were, in order of sequence read abundance, kobuviruses (23% of all reads), astroviruses (22.5%), enteroviruses (13.8%), sapoviruses (5.7%), sapeloviruses (1.5%), coronaviruses (0.69%), bocaviruses (0.22%), and teschoviruses (0.03%). The number of reads for all the porcine viruses combined amounted to 64% of all reads from healthy animals and 68% from diarrheic animals. The fraction of diarrheic versus healthy animals shedding any one of these eight mammalian viruses was identical except for bocaviruses (5/12 versus 3/24) and coronaviruses (11/12 versus 13/24), which were more prevalent in diarrheic than in healthy animals (chi-square test, P < 0.05). Out of 36 animals tested, kobuviruses were found in 83%, astroviruses in 75%, enteroviruses in 80%, sapoviruses in 42%, sapeloviruses in 53%, coronaviruses in 67%, bocaviruses in 22%, and teschovirus in 36%. The average percentages of sequence reads for each virus per infected piglet were compared between healthy and diarrheic piglets. Teschoviruses and bocaviruses were excluded due to the limited number of reads (<1,500). Except for kobuviruses, less than a 2.5-fold difference was measured between infected healthy and sick animals in their percentages of sequence reads to these eight viruses. Unexpectedly, the percentage of kobuviruses reads per animal was 12 times higher in healthy than in diarrheic piglets. Overall, except for the higher rate of detection of bocaviruses and coronaviruses, the fraction of animals shedding any particular virus, the fraction of viral sequence reads per infected pig, and the overall fraction of all viral reads combined were not greater in diarrheic than in healthy piglets.
The average number of porcine viruses per feces samples (coinfections) was 4.2 for healthy and 5.4 for diarrheic piglets. A statistically greater number of diarrheic piglets shed 6 or more viruses (8/12) than did healthy piglets (6/24) (chi-square test, P < 0.05). The healthy animals showing the fewest coinfections were the youngest (19 days old) and still unweaned animals, shedding an average of 1.5 different viruses. Kobuviruses were the only virus type found in all 6 still-unweaned healthy piglets.
New porcine viruses.
Several long contigs with highly significant E scores to eukaryotic viruses were identified. In samples where a high number of virus-specific reads (>1,000) were generated, nearly complete viral genomes were obtained. Full or nearly complete genomes were then acquired by bridging sequence gaps by PCR or RT-PCR, and the extremities of linear genomes were amplified using 5′ and 3′ RACE. Genomes were then resequenced directly from overlapping PCR or RT-PCR fragments. Circular genomes were amplified by inverse PCR.
Porcine astroviruses.
The family Astroviridae consists of positive single-stranded RNA (ssRNA) virus whose genomes range in length from 6.4 to 7.3 kb and contain three open reading frames (ORFs) designated ORF1a (nonstructural proteins), ORF1b (RdRp), and ORF2 (capsid). Astroviruses (AstV) can cause gastroenteritis in mammalian and avian species and have been identified in numerous mammalian species (9, 40, 41, 53, 57, 69, 81, 95, 99). Astroviruses have also been recently associated with encephalitis in a child with agammaglobulinemia (80) and with a shaking syndrome in farmed minks (9).
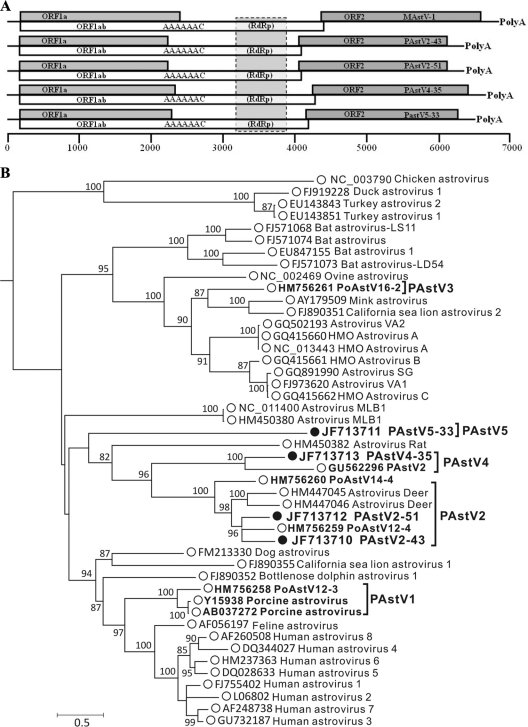
In this study, astrovirus sequences were detected in feces from 10/12 diarrheic and 17/24 healthy piglets. Four viral strains were selected for full genome sequencing based on a high number of sequence reads and high divergence from previously reported porcine astrovirus sequences. The nearly complete genomes of PAstV5-33, PAstV2-43, PAstV2-51, and PAstV4-35 were 6,457 bp, 6,293 bp, 6,333 bp, and 6,609 bp in length, respectively, excluding their 3′ poly(A) tail (Fig. 2 A). Using the capsid proteins of these and other astroviruses, phylogenetic analysis indicated that the four viruses fell into three distinct genetic lineages within the genus Mamastrovirus (Fig. 2B). The clusters of PAstV were labeled 1 through 5 based on the earliest dates of publications describing the first members of these clusters. The same 1-to-5 cluster labeling was applied to the four new genomes. PAstV2-43 and PAstV2-51 clustered with the recently characterized group 2 partial astrovirus genomes PoAstV12-4 and more distantly with PoAstV14-4, both from Canada (69), as well as the deer astroviruses CcAstV-1 and CcAstV-2 (95). PAstV4-35 clusters with group 4 virus PAstV2 from Hungary (81) (Fig. 2). PAstV5-33 formed its own deep branch in the astrovirus tree, likely representing a fifth porcine astrovirus group. A distance matrix of the capsid proteins of astroviruses is provided in Table S1 in the supplemental material. Divergent members of two preexisting clades plus a new clade were therefore characterized, showing the high genetic diversity of porcine astroviruses on this single farm.
