Abstract
The influenza virus nucleoprotein (NP) is believed to play a central role in directing a switch from RNA genome transcription to replication by the viral RNA polymerase. However, this role has recently been disputed with the proposal of alternative regulatory mechanisms. It has been suggested that the expression of viral polymerase and NP allows genome replication by stabilization of cRNA replication intermediates and complementary ribonucleoprotein (cRNP) assembly. Here, we demonstrate that the RNA-binding activity of NP is necessary for stabilization of cRNA, whereas, surprisingly, homo-oligomerization of NP is not essential. However, both RNA binding and homo-oligomerization activities are essential for genome replication.
TEXT
Influenza virus, a member of the Orthomyxoviridae family, contains a segmented, negative-sense, single-stranded RNA (ssRNA) genome. Each viral RNA (vRNA) segment is bound by the viral RNA-dependent RNA polymerase (RdRp) and nucleoprotein (NP) to form viral ribonucleoprotein (vRNP) complexes. Within the vRNP, the RdRp binds to the corkscrew structure formed by the partially complementary conserved 5′ and 3′ ends of the vRNA representing the vRNA promoter. The rest of the vRNA interacts with multiple copies of NP, each molecule covering approximately 24 nucleotides (reviewed in reference 17). NP binds ssRNA with high affinity but little or no sequence specificity (2, 9, 22) and has been shown to homo-oligomerize and interact with the PB1 and PB2 subunits of the viral RdRp (4, 8, 14, 15, 23). Although cryo-electron microscopy reconstitution images of a mini-RNP containing nine NP molecules have been obtained (6), a detailed high-resolution structure of the vRNP is still lacking.
During the viral life cycle, the vRNA is transcribed into mRNA and replicated through a cRNA intermediate into more copies of vRNA by the viral RdRp (reviewed in references 10, 17, and 18). Viral mRNAs associate with cellular factors normally associating with host mRNAs, e.g., nuclear and cytoplasmic cap-binding proteins, leading to the stabilization of viral mRNAs and their processing, nuclear export, and translation (3, 16, 21). In contrast, cRNAs are stabilized by the association of viral factors, i.e., the viral RdRp and NP, which together form a cRNP structure similar to that of vRNPs. During infection, viral mRNAs can be detected initially, while cRNAs become detectable only after the onset of viral protein synthesis (12). This has been interpreted to mean that a switch of polymerase function from a transcriptase to a replicase is required for cRNA synthesis, and various models have been proposed that implicate viral and host factors, as well as small viral RNAs (svRNAs), in the switching (reviewed in reference 17). However, an alternative interpretation is that both mRNA and cRNA are synthesized from early on in a stochastic manner but cRNA is degraded by host nucleases until sufficient amounts of viral RdRp and NP accumulate to stabilize it (20). In agreement with this interpretation, the overexpression of catalytically inactive RdRp and NP prior to viral infection allows the accumulation of cRNA in the presence of cycloheximide, an inhibitor of protein translation (19, 20). The expression of RdRp is absolutely essential for the stabilization of cRNA, while NP, although not essential, greatly increases the amounts of accumulated cRNA. In this study, we address the question of which properties of NP play a role in the stabilization of cRNA and the replication of cRNA to vRNA.
