Abstract
Genetic factors, as well as antigenic stimuli, can influence antibody repertoire formation. Moreover, the affinity of antigen for unmutated naïve B cell receptors determines the threshold for activation of germinal center antibody responses. The gp41 2F5 broadly neutralizing antibody (bNAb) uses the VH2-5 gene, which has 10 distinct alleles that use either a heavy-chain complementarity-determining region 2 (HCDR2) aspartic acid (DH54) or an HCDR2 asparagine (NH54) residue. The 2F5 HCDR2 DH54 residue has been shown to form a salt bridge with gp41 665K; the VH2-5 germ line allele variant containing NH54 cannot do so and thus should bind less avidly to gp41. Thus, the induction of 2F5 bNAb is dependent on both genetic and structural factors that could affect antigen affinity of unmutated naïve B cell receptors. Here, we studied allelic variants of the VH2-5 inferred germ line forms of the HIV-1 gp41 bNAb 2F5 for their antigen binding affinities to gp41 linear peptide and conformational protein antigens. Both VH2-5 2F5 inferred germ line variants bound to gp41 peptides and protein, including the fusion intermediate protein mimic, although more weakly than the mature 2F5 antibody. As predicted, the affinity of the NH54 variant for fusion-intermediate conformation was an order of magnitude lower than that of the DH54 VH2-5 germ line antibody, demonstrating that allelic variants of 2F5 germ line antibodies differentially bind to gp41. Thus, these data demonstrate a genetically determined trait that may affect host responses to HIV-1 envelope epitopes recognized by broadly neutralizing antibodies and has implications for unmutated ancestor-based immunogen design.
INTRODUCTION
The human immunoglobulin (Ig) VH (heavy-chain variable region) genes are grouped into 7 families (VH1 to VH7) (4, 24, 30) that are not equally represented in the human B cell repertoire (7, 8, 39). While VH gene usage in cord blood lymphocytes reflects the relative frequency of VH family size, some VH subfamily genes are expressed at higher frequency (14, 20, 38). The control of VH expression by genetic factors has been described in studies on monozygotic twins (19, 41), and VH polymorphisms have been associated with autoimmune diseases (40, 41, 45, 47). VH gene expression, shaped by both genetic and antigenic stimulation, can also regulate the ability of the host to induce an antibody response against a pathogen. The HIV-1 envelope protein has conserved regions in gp41 to which rare broadly neutralizing human antibodies like 2F5 and 4E10 have been isolated (5, 27), However, only ∼10 to 20% of chronically infected subjects make broadly neutralizing antibodies (bNAbs) (32, 43). Even when bNAbs are made, they are made not during the acute infection but rather only after months of chronic infection (15, 32, 34).
Viral factors contributing to difficulty in inducing bNAbs include conformational masking of transient epitopes (3, 21) as well as glycan shielding (31, 44). HIV-1 gp41 bNAbs are more uncommon than those that target gp120, with one hypothesis being that lack of induction of gp41 neutralizing antibodies is due to viral mimicry of the gp41 membrane proximal external region (MPER), with host molecules leading to tolerance to gp41 bNAb induction (3, 16, 17, 42). However, the nature of host factors regulating the ability to make HIV-1 Env bNAbs remains unknown. Xiao et al. have suggested the lack of antigen recognition by unmutated receptors on naïve B cells, therefore implying that there may be “holes” in the human germ line B cell repertoire for antigens that stimulate naïve B cells that can make broadly neutralizing antibodies, and in particular those that can make 2F5-like gp41 bNAbs (46).
The 2F5 bNAb (25, 29) uses the VH2-5 gene, for which 10 distinct alleles are known. Each uses either a D or an N residue at position H54 (22), and the relative frequencies of human VH2-5-bearing antibodies that use N or D alleles are 54% and 46%, respectively (Table 1). The mature 2F5 bNAb usage of VH2-5 includes the DH54 residue, which resides in the heavy-chain complementarity-determining region 2 (HCDR2) (27) (Fig. 1). Since the 2F5 HCDR2 DH54 residue makes an important contact (salt bridge with the 2F5 core epitope residue 665K) with the antigen, structural considerations suggested that the variant with HCDR2 NH54 is likely to have a weaker affinity for antigen. Thus, genetic and structural factors could affect antigen affinity of unmutated naïve B cell receptors (BCR) and induction of 2F5-like antibodies. In this study, we describe the antigen reactivity of the two allelic variants of the VH2-5 inferred germ line variant of the 2F5 bNAb and show that, while both the variant putative germ line antibodies bind to gp41 antigens, their affinities to the gp41-inter protein (13), a mimic of the putative fusion intermediate conformation, differed by an order of magnitude and were determined by the HCDR2 residues NH54/DH54, which define allelic variants.
Table 1.