Fig. 2.
(A) Genomic organization of novel porcine astroviruses relative to the mink astrovirus (MAstV) genome. (B) Maximum-likelihood phylogenetic analysis of ORF2 of novel porcine astroviruses. Bootstrap values of ≥70% are indicated at each branching point.
Porcine bocaviruses.
The genus Bocavirus, within the Parvoviridae family, consist of linear positive-strand DNA genomes 4 to 6 kb in length. Bocaviruses are distinguished from other parvoviruses by the presence of a third ORF in the middle of their genomes. Bocaviruses originally identified in bovine and canine stool samples have since been identified in numerous mammalian species, including in human (3, 5, 55, 58), gorilla (58, 91), chimpanzee (91), and three lineages in pigs, including one with only partially described genomes 6V and 7V (10, 20, 22, 90, 110). Bocaviruses have been associated with diverse symptoms, most notably respiratory problems and diarrhea (2, 15, 54, 70, 86, 101).
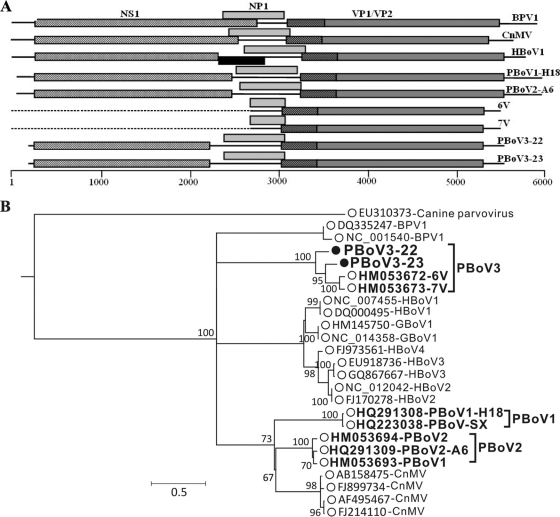
We identified 5 diarrheic and 3 healthy piglets with sequences related to those of known bocaviruses and proceeded to derive viral genomes from two diarrhea samples (Fig. 3 A). The two genomes clustered with one another and more distantly with the partial 6V and 7V genomes from China (20) (Fig. 3B). The full genomes of representatives of the third major clade of porcine bocaviruses were therefore generated. The genetic distances between different bocaviruses are shown in Table S2 in the supplemental material.
Fig. 3.
(A) Gene organization of novel porcine bocaviruses compared with other bocaviruses. BPV1, bovine; CnMV, canine; HBoV1, human; PBoV1-H18, PBoV-A2, 6V, and 7V, porcine. The black box for HBoV1 represents the known spliced exon of NS1 transcripts. (B) Maximum-likelihood phylogenetic analysis of VP1 proteins of novel porcine bocaviruses. Bootstrap values of ≥70% are indicated at each branching point.
Porcine circovirus-like viruses.
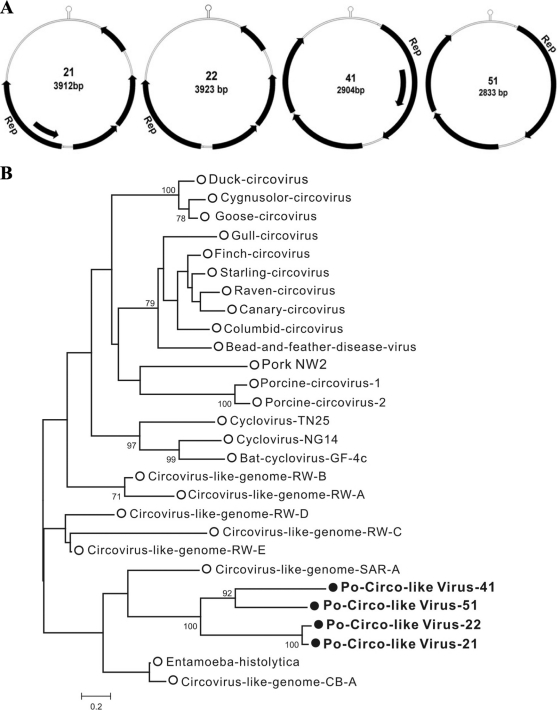
Rolling-circle replication initiator proteins/replicase proteins (Rep) catalyze a break in a stem-loop origin of replication from which a host-encoded DNA polymerase extends the 3′-OH end to replicate the circular viral genome (21, 96). Feces from two healthy and six diarrheic pigs showed the presence of sequences with similarity to Rep. Because Rep genes are located on small circular viral DNA genomes, we used inverse PCR to amplify and sequence four circular DNA genomes.
The gene organization of these genomes was distinct from those of circoviruses or the ChiSCV from wild chimpanzee feces, which contain two back-to-back ORFs (8, 64). Two genomes (21 and 22) were closely related with four or five ORFs, while the other genomes contained three or four ORFs (Fig. 4 A). For two genomes the Rep binding stem-loop structure was located downstream of the Rep genes, while for two genomes the stem-loop was upstream. Phylogenetically these four Rep clustered together and were most closely related to Rep genes found integrated in the genomes of the protozoan parasites Entamoeba histolytica and Entamoeba dispar, with a protein identity of 33 to 34% (Fig. 4B). Like the Entamoeba Rep, these proteins contained N-terminal viral Rep superfamily and C-terminal P-loop nucleoside triphosphatase (NTPase) superfamily domains. The genomic properties of the viruses that we temporarily named porcine circovirus-like viruses are shown in Table S3 in the supplemental material. A new clade of circular DNA viral genomes, representing a potential new viral family, was therefore found in pig feces.