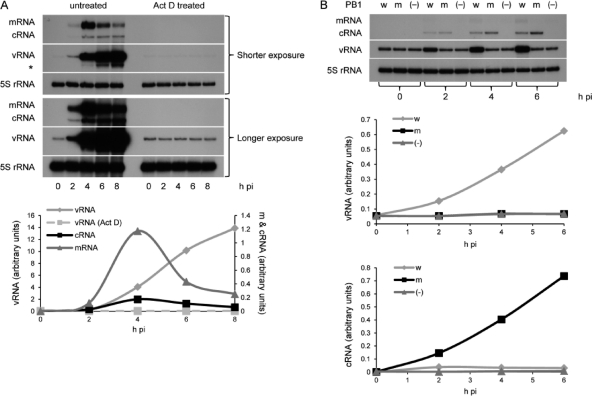
In order to address these questions, we set up an assay to analyze the effect of mutations in NP on the accumulation of cRNA and its replication in viral infection. 293T cells were infected with influenza A/WSN/33 virus in the presence of actinomycin D, an inhibitor of DNA-templated RNA synthesis. As the viral RdRp requires host pre-mRNAs as a source of 5′ capped primers for the initiation of viral mRNA synthesis, treatment with actinomycin D resulted in the inhibition of the accumulation of all three types of viral RNAs (Fig. 1A), as expected (1, 13). Only vRNAs introduced into the cell by the infecting virions could be detected. However, preexpression of NP, together with a catalytically inactive viral RdRp (containing D445A/D446A mutant PB1) (20), resulted in the linear accumulation of cRNA (Fig. 1B). These results are in agreement with previous findings that actinomycin D does not affect viral RNA replication directly and that the accumulation of viral replication products is dependent on viral protein expression (1, 12, 13). Preexpression of NP and wild-type catalytically active viral RdRp resulted not only in cRNA accumulation but also in genomic replication (linear accumulation of vRNA) (Fig. 1B). No cRNA and no replication into vRNA were detected if NP was preexpressed without trimeric RdRp (Fig. 1B). Therefore, the presence of NP alone is not sufficient for cRNA stabilization. These results are in agreement with the previously proposed stabilization model for replication (20). Intriguingly, in the presence of preexpressed inactive RdRp, cRNA continues to accumulate instead of reaching a plateau as seen in the presence of preexpressed wild-type RdRp (Fig. 1B) or during viral infection (Fig. 1A). We speculate that this may relate to instability of functional cRNPs (7), possibly by the cRNA template being degraded following replication, whereas nonfunctional cRNPs are presumably more stable and form a sink for accumulating cRNA.
Fig. 1.
Actinomycin D-mediated inhibition of influenza A/WSN/33 virus RNA replication can be rescued by the expression of PB1, PB2, PA, and NP. (A) Time course of viral infection (multiplicity of infection of 5) in the absence (untreated) or presence (Act D treated) of 5 μg of actinomycin D/ml. The experiment shown in the lower panel is a longer exposure of the experiment whose results are shown in the upper panel, showing the presence of vRNA introduced by the infecting virions in actinomycin D-treated cells. (B) Time course of viral infection (multiplicity of infection of 5) in the presence of 5 μg of actinomycin D/ml after prior (15 to 16 h) transfection of plasmids (11) expressing viral PB2, PA, and NP and wild-type PB1 (w), D445A/D446A mutant PB1 (m), or no PB1 (−). Viral RNA species were analyzed by NA gene-specific primer extension assays (20). Weak mRNA-specific signals detected are due to preexpressed wild-type RdRp being able to carry out residual mRNA synthesis in the presence of actinomycin D. A primer specific for 5S rRNA (TCCCAGGCGGTCTCCCATCC) was used as an internal control. An asterisk indicates an unidentified band possibly related to vRNA. Quantitation was by phosphorimage analysis using Aida software. It should be noted that the intensities of bands between gels from different experiments are not comparable due to variations in labeling intensities of the primers used. pi, postinfection.
