Abstract
Bamboo mosaic virus (BaMV) is a positive-sense RNA virus belonging to the genus Potexvirus. Open reading frame 1 (ORF1) encodes the viral replication protein that consists of a capping enzyme domain, a helicase-like domain (HLD), and an RNA-dependent RNA polymerase domain from the N to C terminus. ORF5 encodes the viral coat protein (CP) required for genome encapsidation and the virus movement in plants. In this study, application of a yeast-two hybrid assay detected an interaction between the viral HLD and CP. However, the interaction did not affect the NTPase activity of the HLD. To identify the critical amino acids of CP interacting with the HLD, a random mutational library of CP was created using error-prone PCR, and the mutations adversely affecting the interaction were screened by a bacterial two-hybrid system. As a result, the mutations A209G and N210S in CP were found to weaken the interaction. To determine the significance of the interaction, the mutations were introduced into a BaMV infectious clone, and the mutational effects on viral replication, movement, and genome encapsidation were investigated. There was no effect on accumulations of BaMV CP and genomic RNAs within protoplasts; however, the virus cell-to-cell movement in plants was restricted. Sequence alignment revealed that A209 of BaMV CP is conserved in many potexviruses. Mutation of the corresponding residue in Foxtail mosaic virus CP also reduced the viral HLD-CP interaction and restricted the virus movement, suggesting that interaction between CP and a widely conserved HLD in the potexviral replication protein is crucial for viral trafficking through plasmodesmata.
INTRODUCTION
To spread throughout hosts, plant viruses have evolved a number of pathways to allow their progeny to pass across plasmodesmata into neighboring cells and travel along the vascular system (8, 26). The virus-encoded movement proteins play a pivotal role through diverse mechanisms in these cell-to-cell and vascular transports. Ancillary proteins, for example, the viral coat proteins (CPs) in some cases, and host factors may also participate in these processes. Numerous studies have been conducted to elucidate the movement mechanisms. Many of the results have been summarized in a number of recent reviews (23, 28, 30). They provided in-depth discussions on issues such as the identification and characterization of the involved viral and host proteins and the transport models for some exemplified viruses, such as Tobacco mosaic virus (TMV) and Potato virus X (PVX). Despite these efforts, many details of the processes remain elusive.
Members of the genus Potexvirus have a positive-strand RNA genome that contains five open reading frames (ORFs), a 5′ methyl cap, and a 3′ poly(A) tail. ORF1 encodes the viral replication protein, consisting of a capping enzyme domain, a helicase-like domain (HLD), and an RNA-dependent RNA polymerase domain (RdRp) from the N terminus to the C terminus (16, 17). The HLD has RNA 5′-triphosphatase and nucleoside triphosphatase (NTPase) activities (18). With the concerted actions of the HLD and the capping enzyme domain, the cap structure can be formed at the 5′ end of the viral RNA (17). ORF2 to ORF4 overlap. They encode the viral movement proteins, called triple gene block protein 1 (TGBp1), TGBp2, and TGBp3. ORF5 is responsible for coat protein production. Based on studies of movement of many potexviruses, such as PVX and White clover mosaic virus, viruses of this group need TGB proteins and CP for their own transport (3, 7, 22, 25). TGBp1 is able to increase plasmodesmal size exclusion limits (1, 11). In addition, it has RNA helicase activity (12) and acts as an RNA-silencing suppressor (31). TGBp2 and TGBp3 are endoplasmic reticulum (ER)-associated proteins (14, 24). Two models have been proposed to describe the cell-to-cell movement of potexviruses (22, 27). The basic difference between these two models concerns whether a virion or a ribonucleoprotein (RNP) complex is transported across plasmodesmata. In the RNP model, the complex consists of viral RNA, TGBp1, and CP, and the viral CP was proposed to act in concert with the TGB proteins to mediate the complex translocation into neighboring cells. Recently, a more elaborate cell-to-cell transport model with RNA silencing of potexviruses was proposed (30).
Bamboo mosaic virus (BaMV) is a potexvirus commonly found in many bamboo varieties and artificially infectious to Nicotiana benthamiana. The functions of TGB proteins in BaMV movement in N. benthamiana have been studied. Alanine substitution for R16 or R21 of TGBp1 diminished the RNA-binding and NTPase activities of the protein and consequently restrained the virus from moving (19). Mutations of the conserved C109 or C112 of TGBp2 reduced cell-to-cell movement and severely inhibited systemic transport of the virus (29). A sorting signal in TGBp3 is necessary and sufficient for the protein to target to cortical ER tubules, a step required for BaMV cell-to-cell movement (32). To search for host factors involved in BaMV replication, each enzymatic domain of the viral replication protein has been used as bait to hunt for its interacting proteins from cDNA libraries prepared from both healthy and BaMV-infected N. benthamiana leaves. A putative plant methyltransferase was found to be associated with the viral RdRp, and it had the potential to suppress the virus replication (5). In this report, we describe another finding: that the viral HLD has a high affinity for the virus's own CP. Moreover, CP mutations that decreased the protein interaction restricted the virus from movement. No effect on virus replication or virion formation was observed in response to these mutations. Similar mutational effects were found in Foxtail mosaic virus (FoMV), suggesting a general role of the HLD-CP interaction in cell-to-cell movement of potexviruses.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The yeast Hybrid Hunter system (Invitrogen) was employed to search for BaMV HLD-interacting proteins from a BaMV-infected leaf cDNA library of N. benthamiana. An interaction between bait and prey in the system would result in the induction of his3 and lacZ genes. Construction of the pYESTrp2-based library was described previously (5), unless the leaves 3 days postinoculation with BaMV were used for mRNA isolation. In brief, the region within ORF1, encoding amino acids 513 to 900 of the viral replication protein, was added with EcoRI and PstI sites at the 5′ and 3′ ends, respectively, by PCR. The amplified DNA fragment was inserted into EcoRI-PstI-cleaved pHybLex/Zeo to become the bait plasmid. Saccharomyces cerevisiae strain L40 that harbored the bait plasmid was transformed with the library of prey plasmid pYESTrp2 cDNAs. The colonies grown on YC-WHUK/Z300 selection agar plates were further analyzed for their β-galactosidase activities based on both filter and quantitative assays. Plasmids were isolated from those exhibiting significant β-galactosidase activities and transformed into Escherichia coli Top10F′. Prey plasmids were isolated from the E. coli transformants and transformed back into the bait-expressing L40 strain to confirm the bait-prey interaction. Nucleotide sequences of the cDNA fragments from the screened prey plasmids were determined with an ABI Prism 3100 genetic analyzer.