Fig. 4.
(A) Genomic organizations of porcine circovirus-like viruses. The locations of putative Rep genes and other ORFs (encoding proteins greater than 100 amino acids long) are indicated by arrows. The stem-loop is also shown. (B) Maximum-likelihood phylogenetic analysis of the Reps of novel porcine circovirus-like viruses and Reps from small circular ssDNA genomes or integrated in Entamoeba histolytica. Circovirus-like genomes are from various environmental sources.
Highly divergent porcine picornavirus-like genomes.
Three feces samples showed the presence of sequences with detectable similarity to the RNA-dependent RNA polymerases (RdRp) of members of the order Picornavirales. Using the same chromosome-walking methods used for the other single-stranded RNA genomes, the entire genomes of two of these highly divergent viruses were acquired, and we provisionally named the viruses posavirus 1 and posavirus 2 (porcine stool-associated RNA viruses 1 and 2). Both genomes consisted of single long ORFs encoding 2,952- and 3,347-amino-acid (aa) polyproteins, respectively. When posavirus polyproteins were used in BLASTx searches of GenBank's NR database, a series of recently (March 2011) deposited cDNA-derived protein sequences from an adult Ascaris suum nematode (the long roundworm of pigs) isolated from a pig on a U.S. farm yielded E scores that were more significant (3 × 10−108 to 10−71) than the next closest E scores to dicistroviruses (10−18 to 10−24). Six sequences of 2,961 to 3,170 amino acids in length (and three shorter, presumably partial sequences) were annotated as A. suum polyproteins or replicase polyproteins (GenBank accession numbers ADY39832.1, ADY39824.1, ADY39838.1, ADY39842.1, ADY39835.1, and ADY39828.1). Amino acid sequence identities of 24 and 28% (39 and 44% similarities) were seen between the more conserved 3′ halves of the posaviruses and the roundworm cDNA-derived polyproteins. A conserved protein domain analysis of the polyproteins from roundworm cDNA, posaviruses, caliciviruses, and the first ORF of a dicistrovirus showed a conserved domain order between the roundworm cDNA and the feces-derived posavirus polyproteins (see Fig. S2 in the supplemental material). Only the roundworm cDNA-derived and the posavirus polyproteins possessed two distinct picornavirus capsid domains downstream of the RT-like superfamily (RdRp) domain. Relative to the roundworm polyproteins, a peptidase domain was missing in the posaviruses, whose P-loop NTPase domain was located further upstream. In order to determine if these virus-like cDNAs were encoded by the A. suum genome or represented infection with RNA viruses, we tested genomic DNA directly extracted from A. suum eggs using 3 sets of nested PCR primers targeting these cDNAs and another PCR primer set targeting a single-copy chromosomal A. suum gene. The single-copy A. suum gene PCR was positive in both the first and nested PCRs, while none of the three polyprotein-specific sets of primers yielded an amplification product. The virus-like cDNAs from A. suum are therefore not encoded by the A. suum genome and likely represent infections with posavirus-like viruses.
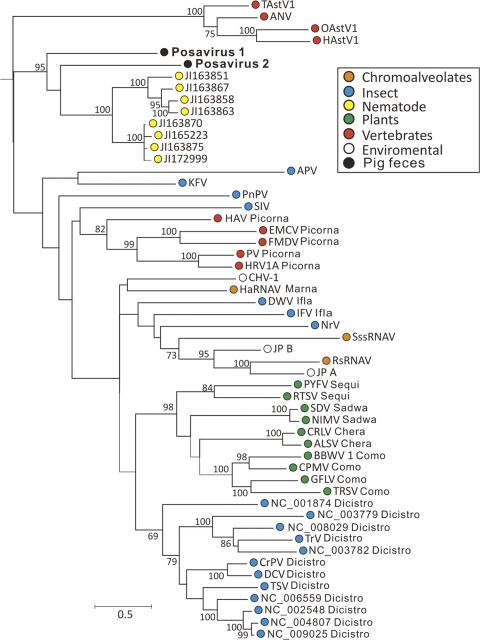
Phylogenetic analysis of the most conserved region of the Picornavirales RdRp region indicated that the two posaviruses were related and closest to the virus-like sequences from A. suum cDNA (Fig. 5). The next closest sequences were from members of the Dicistroviridae family known to infect arthropods. Dicistroviruses consist of two tandem ORFs encoding replicative functions and structural proteins, while both posaviruses and the A. suum cDNA consist of long single ORF.
Fig. 5.
Maximum-likelihood phylogenetic analysis of the RdRps of posaviruses and A. suum cDNA-derived polyproteins and of clades 1 and 6 and astroviruses in the previously defined picornavirus-like supergroup (61) (abbreviations are as used previously). Bootstrap values of ≥70% are indicated at each branching point.