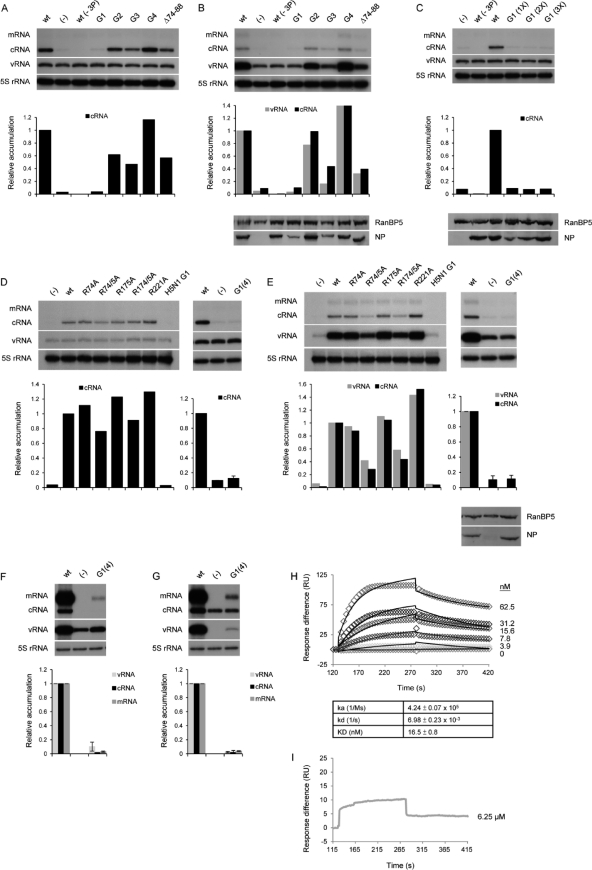
Having established an assay to evaluate the functional role of NP in the context of viral infection, we next examined the effect of mutations in the preexpressed NP on the accumulation and replication of cRNA. The high-resolution structures of H1N1 and H5N1 NP revealed potential RNA-binding sites involving a large number of basic amino acid residues (14, 23). We initially analyzed the effect of previously characterized RNA-binding mutants of H5N1 NP (14) on the accumulation and replication of cRNA in the presence of catalytically inactive mutant or wild-type H1N1 RdRp (Fig. 2A and B, respectively). Preexpression of viral RdRp and the NP-G1 mutant (including amino acid changes R74A, R75A, R174A, R175A, and R221A) yielded low but detectable levels of cRNA similar to the levels detected in the absence of NP (Fig. 2A and B); no replication of cRNA to vRNA by the wild-type RdRp could be detected (Fig. 2B). The other mutant NPs examined (NP-G2, NP-G3, NP-G4, and NP-Δ74-88) yielded detectable levels of cRNA accumulation and replication (Fig. 2A and B), with efficiencies broadly corresponding to the reported RNA binding affinities of the respective NP mutants (14). The exception was NP-G3, which inhibited cRNA accumulation and replication compared to the cRNA accumulation and replication with wild-type NP despite having only a negligible effect on RNA binding. Preliminary data suggest that this is not due to the lower-than-wild-type expression levels of NP-G3 (Fig. 2B and results not shown), but this is currently under further investigation. Although the expression of NP-G1 was also reduced compared to that of the wild type, increasing the expression to wild-type levels did not yield increased levels of detectable cRNA (Fig. 2C).
Fig. 2.