For the β-galactosidase activity assay, yeast cells adhering to a filter paper were disrupted by repeatedly freezing and thawing. The paper was soaked in buffer that contained 100 mM phosphate (pH 7.0), 10 mM KCl, 1 mM MgSO4, and 1 mg/ml X-Gal until color developed. For the quantitative activity assay, 100 μl of disrupted cells was added to 900 μl reaction buffer as described above, except that X-Gal was replaced with 0.64 mg/ml O-nitrophenyl β-galactoside. After incubation at 30°C for 10 h, the optical density at 420 nm was determined with a spectrophotometer, and the activity was calculated according to the manufacturer's manual.
Bacterial two-hybrid screening.
The bacterial adenylate cyclase two-hybrid (BACTH) system (Euromedex) was used to screen BaMV CP mutations that decreased the binding affinity of the viral CP to the HLD. In this system, an interaction between bait and prey could regenerate the adenylate cyclase activity, resulting in the activation of lac and mal operons. The BaMV HLD-coding region was added with PstI and BamHI sites at the 5′ and 3′ ends, respectively, by PCR and inserted into PstI-BamHI-opened pKT25 to become pKT25-HLDBaMV for the production of the T25-HLDBaMV fusion protein. Similarly, the BaMV CP-coding region was added with SphI and BamHI sites at the 5′ and 3′ ends, respectively, and inserted into SphI-BamHI-opened pUT18 to become pUT18-CPBaMV for the production of the CPBaMV-T18 fusion protein. The interaction strength between the BaMV HLD and CP was assayed by growing the recombinant E. coli BTH101 in M63 minimal medium supplemented with 0.2% maltose, 40 μg/ml X-Gal, 50 μg/ml kanamycin, 50 μg/ml streptomycin, 100 μg/ml ampicillin, and 0.5 mM isopropylthiogalactoside (IPTG).
Mutagenesis.
Random mutagenesis of the CP-coding region in pUT18-CPBaMV was performed by error-prone PCR using the GeneMorph II random mutagenesis kit (Stratagene). In brief, 500 ng pUT18-CPBaMV was used as a template in a 50-μl PCR solution for 23 cycles. After sequencing the PCR-amplified DNA fragments, the average mutation frequency was calculated to be 2 mutations per 1 kb of DNA. The amplified CP-coding region was inserted into pUT18 again by using the cut sites of SphI and BamHI to obtain the mutation library of pUT18-CPBaMV. Site-directed mutagenesis was performed with specific primers according to the protocol of the QuikChange site-directed mutagenesis kit (Stratagene).
Preparation of E. coli-expressed proteins.
The BaMV HLD-coding region was added with BamHI and SacI at the 5′ and 3′ ends, respectively, by PCR and inserted into BamHI-SacI-opened pETDuet for the production of the His-tagged BaMV HLD. The viral protein was expressed as inclusion bodies in E. coli BL21(DE3). To purify the viral protein, inclusion bodies were dissolved and refolded according to a protocol described previously (10). In detail, inclusion bodies were dissolved in protein buffer that contained 50 mM Tris (pH 7.5), 20 mM KCl, 0.1% Brij-35, 10% glycerol, and 10 mM β-mercaptoethanol supplemented with 8 M urea. The dissolved protein solution was added drop by drop into 40 volumes of gently stirred protein buffer for protein renaturation. The supernatant of the protein solution was mixed with Ni2+- nitrilotriacetic acid (NTA) resin. After an extensive wash with protein buffer supplemented with 1 M KCl and 20 mM imidazole, the refolded BaMV HLD was eluted with protein buffer supplemented with 500 mM imidazole. Similarly, the PCR-amplified cDNA fragment encoding amino acids 451 to 857 of the FoMV replication protein was inserted into pETDuet by using the cut sites of BamHI and EcoRI, and the plasmid construct was transformed into E. coli BL21(DE3). Preparation of the His-tagged FoMV HLD was like that of BaMV HLD, described above.
To produce the CPBaMV-T18 fusion protein, the culture of the recombinant E. coli BL21 that harbored pUT18-CPBaMV was treated with 0.4 mM IPTG for 16 h at 37°C. The cells were disrupted by sonication, and the protein in the supernatant was collected after a 10-min centrifugation at 15,000 × g. To produce the CPFoMV-T18 fusion protein, the PCR-amplified ORF5 of FoMV was inserted into pUT18 by using the cut sites HindIII and EcoRI, and the plasmid construct pUT18-CPFoMV was transformed into E. coli BL21. Preparation of the fusion protein was as described for CPBaMV-T18.
For the production of BaMV CP in E. coli, BaMV ORF5 was added with NdeI and EcoRI at the 5′ and 3′ ends, respectively, by PCR and inserted into NdeI-EcoRI-opened pET29. The plasmid construct was then transformed into E. coli BL21(DE3). To induce protein expression, 0.5 mM IPTG was added into the culture of the recombinant E. coli, and the cultivation was continued at 30°C for 10 h. The harvested cells were disrupted by sonication in protein extraction buffer that contained 50 mM Tris (pH 8.0), 200 mM NaCl, 10 mM β-mercaptoethanol, and 1 mM EDTA supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). The extract was adjusted to contain 300 mM (NH4)2SO4. After centrifugation, the supernatant was loaded onto a phenyl-Sepharose column (1.6 by 10 cm; GE Healthcare), and the proteins bound to the column were eluted with a linear gradient of (NH4)2SO4 (150 to 0 mM) in protein extraction buffer. The fractions containing BaMV CP were concentrated and dialyzed against NaCl-omitted protein extraction buffer. The protein solution was then loaded onto a DEAE-Sepharose column (1.6 by 10 cm; GE Healthcare), and the elution was performed with a linear gradient of NaCl (0 to 1 M) in protein extraction buffer. Finally, the BaMV CP-containing fractions were concentrated, and the CP within was further purified with a Sephacryl S-300 column (1.6 by 60 cm; GE Healthcare).