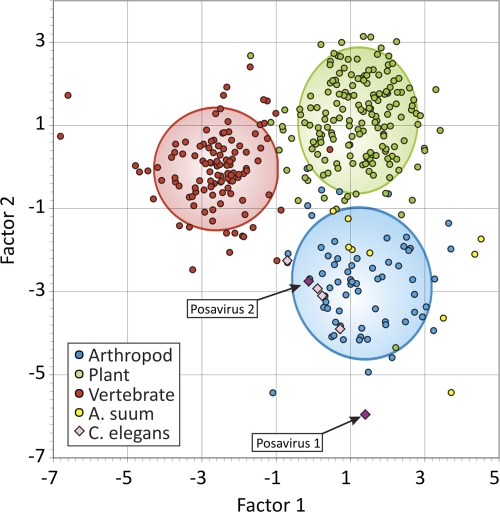
To help determine the cellular host of posaviruses, we then used nucleotide compositional analysis (NCA). Differences in base composition and dinucleotide frequencies of RNA viruses infecting different hosts have been exploited as a means to infer the host origins of uncharacterized viruses (58). A total of 352 representative sequences were used to identify compositional traits among viruses infecting vertebrates, arthropods, and plants by discriminant analysis (see Table S4 in the supplemental material). By this analysis, viruses were correctly assigned to their host origin in 96% of cases using frequencies of each base and ratios of dinucleotide frequencies to those expected from their component base frequencies (see Table S5 in the supplemental material). Since only two ssRNA viruses (both nodaviruses in the Picornavirales order) are known to infect nematodes (Caenorhabditis elegans) (38), NCA cannot yet include this group of animals as a separate classification category. However, when NCA was applied to the posaviruses, A. suum cDNA derived virus-like sequences, and the nodaviruses from C. elegans, all clustered within the arthropod group, although with a wider scatter of canonical factor values than was typical of arthropod viruses (Fig. 6). The shared compositional attributes of viruses infecting nematodes and arthropods (primarily insects) may reflect the greater evolutionary relatedness of their hosts compared to the other two classification categories (vertebrates and plants). Based on 18S rRNA gene phylogeny, both arthropoda and nematoda and other molting phyla have been proposed to belong to the Ecdysozoa clade (1, 32), distinct from the Deuterostomia clade, which includes the vertebrates, and from plants (in the highly distant eukaryotic kingdom Plantae). Using posavirus 1- and 2-specific nested RT-PCR, posavirus 1 was detected in 17 of 36 animals and posavirus 2 in 11 of 36 animals, indicating common infections. Nine animals were coinfected with both posaviruses.
Fig. 6.
Discriminant analysis used to classify viral sequences into host groups using the 4 mononucleotide and 16 dinucleotide frequencies. The graph shows separation of groups using the two most influential factors. Points represent values for individual sequences, with confidence ellipses centered on the centroid of each group.
DISCUSSION
High-density pig farming is conducted worldwide, resulting in the large-scale production of porcine excrement and increasing concern about its safe disposal (25, 113). The crowded conditions and frequent movement of pigs also provide a beneficial environment for viral transmission and evolution (92). The recent emergence of the H1N1 “swine” influenza may have resulted from influenza virus recombination on such farms (60, 94, 105). We present here an initial description of the fecal virome of pigs on a high-density farm, using deep sequencing of enriched viral nucleic acids.
Enteric pig viruses in the Picornaviridae, Astroviridae, Caliciviridae, Coronaviridae, and Parvoviridae families were found, including, in decreasing relative concentration, kobuviruses, astroviruses, enteroviruses, sapoviruses, sapeloviruses, coronaviruses, bocaviruses, and teschoviruses. All feces samples contained at least one BLASTx-recognizable mammalian virus, with averages of 4.2 and 5.4 distinct viruses in healthy and diarrheic pigs, respectively. A higher load (as estimated by the fraction of viral sequence reads in infected animals) of any single virus was not associated with diarrhea. The prevalence of detection was higher in diarrheic pigs only for bocaviruses and coronaviruses. Shedding of six or more distinct viruses was also associated with diarrhea. Coinfections with a large number of viruses may therefore overwhelm piglet innate and immune defenses. In piglets, maternal antibodies are absorbed from the colostrum through their immature gut (17), providing protection against infections to which the sows were exposed (17, 34, 35, 76, 79, 87). The absence of symptoms in some heavily virus-coinfected piglets may be due to such maternal antibodies, especially early after suckling has begun, when piglet serum titers are highest (79, 87); this is consistent with the detection of an average of 1.5 viral coinfections in the unweaned 19-day-old piglets versus 5.2 coinfections in older animals.
Studies characterizing viromes in animal feces have been performed on horses (18), humans (12, 39, 56, 103, 112), bats (31, 65), turkeys (29), and California sea lions (66), providing novel viral genomes for phylogenetic analyses and disease association studies. The high degree of coinfections with distinct mammalian viruses seen here in piglets is greater than reported in these prior fecal virome studies, which is possibly a consequence of the very young age of the animals analyzed here and/or of their high-density living conditions. Whether the high rate of viral infections reflects long-term fecal shedding in chronic infections or frequent reinfections with rapidly cleared viruses will require the analysis of longitudinally collected samples. Long-term fecal shedding of human bocavirus has been seen in children (72), and coinfections with distinct porcine bocaviruses and possible recombination have been recently reported (62).
Beside the previously characterized RNA and DNA viruses, several previously unknown or only partially characterized viral genomes were also identified and their full or near full genomes characterized. Pig astroviruses that may be broadly grouped into 4 major phylogenetic clades have recently been described (51, 52, 69, 81, 100, 104). We sequenced three divergent variants within two of these clades and characterized the first member of a fifth clade (PAstV5-33). The detection of such a high diversity of pig astroviruses on a single farm indicates ongoing viral transmission from multiple sources rather than a single-point introduction. A PCR study of astroviruses on multiple Canadian farms showed that 80% of healthy young pigs were infected with diverse porcine astroviruses (69). The detection of multiple astrovirus species in pigs as well as humans may reflect multiple past cross-species transmissions from other animal sources followed by adaptation to new hosts (40, 41, 57). A recent viral metagenomics analysis of feces from wild California sea lions also detected a large diversity of astroviruses in 51% of animals, consisting of 30% of the mammalian virus reads, versus detection of diverse astroviruses in 75% of piglets, consisting of 22.5% of mammalian viral reads. Further sampling should determine if such high astrovirus diversity, prevalence, and viral loads are a general phenomenon of wild and domesticated mammals. Humans are also infected with several distinct astroviruses (40, 41, 57).