Effects of RNA-binding mutations in NP on cRNA stabilization and replication. (A to E) Cells were infected in the presence of 5 μg of actinomycin D/ml after prior (15 to 16 h) transfection of plasmids (11) expressing wild-type or mutant viral proteins as indicated. (A and B) Wild-type (wt), none (−), or mutant H5N1 NPs (14) were coexpressed with or without (−3P) catalytically inactive (A) or wild-type (B) RNA polymerase. (C) Wild-type (wt), none (−), or increasing amounts of G1 mutant H5N1 NP were coexpressed with or without (−3P) catalytically inactive RNA polymerase. (D and E) Wild-type, none (−), or mutant H1N1 NPs were coexpressed with catalytically inactive (D) or wild-type (E) RNA polymerase. H5N1 NP-G1 (14) was included as a control. Weak mRNA-specific signals detected (B and E) are due to preexpressed wild-type RdRp being able to carry out residual mRNA synthesis in the presence of actinomycin D. (F and G) Ribonucleoprotein reconstitution assays were performed in 293T cells transfected with expression plasmids for the RNA polymerase, wild-type (wt), none (−), or G1(4) mutant H1N1 NP, and an NA gene-specific vRNA (F) or cRNA (G) template. Viral RNA species were analyzed by NA gene-specific primer extension assays (20), and the accumulation relative to the accumulation with wild-type NP was quantitated by phosphorimage analysis using Aida software. The vRNA (F) and cRNA (G) values obtained in the absence of NP (−), corresponding to input RNA generated by RNA polymerase I transcription, were subtracted from the values obtained in the presence of wild-type (wt) or G1(4) mutant NP. The quantitations shown below the panels with NP-G1(4) represent the average and range of two independent experiments. A primer specific for 5S rRNA was used as an internal control. The expression of NP was analyzed by Western blotting using a rabbit polyclonal antibody (a gift of O. Haller, University of Freiburg). RanBP5 detected with a rabbit polyclonal antibody (Santa Cruz) was used as a loading control. (H and I) Surface plasmon resonance results for wild-type (H) or G1(4) mutant (I) H1N1 NP against immobilized RNA suitable for binding one NP molecule (24 nucleotides in length) (14). Different concentrations of purified proteins were injected as shown, and the signal (resonance units [RU]) was plotted over time. Diamonds (H) and the gray line (I) are experimental data, while black lines (H) are fitted curves. ka, association rate; kd, dissociation rate; KD, dissociation constant.
Next we aimed to identify the critical residues in NP-G1 responsible for the inhibitory phenotype while simultaneously confirming their inhibitory effect in an H1N1 NP. We therefore introduced the mutations from H5N1 NP-G1 singly (R74A, R175A, and R221A) or doubly (R74A/R75A and R174A/R175A) into an H1N1 NP and analyzed them for cRNA stabilization and replication. Single mutations showed little effect, and double mutations only partially inhibited the accumulation and replication of cRNA compared to the accumulation and replication of cRNA with the wild type (Fig. 2D and E). Therefore, the two double mutations (R74A/R75A and R174A/R175A) were combined to generate a quadruple mutant, NP-G1(4). When NP-G1(4) was coexpressed with inactive or wild-type RdRp, the levels of cRNA accumulation and replication were similar to the levels detected in the absence of NP (Fig. 2D and E). These four mutations were found to reduce transcription and replication activity to less than 10% in RNP reconstitution experiments (Fig. 2F and G). Finally, the RNA-binding properties of wild-type NP and NP-G1(4) proteins expressed in Escherichia coli and purified to homogeneity were studied by surface plasmon resonance (14). Wild-type NP showed a high RNA-binding affinity, with a dissociation constant (KD) of 16.5 nM (Fig. 2H), which is similar to that of H5N1 NP (14). The RNA-binding affinity of NP-G1(4), however, was too low to be determined. Only a weak response was observed even when a high concentration (6.25 μM) of purified NP-G1(4) protein was applied to the immobilized RNA on the chip surface (Fig. 2I). Therefore, these data are in agreement with the results obtained with H5N1 NP-G1 described above and underline the importance of the RNA binding activity of NP in cRNA expression and replication. Moreover, they support a model of RNA binding involving a number of positively charged residues in the proposed G1 region of NP (14). Overall, these results show that RdRp and NP competent in RNA binding are crucial for the accumulation and replication of cRNA.