Electrophoretic mobility shift assay (EMSA).
Indicated amounts of the E. coli-expressed BaMV CP were mixed with 1 fmol 32P-labeled RNA probe in 12 μl buffer that also contained 20 mM Tris (pH 8.0), 3 mM MgCl2, 10 mM KCl, 2 mM dithiothreitol (DTT), and 4% glycerol. After incubation at room temperature for 15 min, the mixture was separated on a native 6% polyacrylamide gel and visualized by autoradiography. The probe was prepared by incubating 5 μg template DNA, the BaMV 5′-untranslated region (UTR) fragment preceded by the T7 promoter, with 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.5 μm UTP, 100 μCi [α-32P]UTP (6,000 Ci/mmol), 1 μl RNase inhibitor (40 U/μl), and 3 μl T7 polymerase (14 U/μl) in 50 μl 1× T7 transcription buffer. After incubation at 37°C for 2 h, template DNA was removed by RNase E, and the radiolabeled probe was purified with a MicroSpin G-50 column (GE Healthcare).
Circular dichroism spectrum.
The E. coli-expressed BaMV CP was purified and adjusted to a concentration of 25 μg/ml in 2 mM phosphate buffer (pH 7.2). The spectrum within the range of 190 to 250 nm was recorded at room temperature using a Jasco J-815 CD spectrometer fitted with a quartz cell of a 5-mm path length.
Agrobacterium-mediated expression of BaMV replicase complex.
The cDNA of BaMV satellite RNA SF4 was added with HindIII and ClaI recognition sequences at the 5′ and 3′ ends, respectively, by PCR and inserted into the binary plasmid pKn. This construction placed the SF4 cDNA downstream of the Cauliflower mosaic virus (CMV) 35S promoter. Similarly, BaMV ORF5 was inserted into pKn by using the cut sites XhoI and KpnI for the production of viral CP in plants. BaMV ORF1, which had been tagged with a hemagglutinin (HA)-coding sequence at the 3′ end, was inserted into the binary plasmid pEpyon by using the cut sites of SmaI and SacI. pEpyon contains a TMV U1 ω sequence preceded by the CMV 35S promoter so that the translation of BaMV replication protein could be enhanced. Agrobacterium tumefaciens LBA4404 carrying specified binary plasmids were infiltrated into 3-week-old N. benthamiana plants. The leaves were harvested 5 days postinoculation and homogenized in buffer that contained 50 mM Tris (pH 8.0), 15 mM MgCl2, 120 mM KCl, 0.1% β-mercaptoethanol, 20% glycerol, 1 μM pepstain, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). After centrifugation at 30,000 × g for 1 h, the membrane fraction (P30) was collected and solubilized in detergent buffer that contained 50 mM Tris (pH 8.0), 150 mM NaCl, and 1.5% NP-40 for coimmunoprecipitation assay and endogenous RdRp activity assay.
Endogenous RdRp activity assay.
The solubilized P30 fraction that contained specified BaMV proteins and RNA was added into reaction buffer that contained 30 mM Tris (pH 8.8), 10 mM MgCl2, 20 mM DTT, 2 mM ATP, 2 mM CTP, 2 mM GTP, 2 μM UTP, and 100 μCi [α-32P]UTP (6,000 Ci/mmol), and the mixture was incubated at room temperature for 3 h. The reaction product was extracted with an equal volume of phenol-chloroform, and the nucleic acids within were precipitated with ethanol. The radiolabeled RNA products were separated on a 0.8% agarose gel and visualized by autoradiography.
Virus replication assay in protoplasts.
Preparation of N. benthamiana protoplasts and plasmid transfection were performed as described previously (5). In brief, 3 μg pCBG, a green fluorescent protein (GFP) gene-carrying BaMV infectious clone, or its derivative was mixed with 1 × 105 protoplasts in the presence of final 20% polyethylene glycol 4000. Protoplasts were disrupted 2 days posttransfection, and accumulations of the viral CP and genomic RNAs were analyzed by Western blotting and Northern blotting, respectively.
Virus infectivity assay in plants.
A specified infectious clone or its derivative was mechanically inoculated into leaves of 3-week-old N. benthamiana or Chenopodium quinoa at a dosage of 2 μg DNA per leaf, as described previously (21). The expression of GFP, represented by fluorescent spots, in infected plants at the indicated days postinoculation was visualized with Kodak Image Station 2000MM.
Northern blotting.
Total RNA was isolated from 1 × 105 transfected protoplasts using the TriSolution Reagent Plus kit (GeneMark). Each RNA sample (17 μg) was treated with 3 M glyoxal and separated on a 0.8% agarose gel. After transfer and fixation onto a nylon membrane, the samples were hybridized with a radiolabeled probe complementary to the 3′-UTR of BaMV. The probe was produced from an in vitro transcription reaction that contained 5 μg HindIII-cleaved pBaMV-O/SB2.6, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.5 μM UTP, 100 μCi [α-32P]UTP (6,000 Ci/mmol), 40 U of RNase inhibitor, and 3 μl SP6 polymerase (14 U/μl) in 50 μl 1× SP6 transcription buffer. After incubation at 37°C for 30 min, template DNA was removed by RNase E, and the radiolabeled probe was purified with a MicroSpin G-50 column (GE Healthcare).
Transmission electron microscopy.
Full-length BaMV cDNA was amplified from pCBG by PCR and inserted into pKn by using the cut sites SbfI and SacI. A. tumefaciens LBA4404 carrying pKn-based infectious clones was infiltrated into the leaves of 3-week-old N. benthamiana. The leaves were harvested 5 days postinfiltration, and the virus particles within were purified according to the protocol described previously (20). The purified virus particles on a copper grid were negatively stained with 3% uranyl acetate and observed under a transmission electron microscope (JEM-100CX II).
RESULTS
Yeast two-hybrid screening.