Several porcine bocavirus species have been recently characterized (10, 20, 22, 90, 110). The bocavirus genome sequences obtained here were related to the partially sequenced 6V and 7V porcine bocaviruses from China originally reported by Cheng et al. (20) and the nearly full genomes recently reported by Lau et al. (62). The 25% rate of detection of bocavirus shedding seen here is lower than the 46% rate of porcine bocavirus-like virus infection in feces of healthy Swedish pigs (11) and the 70% detection in tissues and sera of Chinese pigs with respiratory symptoms (111). This lower apparent rate of infection may be due to differences in the age of animals analyzed, tissues analyzed, and farming methods and/or a higher sensitivity for PCR-based virus detection. Overall, the genetic diversity of porcine bocaviruses is greater than that of the so far monophyletic bocaviruses found in humans, cows, and dogs. Determination of whether the genetic diversity of bocaviruses infecting other wild or farm animals will be as high as that found in pigs or whether modern farming is associated with a higher viral diversity will require further investigations.
A series of highly divergent viruses with small circular DNA genomes encoding Rep proteins were characterized. Rep proteins are components of many viral species with single-stranded circular DNA genomes. These viruses include circoviruses infecting birds and pigs (42, 98) and the related cycloviruses infecting farm animals and bats (67) and possibly humans (64). Unexpectedly, circoviruses and cycloviruses have also recently been reported in dragonflies (83). Rep proteins encoded on circular DNA genomes have also been found in chimpanzee stools (8) and in environmental samples (84). Plant viruses of the Geminiviridae and Nanoviridae families with small circular ssDNA genomes also encode Rep proteins (37), and Rep homologues have been found encoded within the genome of canarypox virus, on chromosomes of the parasitic protozoans Entamoeba histolytica and Giardia duodenalis, and on a plasmid, p4M, from a Gram-positive bacterium, possibly the result of integration by virus-like extrachromosomal elements (44). Largely defective Rep genes from ancient circovirus integration events have recently been described in mammalian genomes (7, 59).
The source of the Rep-encoding circular DNA viruses found in the current study may be infected pig cells, other organisms in pig gut, or a dietary source. Based on their closest sequence similarity to the Rep sequence in the genome of the human protozoan parasitic Entamoeba histolytica (E = 10−31), with which they cluster phylogenetically, we suggest that these circular DNA genomes replicate in E. histolytica or another, more pig-specific Entamoeba species such as E. polecki or E. suis. Sequence similarity searches (www.amoebadb.org) within the genome of the human commensal E. dispar (E = 10−27) and the reptile-infecting E. invandens (E = 10−7) also showed the presence of Rep sequences in these protozoans. Based on sequence similarity between virus-like sequence in Entamoeba and the circular viral genomes from pig feces, we propose that these circular DNAs originate from parasitic protozoans commonly found in the guts of pigs.
The order Picornavirales includes viruses replicating in all the major divisions of eukaryotes, including protozoans, plants, insects, and mammals, which may reflect the origin of this diverse group of viruses prior to the radiation of eukaryotes (61). Two novel viral RNA genomes with distant sequence relatedness to other Picornavirales were characterized and provisionally named posaviruses. Several recently reported polyproteins originating from cDNA of the helminth A. suum (the long roundworm of pigs), but shown here by PCR not to be encoded in its genome, showed the highest degree of sequence similarity to posaviruses and a conserved protein domain organization, including the presence of helicase, RdRp, and rhinovirus capsid-like domains. By genetic distance criteria, posaviruses were therefore more closely related to the A. suum cDNA-derived sequences than to any other described Picornavirales. When their nucleotide compositions were analyzed, both posaviruses and A. suum polyproteins fell within the arthropod grouping, although with a wide scatter. Evolutionarily, nematodes and arthropods are more closely related than they are to vertebrates and plants (1, 32). The similar nucleotide compositions of Picornavirales known to infect arthropods and the nematode C. elegans-infecting nodaviruses was therefore expected. The similar nucleotide compositions of the posaviruses, the A. suum virus-like polyprotein-encoding cDNA, C. elegans, nodaviruses, and arthropod-infecting Picornavirales therefore support the possibility that the posaviruses (and the A. suum cDNA) represent new Picornavirales capable of infecting nematodes, frequent parasites in pig guts (48). The similar genomic organization and polyprotein lengths and strong sequence conservation between posaviruses and Ascaris suum cDNA-derived polyproteins, plus the similar NCA characteristics of Ecdysozoa-infecting Picornavirales, posaviruses, and A. suum cDNA, all support the possibility that the posavirus sequences detected in the porcine feces originated from infections of parasitic nematodes.
A high rate of enteric viral coinfections with mammalian RNA and DNA viruses and viruses that may replicate in protozoans and nematodes was measured here in piglets raised on a high-density farm and may reflect farming conditions favorable to frequent virus transmissions. Recombination is an integral part of the evolution of the picornaviruses (kobuviruses, enteroviruses, sapeloviruses, and teschoviruses) making up the majority of the piglet fecal viral sequences. Recombination has also been reported within the Caliciviridae, Astroviridae, Parvoviridae, and Circoviridae-like viral families detected here (16, 45, 47, 50, 58, 63, 68, 77, 78, 82, 88, 89, 93, 104, 108). The high rate of coinfections seen in this high-density farm therefore provides favorable conditions for recombination and accelerated viral evolution. Whether high rates of coinfections with the same group of viruses also occur on smaller farms or in different countries or vary seasonally will require further animal sampling. Determining the extent of viral zoonoses occurring on mixed-animal farms may also be of interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard E. Davis and Jianbin Wang for information on the source of the A. suum cDNA sequences.