Next, we addressed the role of oligomerization of NP in cRNA stabilization and replication. Homo-oligomerization of NP, in which the tail loop of one NP is inserted into a groove of the neighboring NP molecule, is an important factor in maintaining RNP structure. The amino acid residues involved in these interactions have been identified and well characterized (6, 8, 23), including in our laboratory (5, 14). The expected inhibitory effects of replacing amino acid residues involved in the intra- and interchain interactions of the tail loop (E339A, V408S/P410S, R416A, and L418S/P419S) on viral replication and transcription (5) were confirmed by vRNP (Fig. 3A) and cRNP (Fig. 3B) reconstitution experiments. To determine whether homo-oligomerization is important for the accumulation of cRNA, NP mutants were preexpressed with viral RdRp in actinomycin D-treated cells infected with influenza virus (Fig. 3C and D). Surprisingly, substantial (50 to 80%) accumulation of cRNA could be detected in the presence of catalytically inactive viral RdRp and mutant NPs compared to the accumulation with wild-type NP (Fig. 3C). However, the replication of cRNA in the presence of wild-type viral RdRp and mutant NPs was severely inhibited (Fig. 3D). The reduced relative accumulation of cRNA (25 to 45%) in the presence of wild-type RdRp and mutant NPs compared to the accumulation with wild-type NP (Fig. 3D) is presumably due to the inability of the mutant NP to fully support replicative synthesis of vRNA and cRNA. As oligomerization NP mutants have previously been found to possess reduced RNA binding affinity compared to that of the wild type in vitro (5, 9), probably due to their reduced oligomeric state, we analyzed the association of oligomerization NP mutants with cRNA. Consistent with the model that cRNA stabilization requires NP to bind RNA, the cRNA detected in the presence of inactive RdRp (Fig. 3C) was shown to be bound by the wild-type or mutant NP through primer extension analyses of RNA coimmunoprecipitated with NP (Fig. 3E). Overall, we conclude that homo-oligomerization of NP, although essential for replication, is not essential for stabilization of cRNA.
Fig. 3.
Effects of homo-oligomerization mutations in NP on cRNA stabilization and replication. (A and B) Ribonucleoprotein reconstitution assays were performed in 293T cells transfected with expression plasmids for the RNA polymerase, wild-type (wt), none (−), or mutant H1N1 NP, and an NA gene-specific vRNA (A) or cRNA (B) template. Viral RNA species were analyzed by NA gene-specific primer extension assays (20), and the accumulation relative to the accumulation with wild-type NP was quantitated by phosphorimage analysis using Aida software. The vRNA (A) and cRNA (B) values obtained in the absence of NP (−), corresponding to input RNA generated by RNA polymerase I transcription, were subtracted from the values obtained in the presence of wild-type (wt) or mutant NPs. The results shown represent the average and range of two independent experiments. (C and D) Cells were infected in the presence of 5 μg of actinomycin D/ml after prior (15 to 16 h) transfection of plasmids (11) expressing wild-type (wt), none (−), or mutant NPs together with catalytically inactive (C) or wild-type (D) RNA polymerase. Viral RNA species were analyzed and quantitated as described above. Weak mRNA-specific signals detected (D) are due to preexpressed wild-type RdRp being able to carry out residual mRNA synthesis in the presence of actinomycin D. A primer specific for 5S rRNA was used as an internal control. Expression of NP was analyzed by Western blotting. RanBP5 was used as a loading control. (E) Immunoprecipitations of cell lysates from the experiment whose results are shown in panel C were performed with or without antibodies specific for NP. RNA was isolated from cell lysates (left) or immunoprecipitates (right) and analyzed by NA gene-specific primer extension. Expression and immunoprecipitation of NP was analyzed by Western blotting.
It is not currently known what regulates the relative rates and levels of vRNA and cRNA accumulation during viral infection (reviewed in reference 17). Whereas vRNA continues to accumulate at late time points and to high levels, cRNA accumulation flattens out at low levels early during replication (Fig. 1A). As discussed earlier, our data (Fig. 1B) suggest that cRNA may achieve equilibrium between vRNA-templated synthesis and degradation of the cRNA template following replication to vRNA. We speculate that in RNPs assembled with wild-type polymerase and NP oligomerization mutants (Fig. 3D), this state of equilibrium would be disrupted due to both the reduction in cRNA stabilization compared to the stabilization with wild-type NP (Fig. 3C) and the replication-deficient phenotype (Fig. 3A and B). According to this model, both the rate of cRNA degradation (following cRNA-templated replication to vRNA) and rate of vRNA-templated synthesis of cRNA would be reduced compared to the cRNA degradation and synthesis during replication of wild-type RNPs.