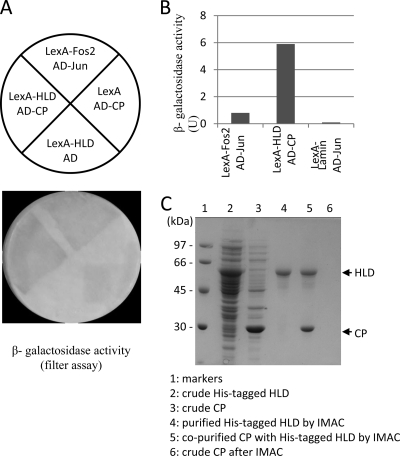
Viruses recruit or hijack host factors to complete their replication cycle and evade host defense mechanisms. To continue searching for plant proteins that interact with the BaMV replication protein, the viral HLD was used as bait to screen against a cDNA library of BaMV-infected leaves of N. benthamiana. Initially, 60 yeast colonies exhibiting the histidine autotrophic phenotype and significant β-galactosidase activity were identified. DNA sequencing found that 48 out of these 60 colonies contained the BaMV CP gene in their prey plasmid (Fig. 1A). The high frequency of finding CP in the prey plasmid suggests a strong interaction between the two viral proteins and/or the abundance of CP mRNA in the viral-infected leaves. Quantitative β-galactosidase activity assay indicated a stronger interaction between the BaMV HLD and CP than that of the positive control (Fig. 1B). To carry out an in vitro pulldown assay, the His-tagged HLD was expressed in E. coli, which formed inclusion bodies, refolded, and purified by immobilized metal affinity chromatography (IMAC). The purified HLD was mixed with the crude E. coli-expressed BaMV CP and Ni2+-NTA resin. BaMV CP could be found with HLD in the elution fraction (Fig. 1C), further confirming the HLD-CP interaction.
Fig. 1.
Interaction between the BaMV HLD and CP. (A) S. cerevisiae L40 expressing the various combinations of bait and prey proteins were grown on YC-WU/Z300 plates and then transferred to a filter paper for β-galactosidase activity assays. The cells expressing LexA-Fos2 and AD-Jun were used as a positive control. (B) Yeast cells expressing the indicated bait and prey proteins were disrupted and incubated with O-nitrophenyl β-galactoside for 10 h. The activity was determined based on the optical density measured at 420 nm. (C) Copurification of BaMV CP with the refolded His-tagged HLD by IMAC. The renaturation and purification of BaMV HLD were described in Materials and Methods. Lanes 2 and 3 are crude extracts of E. coli that expressed BaMV HLD and CP, respectively. Lane 4 shows the refolded HLD purified from lane 2 by IMAC. Lane 5 shows the copurification of HLD and CP by IMAC. It should be noted that BaMV CP alone could not be purified by IMAC (lane 6).
Agroinfiltration-immunoprecipitation assay.
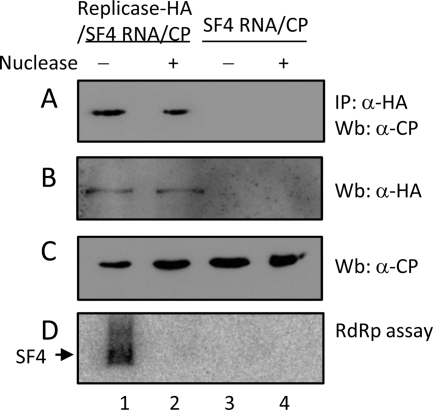
BaMV replicase complex with endogenous RdRp activity in the P30 fraction was prepared by transiently expressing the HA-tagged viral replication protein with satellite RNA SF4 in N. benthamiana leaves via Agrobacterium infiltration. Incubation of the P30 fraction with isotope-labeled nucleoside triphosphates resulted in the formation of the isotope-labeled SF4 (Fig. 2, lane 1). To determine the in vivo interaction between the BaMV replication protein and CP, the two viral proteins were coexpressed in N. benthamiana. Coimmunoprecipitation by using anti-HA antisera showed that CP could be detected in the precipitate if the sample contained the viral replication protein (Fig. 2, lanes 1 and 2). Because both viral proteins have RNA-binding activity, it was unclear whether the two proteins were indirectly associated through RNA molecules. Removal of the endogenous satellite RNA SF4 by micrococcus nuclease did not interrupt the protein interaction (Fig. 2, lane 2), suggesting that BaMV HLD and CP were in direct contact.
Fig. 2.
In vivo interaction between BaMV replication protein and CP. Recombinant strains of A. tumefaciens carrying the indicated protein- and RNA-expressing plasmids were infiltrated into N. benthamiana leaves as described in Materials and Methods. The P30 fraction, isolated from the leaves, was solubilized and treated with or without micrococcus nuclease prior to coimmunoprecipitation using anti-HA antiserum (A). Western blotting indicates the presence of CP in the immune precipitate (A) and BaMV replication protein (B) and CP (C) in the P30 fraction. Endogenous RdRp activity assay (D) was used to ensure that the satellite RNA SF4 in the P30 fraction was removed after treatment with micrococcus nuclease.
Bacterial two-hybrid screening.