This work was supported by NHLBI grant R01HL083254 and the Blood Systems Research Institute (E.D.), Shanghai Jiao Tong University (T.S.), and NSF award CNS-0619926 to the Bio-X2 cluster at Stanford University for computing resources.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 7 September 2011.
REFERENCES
- 1. Aguinaldo A. M., et al. 1997. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387: 489–493 [DOI] [PubMed] [Google Scholar]
- 2. Allander T. 2008. Human bocavirus. J. Clin. Virol. 41: 29–33 [DOI] [PubMed] [Google Scholar]
- 3. Allander T., et al. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102: 12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allander T., Emerson S. U., Engle R. E., Purcell R. H., Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 98: 11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arthur J. L., Higgins G. D., Davidson G. P., Givney R. C., Ratcliff R. M. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 5: e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bank-Wolf B. R., König M., Thiel H. J. 2010. Zoonotic aspects of infections with noroviruses and sapoviruses. Vet. Microbiol. 140: 204–212 [DOI] [PubMed] [Google Scholar]
- 7. Belyi V. A., Levine A. J., Skalka A. M. 2010. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the Parvoviridae and Circoviridae are more than 40 to 50 million years old. J. Virol. 84: 12458–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blinkova O., et al. 2010. Novel circular DNA viruses in stool samples of wild-living chimpanzees. J. Gen. Virol. 91: 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blomström A. L., Widén F., Hammer A. S., Belák S., Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 48: 4392–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blomström A. L., et al. 2009. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 146: 125–129 [DOI] [PubMed] [Google Scholar]
- 11. Blomström A. L., et al. 2010. Studies of porcine circovirus type 2, porcine boca-like virus and torque teno virus indicate the presence of multiple viral infections in postweaning multisystemic wasting syndrome pigs. Virus Res. 152: 59–64 [DOI] [PubMed] [Google Scholar]
- 12. Breitbart M., et al. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185: 6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Breitbart M., et al. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99: 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Briese T., et al. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 5: e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown K. E. 2010. The expanding range of parvoviruses which infect humans. Rev. Med. Virol. 20: 231–244 [DOI] [PubMed] [Google Scholar]
- 16. Bull R. A., Tanaka M. M., White P. A. 2007. Norovirus recombination. J. Gen. Virol. 88: 3347–3359 [DOI] [PubMed] [Google Scholar]
- 17. Butler J. E., Zhao Y., Sinkora M., Wertz N., Kacskovics I. 2009. Immunoglobulins, antibody repertoire and B cell development. Dev. Comp. Immunol. 33: 321–333 [DOI] [PubMed] [Google Scholar]
- 18. Cann A. J., Fandrich S. E., Heaphy S. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30: 151–156 [DOI] [PubMed] [Google Scholar]
- 19. Chen L. H., et al. 2009. Illness in long-term travelers visiting GeoSentinel clinics. Emerg. Infect. Dis. 15: 1773–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng W. X., et al. 2010. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One 5: e13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheung A. K. 2007. A stem-loop structure, sequence non-specific, at the origin of DNA replication of porcine circovirus is essential for termination but not for initiation of rolling-circle DNA replication. Virology 363: 229–235 [DOI] [PubMed] [Google Scholar]
- 22. Cheung A. K., et al. 2010. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 155: 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Childs J. E., Gordon E. R. 2009. Surveillance and control of zoonotic agents prior to disease detection in humans. Mt. Sinai J. Med. 76: 421–428 [DOI] [PubMed] [Google Scholar]
- 24. Chiu C. Y., et al. 2008. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc. Natl. Acad. Sci. U. S. A. 105: 14124–14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Costantini V. P., et al. 2007. Effects of different animal waste treatment technologies on detection and viability of porcine enteric viruses. Appl. Environ. Microbiol. 73: 5284–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Culley A. I., Lang A. S., Suttle C. A. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424: 1054–1057 [DOI] [PubMed] [Google Scholar]
- 27. Culley A. I., Lang A. S., Suttle C. A. 2006. Metagenomic analyis of coastal RNA virus communities. Science 312: 1795–1798 [DOI] [PubMed] [Google Scholar]
- 28. Curns A. T., et al. 2010. Reduction in acute gastroenteritis hospitalizations among US children after introduction of rotavirus vaccine: analysis of hospital discharge data from 18 US states. J. Infect. Dis. 201: 1617–1624 [DOI] [PubMed] [Google Scholar]
- 29. Day J. M., Ballard L. L., Duke M. V., Scheffler B. E., Zsak L. 2010. Metagenomic analysis of the turkey gut RNA virus community. Virol. J. 7: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Domingo E. 2010. Mechanisms of viral emergence. Vet. Res. 41: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donaldson E. F., et al. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 84: 13004–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dunn C. W., et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749 [DOI] [PubMed] [Google Scholar]
- 33. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elahi S., Buchanan R. M., Babiuk L. A., Gerdts V. 2006. Maternal immunity provides protection against pertussis in newborn piglets. Infect. Immun. 74: 2619–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elvinger F., Baldwin C. A., Liggett A. D., Tang K. N., Dove C. R. 1996. Protection of pigs by vaccination of pregnant sows against eastern equine encephalomyelitis virus. Vet. Microbiol. 51: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. FAO, OIE, WHO, UNICEF, UNSIC, and World Bank 2008. Contributing to one world, one health: a strategic framework for reducing risk of infectious diseases at the animal-human-ecosystem interphase. http://un-influenza.org/files/OWOH_14Oct08.pdf