In summary, our results show that the viral RdRp is essential, while NP plays an important role in the stabilization of cRNA replication intermediates. The RNA-binding activity of NP was found to be essential for stabilizing cRNA; surprisingly, however, the oligomerization activity was not critical. This raises the question of how NP enhances cRNA stability. We envisage that single NP molecules bound to cRNA and the RdRp near the cRNA termini could stabilize the association of the RdRp with the promoter sequences comprising the 5′ and 3′ ends of cRNA. This might be sufficient for enhancing cRNP stability. Additionally or alternatively, nonoligomeric NP molecules may be bound along the length of the cRNA, contributing to cRNP stability. An NP mutant unable to bind RdRp would help to distinguish between these possibilities. In contrast to the stabilization of cRNA, both the RNA-binding and homo-oligomerization activities of NP were found to be essential for the assembly of replication-competent cRNPs.
Acknowledgments
We thank Otto Haller for antibodies and Koyu Hara for the 5S rRNA-specific primer.
This work was supported by a grant from the MRC (grant G0700848) to E. Fodor and a Hong Kong Research Grants Council General Research Fund grant (CUHK 472808) to P.-C. Shaw.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Barrett T., Wolstenholme A. J., Mahy B. W. 1979. Transcription and replication of influenza virus RNA. Virology 98: 211–225 [DOI] [PubMed] [Google Scholar]
- 2. Baudin F., Bach C., Cusack S., Ruigrok R. W. 1994. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 13: 3158–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bier K., York A., Fodor E. 2011. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 92(Pt. 7): 1627–1634 [DOI] [PubMed] [Google Scholar]
- 4. Biswas S. K., Boutz P. L., Nayak D. P. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72: 5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan W. H., et al. 2010. Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities. J. Virol. 84: 7337–7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coloma R., et al. 2009. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 5: e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalton R. M., et al. 2006. Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes. Virol. J. 3: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elton D., Medcalf E., Bishop K., Digard P. 1999. Oligomerization of the influenza virus nucleoprotein: identification of positive and negative sequence elements. Virology 260: 190–200 [DOI] [PubMed] [Google Scholar]
- 9. Elton D., Medcalf L., Bishop K., Harrison D., Digard P. 1999. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 73: 7357–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engelhardt O. G., Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16: 329–345 [DOI] [PubMed] [Google Scholar]
- 11. Fodor E., et al. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76: 8989–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hay A. J., Lomniczi B., Bellamy A. R., Skehel J. J. 1977. Transcription of the influenza virus genome. Virology 83: 337–355 [DOI] [PubMed] [Google Scholar]
- 13. Mark G. E., Taylor J. M., Broni B., Krug R. M. 1979. Nuclear accumulation of influenza viral RNA transcripts and the effects of cycloheximide, actinomycin D, and alpha-amanitin. J. Virol. 29: 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng A. K., et al. 2008. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 22: 3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poole E., Elton D., Medcalf L., Digard P. 2004. Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology 321: 120–133 [DOI] [PubMed] [Google Scholar]
- 16. Read E. K., Digard P. 2010. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J. Gen. Virol. 91: 1290–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Resa-Infante P., Jorba N., Coloma R., Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruigrok R. W., Crepin T., Hart D. J., Cusack S. 2010. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 20: 104–113 [DOI] [PubMed] [Google Scholar]
- 19. Vreede F. T., Brownlee G. G. 2007. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 81: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vreede F. T., Jung T. E., Brownlee G. G. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78: 9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang W., et al. 2008. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic Acids Res. 36: 4913–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamanaka K., Ishihama A., Nagata K. 1990. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J. Biol. Chem. 265: 11151–11155 [PubMed] [Google Scholar]
- 23. Ye Q., Krug R. M., Tao Y. J. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444: 1078–1082 [DOI] [PubMed] [Google Scholar]