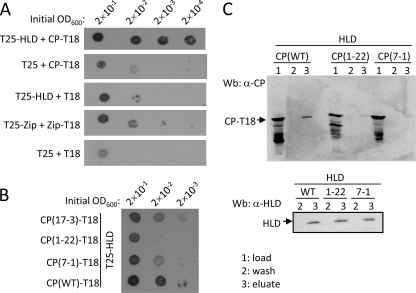
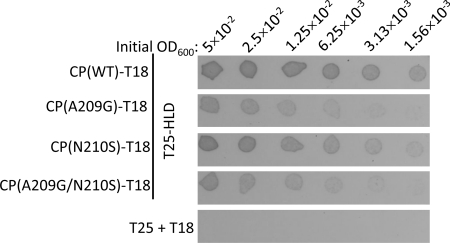
At first, we investigated whether the enzymatic activity of the HLD could be modulated by CP. The presence of CP had no influence on the RNA 5′-triphosphatase/NTPase activity of the HLD (data not shown). To map the regions of CP that contacted the HLD, BaMV ORF5 was subjected to random mutagenesis by error-prone PCR. A bacterial two-hybrid system was exploited for the screening of mutant CPs that had weaker binding affinities to the HLD because of the following reasons: (i) there is no need for transformation back and forth between yeast and E. coli and (ii) the transformation frequency of E. coli is much higher than that of yeast. In this system, adenylate cyclase was split into two parts, T18 and T25; one was fused with bait and the other was fused with prey. The interaction of bait and prey would bring back the enzymatic activity for cAMP production, which in turn would activate the transcription of the lac and mal operons. The usefulness of this screening system in this study was first verified by growing the recombinant E. coli BTH101 strain that produced T25-HLDBaMV and CPBaMV-T18 on M63 selection medium, supplemented with X-Gal and maltose as the sole carbon source. An interaction between the two viral proteins would support the growth of blue colonies on the selection medium plate. The initial cell density minimally required for giving rise to apparent blue colonies of the cells expressing T25-HLDBaMV and CPBaMV-T18 was three orders of magnitude less than that of the cells expressing none or only one of the viral fusion proteins (Fig. 3A). Moreover, the cells expressing the two viral fusion proteins grew much better than those expressing T25-Zip and Zip-T18, used as a positive control, suggesting that the interaction between BaMV HLD and CP was relatively strong in E. coli. The mutant CPs with weaker affinities to HLD were screened from a randomly mutated CP library. Three clones, CP(17-3), CP(1-22), and CP(7-1), were subsequently identified (Fig. 3B). DNA sequencing revealed that CP(1-22) contained four amino acid mutations, which are I130M, D170N, A209G, and N218K, while both CP(17-3) and CP(7-1) had a single N210S mutation. To confirm the differential interaction of the variant CPs to the HLD, E. coli cell extracts containing various CP-T18 fusion proteins were mixed with purified His-tagged HLD and Ni2+-NTA resin, and the bound proteins were subsequently eluted. The amount of wild-type CP copurified with His-tagged HLD was indeed much more abundant than that of the two mutant CPs (Fig. 3C).
Fig. 3.
BaMV CP mutants that had weaker affinities to the viral HLD. (A) A 2-μl aliquot of E. coli BTH101 cell culture at the indicated optical density was added onto an M63 selection agar plate and incubated at 30°C for 5 days. The combinations of bait and prey proteins expressed in the cells were as indicated. The cell culture expressing T25-Zip and Zip-T18 was used as a positive control. (B) The cell cultures expressing T25-HLD and the indicated CP-T18 variants were grown on an M63 selection agar plate as described above. (C) E. coli cell extracts containing the indicated CPBaMV-T18 variants were mixed with purified His-tagged HLD and Ni2+-NTA resin. After being washed with 20 mM imidazole-containing buffer, the proteins were eluted with 500 mM imidazole-containing buffer. CPBaMV-T18 in fractions of load, wash, and eluate and the HLD in wash and eluate were analyzed by Western blotting by using specific antisera.
Biochemical characterization of the mutant CPs.
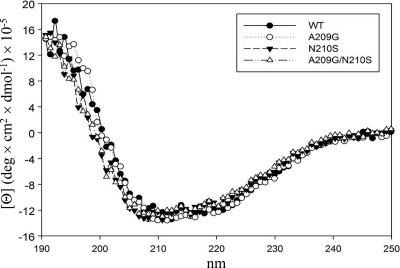
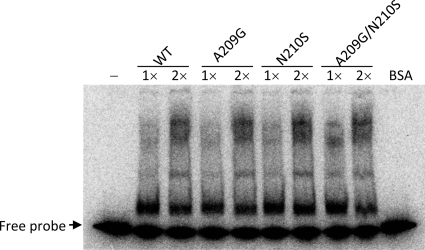
Although there are four mutated residues of CP(1-22), A209 was thought to play a more important role than the others, because it is next to the N210 residue that was changed to serine in CP(17-3) and CP(7-1). This speculation prompted us to propose that the region around A209 and N210 of CP contributes a significant role in the protein interaction. Accordingly, two single mutations, A209G and N210S, and one double mutation, A209G/N210S, were created in a pET29-based CP expression vector. To determine whether the mutations would affect the protein structure, we analyzed the circular dichroism spectra of the various purified CPs (Fig. 4). Nearly identical spectra indicated that the global structure of CP was not altered by the mutations. The prime function of CP is to encapsidate the viral genome. The mutational effects on RNA binding were assayed by EMSA using a probe corresponding to the BaMV 5′-UTR (Fig. 5). The results suggested that the RNA-binding activity of CP was not changed in response to the mutations.
Fig. 4.
Circular dichroism spectra of various BaMV CPs. The protein preparation and CD measurement were as described in Materials and Methods.
Fig. 5.
RNA-binding affinities of various BaMV CPs. The mobility shifts of the RNA probe caused by the CP variants in a native polyacrylamide gel were assayed as described in Materials and Methods. The amount of CP used in the assay was 0.23 μg (1×) or 0.46 μg (2×) per reaction. Bovine serum albumin (BSA; 0.46 μg) was used as a negative control.
Effects of CP mutations on BaMV accumulation in plants.