- 37. Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 38. Félix M. A., et al. 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9: e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Finkbeiner S. R., et al. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4: e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Finkbeiner S. R., Kirkwood C. D., Wang D. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 5: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Finkbeiner S. R., et al. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 83: 10836–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finsterbusch T., Mankertz A. 2009. Porcine circoviruses—small but powerful. Virus Res. 143: 177–183 [DOI] [PubMed] [Google Scholar]
- 43. Forrest H. L., Webster R. G. 2010. Perspectives on influenza evolution and the role of research. Anim. Health Res. Rev. 11: 3–18 [DOI] [PubMed] [Google Scholar]
- 44. Gibbs M. J., Smeianov V. V., Steele J. L., Upcroft P., Efimov B. A. 2006. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 23: 1097–1100 [DOI] [PubMed] [Google Scholar]
- 45. Graham R. L., Baric R. S. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 84: 3134–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haagmans B. L., Andeweg A. C., Osterhaus A. D. 2009. The application of genomics to emerging zoonotic viral diseases. PLoS Pathog. 5: e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hansman G. S., Oka T., Katayama K., Takeda N. 2007. Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med. Virol. 17: 133–141 [DOI] [PubMed] [Google Scholar]
- 48. Haugegaard J. 2010. Prevalence of nematodes in Danish industrialized sow farms with loose housed sows in dynamic groups. Vet. Parasitol. 168: 156–159 [DOI] [PubMed] [Google Scholar]
- 49. Herikstad H., et al. 2002. A population-based estimate of the burden of diarrhoeal illness in the United States: FoodNet, 1996-7. Epidemiol. Infect. 129: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoelzer K., Shackelton L. A., Holmes E. C., Parrish C. R. 2008. Within-host genetic diversity of endemic and emerging parvoviruses of dogs and cats. J. Virol. 82: 11096–11105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Indik S., Valícek L., Smíd B., Dvoráková H., Rodák L. 2006. Isolation and partial characterization of a novel porcine astrovirus. Vet. Microbiol. 117: 276–283 [DOI] [PubMed] [Google Scholar]
- 52. Jonassen C. M., et al. 2001. Comparison of capsid sequences from human and animal astroviruses. J. Gen. Virol. 82: 1061–1067 [DOI] [PubMed] [Google Scholar]
- 53. Jonassen C. M., Jonassen T. Ø. T., Sveen T. M., Grinde B. 2003. Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res. 91: 195–201 [DOI] [PubMed] [Google Scholar]
- 54. Kahn J. 2008. Human bocavirus: clinical significance and implications. Curr. Opin. Pediatr. 20: 62–66 [DOI] [PubMed] [Google Scholar]
- 55. Kapoor A., et al. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199: 196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kapoor A., et al. 2008. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc. Natl. Acad. Sci. U. S. A. 105: 20482–20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kapoor A., et al. 2009. Multiple novel astrovirus species in human stool. J. Gen. Virol. 90: 2965–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kapoor A., et al. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Katzourakis A., Gifford R. J. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6: e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kingsford C., Nagarajan N., Salzberg S. L. 2009. 2009 Swine-origin influenza A (H1N1) resembles previous influenza isolates. PLoS One 4: e6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koonin E. V., Wolf Y. I., Nagasaki K., Dolja V. V. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6: 925–939 [DOI] [PubMed] [Google Scholar]
- 62. Lau S. K., et al. 2011. Co-existence of multiple strains of two novel porcine bocaviruses in the same pig, a previously undescribed phenomenon in Parvoviridae and evidence for inter- and intra-host genetic diversity and recombination. J. Gen. Virol. 92: 2047–2059 [DOI] [PubMed] [Google Scholar]
- 63. Lefeuvre P., Lett J. M., Varsani A., Martin D. P. 2009. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 83: 2697–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li L., et al. 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84: 1674–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li L., et al. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84: 6955–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li L., et al. 27. July 2011. The fecal viral flora of California sea lions. J. Virol. doi:10.1128/JVI.05026-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li L., et al. 2010. Possible cross-species transmission of circoviruses and cycloviruses in farm animals. J. Gen. Virol. 92: 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lukashev A. N. 2010. Recombination among picornaviruses. Rev. Med. Virol. 20: 327–337 [DOI] [PubMed] [Google Scholar]
- 69. Luo Z., et al. 2010. Multiple novel and prevalent astroviruses in pigs. Vet. Microbiol. 149: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manteufel J., Truyen U. 2008. Animal bocaviruses: a brief review. Intervirology 51: 328–334 [DOI] [PubMed] [Google Scholar]
- 71. Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet. Microbiol. 140: 246–255 [DOI] [PubMed] [Google Scholar]
- 72. Martin E. T., et al. 2010. Frequent and prolonged shedding of bocavirus in young children attending daycare. J. Infect. Dis. 201: 1625–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mazet J. A., et al. 2009. A “one health” approach to address emerging zoonoses: the HALI project in Tanzania. PLoS Med. 6: e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meng X. J. 21 February 2011. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. doi:10.1016/j.viruses.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Olival K. J., Daszak P. 2005. The ecology of emerging neurotropic viruses. J. Neurovirol. 11: 441–446 [DOI] [PubMed] [Google Scholar]
- 76. Ostanello F., et al. 2005. Experimental infection of 3-week-old conventional colostrum-fed pigs with porcine circovirus type 2 and porcine parvovirus. Vet. Microbiol. 108: 179–186 [DOI] [PubMed] [Google Scholar]