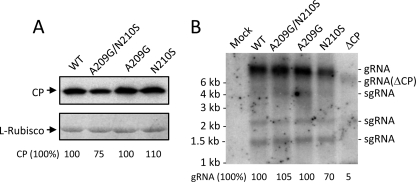
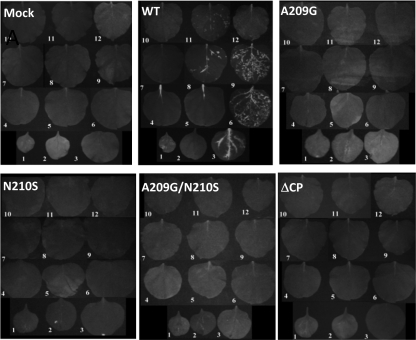
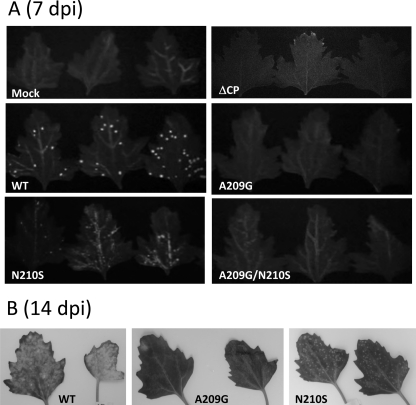
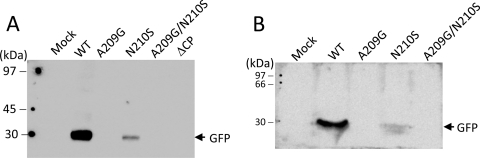
Each of the CP mutations, A209G, N210S, A209G/N210S, and CP deletion (ΔCP), was subcloned into the genetic background of pCBG and transfected into N. benthamiana protoplasts. Accumulations of the viral CP and genomic RNAs were measured 2 days posttransfection. Neither CP nor genomic RNA accumulation was affected by the point mutations (Fig. 6A and B). However, the level of the viral RNAs was significantly reduced when CP was absent (Fig. 6B). The various infectious clones were also used to inoculate N. benthamiana and C. quinoa. In this study, green fluorescence emitted from GFP was a convenient marker for monitoring BaMV replication and movement in plants. In N. benthamiana, inoculation using the wild-type clone resulted in GFP expression in both the inoculated and systemic leaves 20 days postinfection (dpi) (Fig. 7). In contrast, inoculation using any of the mutant clones failed to produce a discernible amount of GFP, even in the inoculated leaves. In C. quinoa, the leaves inoculated with the wild-type clone exhibited clear green fluorescent spots 7 dpi (Fig. 8A). The leaves infected by the N210S mutant seemed to produce few dim fluorescent spots. However, there was no clear sign of fluorescent spots on those leaves inoculated with the A209G, A209G/N210S, or ΔCP mutant. Delayed observation revealed that the N210S mutant indeed caused a mild infection, as distinct lesions appeared in the leaves at the time when the leaves infected by the wild-type clone had already withered (Fig. 8B). GFP expression levels in the inoculated leaves of N. benthamiana and C. quinoa, 20 and 7 dpi, respectively, were measured by Western blotting. An abundance of GFP could be detected in both plant samples that were inoculated by the wild-type clone; however, the GFP expression was rare in the leaves infected by the N210S mutant (Fig. 9). No GFP was detected in leaves inoculated with the A209G or ΔCP mutant. Taken together, the data clearly indicated that the mutations at A209 and N210 of CP did not have a detrimental effect on viral replication within plant cells; however, they did severely delay the virus cell-to-cell movement. The slightly differential effects caused by the A209G and N210S mutations urged us to determine whether the deleterious effects on the virus movement correlated with the strength of the HLD-CP interaction. The relative binding affinities of CP(A209G) and CP(N210S) to the HLD were compared by growing the relevant E. coli BTH101 cells on M63 selection plates with initial cell densities at a 2-fold serial dilution (Fig. 10). The results indicated that CP(N210S) had a slightly stronger binding affinity to HLD than CP(A209G).
Fig. 6.
Effects of CP mutations on BaMV replication in protoplasts. pCBG and the indicated derivatives were transfected into N. benthamiana protoplasts as described in Materials and Methods. (A) BaMV CP accumulation in protoplasts was assayed by Western blotting using anti-BaMV CP antiserum. The amount of Coomassie blue-stained ribulose-1,5-bisphosphate carboxylase (l-RubisCO) was used as the protein loading control. (B) BaMV genomic and subgenomic RNAs accumulated in protoplasts were assayed by Northern blotting by using a probe complementary to the BaMV 3′-UTR. Each lane was loaded with approximately 17 μg total RNA.
Fig. 7.
Effects of CP mutations on BaMV infectivity in N. benthamiana. Plants were inoculated with pCBG or its derivatives as described in Materials and Methods. Fluorescent images of N. benthamiana leaves were taken 20 dpi. Leaves 1 to 3 were the inoculated leaves; others were upper leaves.
Fig. 8.
Effects of CP mutations on BaMV infectivity in C. quinoa. (A) Leaves of C. quinoa were inoculated with pCBG or its derivatives as described in Materials and Methods. Fluorescent images of inoculated leaves of C. quinoa were taken 8 dpi. (B) Effects of the CP mutations on the development of disease symptoms. The photos of the inoculated leaves were taken 14 dpi.
Fig. 9.
GFP expression in inoculated leaves of N. benthamiana (A) and C. quinoa (B). Inoculated leaves in Fig. 7 and Fig. 8A were homogenized in buffer that contained 50 mM Tris (pH 8.0), 2% SDS, 2 mM DTT, and 4% glycerol. The relative amounts of GFP in the samples were analyzed by Western blotting by using anti-GFP antiserum. Each lane was loaded with 1.5 mg total protein.
Fig. 10.
Differential binding of the various BaMV CPs to the HLD. A 2-μl aliquot of E. coli BTH101 expressing the indicated bait and prey proteins was grown on an M63 selection agar plate as described in Materials and Methods. The cells expressing T25 and T18 were used as a negative control.
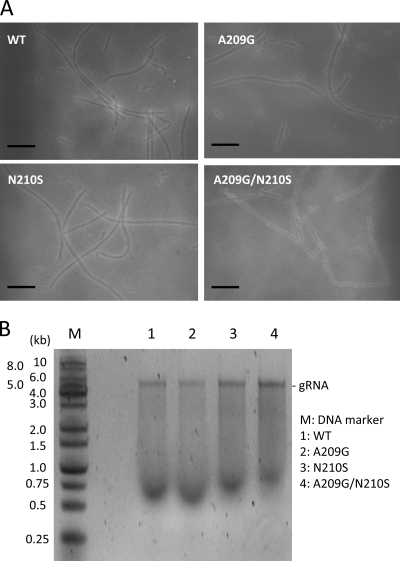
To determine whether the mutant CPs were competent in viral particle formation, we set out to isolate the BaMV virion from the protoplasts transfected with various pCBG clones. The attempt was unsuccessful because the production yield was very low. To overcome this problem, the full-length BaMV transcript was transiently expressed in N. benthamiana through the help of A. tumefaciens. BaMV virions in the agroinfiltrated leaves were isolated and purified. Many flexuous viral particles could be seen under a transmission electron microscope in each preparation, regardless of whether it was prepared from wild-type or mutant virus-infected leaves (Fig. 11A). The purified virions were disrupted, and the RNA molecules inside were analyzed by agarose electrophoresis (Fig. 11B). Apparently, the viral genomic RNA could be encapsidated by the wild-type as well as the mutant CPs.
Fig. 11.