- 77. Pantin-Jackwood M. J., Spackman E., Woolcock P. R. 2006. Phylogenetic analysis of Turkey astroviruses reveals evidence of recombination. Virus Genes 32: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Phan T. G., et al. 2007. Genetic heterogeneity, evolution, and recombination in noroviruses. J. Med. Virol. 79: 1388–1400 [DOI] [PubMed] [Google Scholar]
- 79. Pignatelli J., Grau-Roma L., Jiménez M., Segalés J., Rodríguez D. 2010. Longitudinal serological and virological study on porcine torovirus (PToV) in piglets from Spanish farms. Vet. Microbiol. 146: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Quan P. L., et al. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 16: 918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Reuter G., Pankovics P., Boros A. 2011. Identification of a novel astrovirus in a domestic pig in Hungary. Arch. Virol. 156: 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rivera R., Nollens H. H., Venn-Watson S., Gulland F. M., Wellehan J. F. 2010. Characterization of phylogenetically diverse astroviruses of marine mammals. J. Gen. Virol. 91: 166–173 [DOI] [PubMed] [Google Scholar]
- 83. Rosario K., et al. 2011. Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata:Anisoptera). J. Gen. Virol. 92: 1302–1308 [DOI] [PubMed] [Google Scholar]
- 84. Rosario K., Duffy S., Breitbart M. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90: 2418–2424 [DOI] [PubMed] [Google Scholar]
- 85. Scallan E., Griffin P. M., Angulo F. J., Tauxe R. V., Hoekstra R. M. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg. Infect. Dis. 17: 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schildgen O., et al. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sestak K., Lanza I., Park S. K., Weilnau P. A., Saif L. J. 1996. Contribution of passive immunity to porcine respiratory coronavirus to protection against transmissible gastroenteritis virus challenge exposure in suckling pigs. Am. J. Vet. Res. 57: 664–671 [PubMed] [Google Scholar]
- 88. Shackelton L. A., Parrish C. R., Truyen U., Holmes E. C. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U. S. A. 102: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shackelton L. A., Hoelzer K., Parrish C. R., Holmes E. C. 2007. Comparative analysis reveals frequent recombination in the parvoviruses. J. Gen. Virol. 88: 3294–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shan T., et al. 2011. Genomic characterization and high prevalence of bocaviruses in swine. PLoS One 6: e17292–e17292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sharp C. P., et al. 2010. Widespread infection of chimpanzees and gorillas with homologues of human parvoviruses B19, PARV4, and human bocavirus in the wild. J. Virol. 84: 10289–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shi M., et al. 2010. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J. Virol. 84: 8700–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Simmonds P. 2006. Recombination and selection in the evolution of picornaviruses and other mammalian positive-stranded RNA viruses. J. Virol. 80: 11124–11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Smith G. J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459: 1122–1125 [DOI] [PubMed] [Google Scholar]
- 95. Smits S. L., et al. 2010. Identification and characterization of deer astroviruses. J. Gen. Virol. 91: 2719–2722 [DOI] [PubMed] [Google Scholar]
- 96. Steinfeldt T., Finsterbusch T., Mankertz A. 2001. Rep and Rep′ protein of porcine circovirus type 1 bind to the origin of replication in vitro. Virology 291: 152–160 [DOI] [PubMed] [Google Scholar]
- 97. Taubenberger J. K., Kash J. C. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7: 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Todd D. 2004. Avian circovirus diseases: lessons for the study of PMWS. Vet. Microbiol. 98: 169–174 [DOI] [PubMed] [Google Scholar]
- 99. Toffan A., et al. 2009. Genetic characterization of a new astrovirus detected in dogs suffering from diarrhoea. Vet. Microbiol. 139: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ulloa J. C., Gutiérrez M. F. 2010. Genomic analysis of two ORF2 segments of new porcine astrovirus isolates and their close relationship with human astroviruses. Can. J. Microbiol. 56: 569–577 [DOI] [PubMed] [Google Scholar]
- 101. Ursic T., et al. 2011. Human bocavirus as the cause of a life-threatening infection. J. Clin. Microbiol. 49: 1179–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Victoria J. G., Kapoor A., Dupuis K., Schnurr D. P., Delwart E. L. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 4: e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Victoria J. G., et al. 2009. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 83: 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang Q. H., et al. 2001. Genetic analysis of the capsid region of astroviruses. J. Med. Virol. 64: 245–255 [DOI] [PubMed] [Google Scholar]
- 105. Weingartl H. M., et al. 2010. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J. Virol. 84: 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Whelan S., Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699 [DOI] [PubMed] [Google Scholar]
- 107. Wilson M. E. 2005. Diarrhea in nontravelers: risk and etiology. Clin. Infect. Dis. 41(Suppl. 8): S541–S546 [DOI] [PubMed] [Google Scholar]
- 108. Wolfaardt M., Kiulia N. M., Mwenda J. M., Taylor M. B. 2011. Evidence of a recombinant wild-type human astrovirus strain from a Kenyan child with gastroenteritis. J. Clin. Microbiol. 49: 728–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wu Q., et al. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. U. S. A. 107: 1606–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zeng S., et al. 2011. Complete coding sequences and phylogenetic analysis of porcine bocavirus. J. Gen. Virol. 92: 784–788 [DOI] [PubMed] [Google Scholar]
- 111. Zhai S., et al. 2010. High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch. Virol. 155: 1313–1317 [DOI] [PubMed] [Google Scholar]
- 112. Zhang T., et al. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ziemer C. J., et al. 2010. Fate and transport of zoonotic, bacterial, viral, and parasitic pathogens during swine manure treatment, storage, and land application. J. Anim. Sci. 88: E84–E94 [DOI] [PubMed] [Google Scholar]
- 114. Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. Ph.D. dissertation. The University of Texas, Austin, TX [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.