BaMV virions formed by the various mutants of BaMV CP. (A) Virions were prepared and observed under a transmission electron microscope as described in Materials and Methods. All scale bars indicate 100 nm. (B) BaMV virions were heated in buffer containing 6 mM phosphate (pH 7.2), 0.2 mM EDTA, 1% β-mercaptoethanol, 1% bentonite, and 1% SDS at 60°C for 5 min, followed by phenol extraction. The purified RNA molecules were analyzed by 0.8% agarose electrophoresis.
Effects of CP mutations on FoMV accumulation in plants.
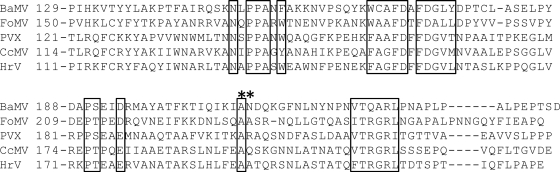
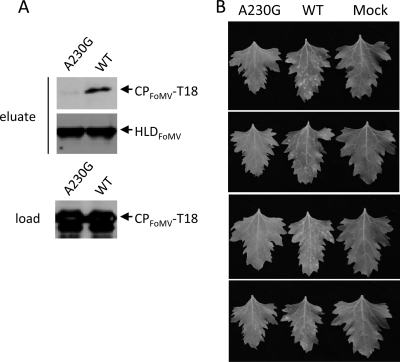
It has been known that CP is required for the movement of potexviruses. According to the aforementioned data, the role of BaMV CP in the virus movement is dictated by its binding ability to the HLD. Thus, it was important to know whether this phenomenon is widely conserved among potexviruses. To answer this question, the amino acid sequences of several potexvirus CPs were aligned, and the results indicated that the residue corresponding to BaMV A209 was conserved (Fig. 12). Accordingly, A230 of FoMV CP was subjected to mutagenesis. The HLD of the FoMV replication protein was expressed with a His tag at its N terminus in E. coli. Similarly to the BaMV HLD, the protein formed inclusion bodies; therefore, it was denatured, refolded, and purified by the protocol that had been used for the preparation of the BaMV HLD. The NTPase activity of the refolded protein (data not shown) suggested that it had a correct conformation. FoMV CP was also expressed in E. coli as a fusion protein (CPFoMV-T18). The interaction between the FoMV HLD and CPFoMV-T18 was verified by copurification of the two proteins by IMAC (Fig. 13A). A significant amount of CPFoMV-T18 could be found in the eluate with the FoMV HLD. However, when the A230G mutation was introduced into CP, the yields of the CP fusion proteins in the eluate drastically decreased. This result suggested that A230 is involved in the HLD-CP interaction. The same mutation was created in the FoMV infectious clone pCF, and the derivative was used to inoculate C. quinoa. Lesions were observed on the leaves inoculated with the wild-type pCF 8 dpi; in contrast, no apparent lesions appeared on the leaves receiving the clone carrying the A230G mutation (Fig. 13B).
Fig. 12.
Alignment of partial amino acid sequences of CP among several members of the genus Potexvirus. The GenBank accession numbers of FoMV, PVX, Cassava common mosaic virus (CcMV), and Hydrangea ringspot virus (HrV) are ABW25052, CAA61265, NP_042699, and YP_224088, respectively. Consensus motifs and residues are boxed. An asterisk denotes the residues that were mutated in this study.
Fig. 13.
A230G mutation of FoMV CP that decreased the CP-HLD interaction and reduced the viral infectivity in C. quinoa. (A) The purified His-tagged FoMV HLD was mixed with the E. coli extract that contained the indicated CPFoMV-T18 fusion proteins and Ni2+-NTA resin. The proteins bound to the resin were purified by IMAC. CPFoMV-T18 in fractions of load and eluate and the HLD in the eluate were analyzed by Western blotting using specific antisera. (B) Lesion developments on the leaves of C. quinoa that were inoculated with the wild type or A230G mutant of FoMV. The photos of the inoculated leaves were taken 8 dpi.
DISCUSSION
CP of the genus Potexvirus is a multifunctional protein, playing a role in virion assembly (15), cell-to-cell movement (7), and regulation of viral translation (2). In addition, CP has been reported to regulate RNA replication in other plant viruses. For example, RNA replication of Alfalfa mosaic virus (AMV) was strongly stimulated at low CP concentrations but progressively decreased at higher concentrations (9). In the protoplast assay, we found that CP deletion in BaMV significantly decreased genomic RNA accumulation. It is not currently known how CP affects RNA accumulation. Perhaps it has a role similar to that in AMV CP in the activation of RNA replication. Alternatively, the presence of CP might protect RNA from RNase degradation, leading to the increase in RNA accumulation.
Understanding the mechanism underlying the CP requirement for potexvirus movement is an important issue. This study determined that BaMV CP interacts with the viral HLD. The A209G and N210S mutations on the CP reduced the affinity of the interaction and restricted the virus from cell-to-cell movement; however, the CP's function in RNA binding and genome encapsidation remained intact. This result is consistent with a previous notion that the C-terminal region of PVX CP plays a role in virus movement independent of encapsidation (6). The importance of the HLD-CP interaction in BaMV movement suggests that the BaMV replication protein is included in the infectious entity that moves to and through plasmodesmata. It is very likely that the infectious entity is an RNP complex comprising TGBp1, CP, RNA, and the replication protein. Involvement of the TMV replication protein in the viral movement complex has been proposed based on the remarkable difference between the time required for TMV to spread from the primary inoculated cells to secondary cells (i.e., 18 to 20 h postinoculation) and that required for spread from secondary to tertiary cells (2 to 4 h) (13). Such an arrangement enables TMV to replicate immediately once it is deposited in the adjacent cell. With the same argument, recruitment of the BaMV replication protein in the RNP complex would allow the virus to reinitiate replication rapidly in the newly invaded cells. Interfering with the CP-HLD interaction of FoMV also deprived the virus of the ability to move, implying that recruitment of the replication protein in the virus movement complex may be a general feature of potexviruses.
The HLD of the BaMV replication protein preferentially binds the cloverleaf-like structure in the 3′-UTR BaMV RNA (4). If inclusion of the viral replication protein in the movement complex is necessary for efficient dissemination of BaMV in plant, why is the interaction between HLD and the viral RNA not sufficient to do the job? The expression amount of CP is overwhelmingly larger than that of the replication protein at the time when the virus moves. Therefore, we speculate that the abundance of CP will interfere with the interaction between replication protein and RNA. To overcome this situation, the virus evokes to carry its replication protein in the movement complex through the binding of CP to the HLD. Once the complex is deposited in the adjacent cells, CP is removed from the viral RNA by a yet-unknown mechanism, and the replication protein can immediately start to replicate the viral RNA.
ACKNOWLEDGMENTS
We thank C.-H. Yang at National Chung Hsing University for providing pEpyon.
This work was supported by grant NSC 98-2321-B-005-004-MY3 from the National Science Council, Taiwan, Republic of China.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Angell S. M., Davies C., Baulcombe D. C. 1996. Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichrome cells of Nicotiana clevelandii. Virology 216: 197–201 [DOI] [PubMed] [Google Scholar]
- 2. Atabekov J. G., et al. 2001. Translational activation of encapsidated Potato virus X RNA by coat protein phosphorylation. Virology 286: 466–474 [DOI] [PubMed] [Google Scholar]
- 3. Beck D. L., Guilford P. J., Voot D. M., Andersen M. T., Forster R. L. S. 1991. Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology 183: 695–702 [DOI] [PubMed] [Google Scholar]
- 4. Chen I.-H., Meng M., Hsu Y.-H., Tsai C.-H. 2003. Functional analysis of the cloverleaf-like structure in the 3′ untranslated region of bamboo mosaic potexvirus RNA revealed dual roles in viral RNA replication and long distance movement. Virology 315: 415–424 [DOI] [PubMed] [Google Scholar]
- 5. Cheng C.-W., et al. 2009. Suppression of Bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana. J. Virol. 83: 5796–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fedorkin O. N., et al. 2001. Cell-to-cell movement of Potato virus X involves distinct functions of the coat protein. J. Gen. Virol. 82: 449–458 [DOI] [PubMed] [Google Scholar]
- 7. Forster R. L. S., et al. 1992. The coat protein of white clover mosaic potexvirus has a role in facilitating cell-to-cell transport in plants. Virology 191: 480–484 [DOI] [PubMed] [Google Scholar]
- 8. Gilbertson R. L., Lucas W. J. 1996. How do viruses traffic on the ‘vascular highway’? Trends Plant Sci. 1: 260–268 [Google Scholar]
- 9. Guogas L. M., Laforest S. M., Gehrke L. 2005. Coat protein activation of Alfalfa mosaic virus replication is concentration dependent. J. Virol. 79: 5752–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han Y.-T., Hsu Y.-H., Lo C.-W., Meng M. 2009. Identification and functional characterization of regions that can be crosslinked to RNA in the helicase-like domain of BaMV replicase. Virology 389: 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howard A. R., et al. 2004. Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328: 185–197 [DOI] [PubMed] [Google Scholar]
- 12. Kalinina N. O., Rakitina D. V., Solovyev A. G., Schiemann J., Morozov S. Y. 2002. RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296: 321–329 [DOI] [PubMed] [Google Scholar]
- 13. Kawakami S., Watanabe Y., Beachy R. N. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. U. S. A. 101: 6291–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krishnamurthy K., et al. 2003. The Potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology 309: 135–151 [DOI] [PubMed] [Google Scholar]
- 15. Kwon S. J., et al. 2005. cis-Acting sequences required for coat protein binding and in vitro assembly of Potato virus X. Virology 334: 83–97 [DOI] [PubMed] [Google Scholar]
- 16. Li Y.-I., et al. 1998. Identification and characterization of the E. coli-expressed RNA-dependent RNA polymerase of bamboo mosaic virus. J. Virol. 72: 10093–10099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y.-I., et al. 2001. The helicase-like domain of plant potexvirus replicase participates in the formation of 5′ cap structure of RNA by exhibiting an RNA 5′-triphosphatase activity. J. Virol. 75: 12114–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y.-I., Chen Y.-J., Hsu Y.-H., Meng M. 2001. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of bamboo mosaic virus replicase. J. Virol. 75: 782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin M.-K., Chang B.-Y., Liao J.-T., Lin N.-S., Hsu Y.-H. 2004. Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 85: 251–259 [DOI] [PubMed] [Google Scholar]
- 20. Lin N.-S., Chen C.-C. 1991. Association of bamboo mosaic virus (BoMV) and BoMV-specific electron-dense crystalline bodies with chloroplasts. Phytopathology 81: 1551–1555 [Google Scholar]
- 21. Lin N.-S., Hsu Y.-H. 1994. A satellite RNA associated with Bamboo mosaic potexvirus. Virology 202: 707–714 [DOI] [PubMed] [Google Scholar]
- 22. Lough T.-J., et al. 2000. Cell-to-cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant Microbe Interact. 13: 962–974 [DOI] [PubMed] [Google Scholar]
- 23. Lucas W. J. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- 24. Mitra R., et al. 2003. The Potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement. Virology 312: 35–48 [DOI] [PubMed] [Google Scholar]
- 25. Morozov S. Y., Solovyev A. G. 2003. Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84: 1351–1366 [DOI] [PubMed] [Google Scholar]
- 26. Oparka K. J. 2004. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 9: 33–41 [DOI] [PubMed] [Google Scholar]
- 27. Santa Cruz S., Roberts A. G., Prior D. A. M., Chapman S., Oparka K. J. 1998. Cell-to-cell and phloem-mediated transport of Potato virus X: the role of virions. Plant Cell 10: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scholthof H. B. 2005. Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10: 376–382 [DOI] [PubMed] [Google Scholar]
- 29. Tseng Y.-H., et al. 2009. The two conserved cysteine residues of the triple gene block protein 2 are critical for both cell-to-cell and systemic movement of Bamboo mosaic virus. Mol. Plant Microbe Interact. 22: 1379–1388 [DOI] [PubMed] [Google Scholar]
- 30. Verchot-Lubicz J. 2005. A new cell-to-cell transport model for potexviruses. Mol. Plant Microbe Interact. 18: 283–290 [DOI] [PubMed] [Google Scholar]
- 31. Voinnet O., Lederer C., Baulcombe D. C. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 10: 157–167 [DOI] [PubMed] [Google Scholar]
- 32. Wu C.-H., Lee S.-C., Wang C.-W. 2011. Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 193: 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]