2F5 VH2-5 HCDR2 UA variants do not neutralize HIV-1a
| Antibody | IC50 (μg/ml) |
||
|---|---|---|---|
| BG1168 | SF162 | MN | |
| 2F5 MAb | 2.347 | 1.163 | 0.103 |
| RUA variant 1 (DH54) | >50 | >50 | >50 |
| RUA variant 2 (NH54) | >50 | >50 | >50 |
HIV-1 pseudovirus neutralization was determined by reduction of luciferase reporter gene expression after a single round of infection by pseudotyped HIV-1 viruses in TZM-bl cells. The pseudotyped viruses containing Envs derived from HIV-1 isolates BG1168.B, SF162.B, and MN.B were preincubated with serial dilutions (3-fold) of the antibodies (150 μg/ml) in 96-well plates. After mixing and incubation with target TZM-bl cells, the concentration of half-maximal inhibition (IC50) was calculated from the luciferase activities determined by luminescence measurements. The pseudotyped virus murine leukemia virus (MLV-SVA), which is not neutralized by 2F5, was used as a control.
Fig. 1.
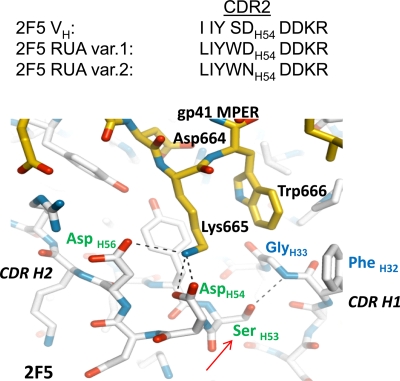
Bonding between 2F5 HCDR2 DH54 and gp41 MPER 665K (27). The contact surface of 2F5 bound to its antigenic peptide shows a strong complementarity of charge, and two CDR H2 residues, DH56 and DH54, interact through hydrogen bonds and salt bridges with K665 of the 2F5 core tripeptide DKW. Based on the CDR H2 substitutions in the two UAs, the following predictions can be made. First, the CDR H2 DH54 residue in variant 1 is retained and, if positioned appropriately, could form a salt bridge with K665 of the 2F5 core DKW. However, the bulky side chain of WH53 (from SH53 to WH53, red arrow) is very likely to perturb the local environment of HCDR2, through potential steric clashes with HCDR1 backbone and FH32 side chain, and disruption of the H bond with GH33. Second, the additional alteration of DH54 to NH54 in UA variant 2 HCDR2 will disrupt the salt bridge with 665K. Also, there is a potential for HCDR2 NH54 to H bond with DH56, which would further preclude establishing bonding with gp41 K665. Critical CDR residues are labeled in green, HCDR1 residues are labeled in blue, and gp41 MPER residues are labeled in black. gp41 MPER is shown in yellow, and 2F5 antibody backbone is shown in white.
MATERIALS AND METHODS
2F5 UA.
The unmutated ancestor (UA) for 2F5 was inferred by Bayesian inference, summing over all gene segments and recombination sites, and computing the posterior probability for each combination using SODA-2, DNAML, and additional in-house software. The two highest-scoring heavy chains, differing from each other by two amino acids, were recombinantly expressed as IgG1 and designated DH54 and NH54. The single highest-scoring light chain was expressed with both UA variants.
Proteins, peptides, antibodies, and phospholipids.
The HIV-1 gp41 peptide containing the 2F5 epitope (QQEKNEQELLELDKWASLWN) was synthesized with an N-terminal biotin tag and purified by reverse-phase high-pressure liquid chromatography (HPLC). The GCN4-gp41-inter proteins were produced as described earlier (12, 13). Chloroform stocks of phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), cholesterol (CH), and bovine heart cardiolipin were obtained from Avanti Polar Lipids. 2F5 monoclonal antibody (MAb) was obtained from Polymun, and Synagis (control antibody) was obtained from Medimmune. 13H11 MAb and Fabs of 2F5 and 13H11 were produced as described earlier (23, 26). Phospholipids used in surface plasmon resonance (SPR) measurements were in the form of POPC-cardiolipin (25:75) or POPC-phosphatidylserine (PS) liposomes as described earlier (10).
SPR measurements of antigen reactivity.
2F5 MAb and UA binding to gp41 MPER peptide and gp41-inter protein was measured using a BIAcore 3000 instrument, and data analyses were performed with BIAevaluation 4.1 software (Biacore/GE Healthcare) as described earlier (2, 13). Biotinylated 2F5 nominal epitope peptide (QQEKNEQELLELDKWASLWN) and a control scrambled version of the same sequence (NKEQDQAEESLQLWEKLNWL) were immobilized (150 resonance units [RU]) on adjacent flow channels of a streptavidin chip (Biacore/GE Healthcare). A scrambled MPER peptide immobilized on the remaining flow channel served as a negative-control surface to subtract out the nonspecific interactions of sample with the chip (1). The GCN4-gp41-inter-specific binding responses were measured by anchoring each antibody on an anti-Fc antibody-immobilized chip as described earlier (1, 13).
Epitope mapping of 2F5 antibody and UA antibodies was carried out on a BIAcore 4000 instrument and using a streptavidin chip (Biacore/GE Healthcare). The biotinylated 2F5 nominal epitope peptide and alanine-substituted mutation scanning the 2F5 nominal epitope were immobilized in duplicate on 16 different spots on four flow channels of the chip. The median spot on each flow channel served as a blank surface to subtract out responses due to nonspecific interactions. SPR antibody binding and epitope mapping analysis was carried out as described earlier (1, 26, 33).
RESULTS
2F5 unmutated ancestor antibodies.
The unmutated ancestor to 2F5 was inferred using Bayesian methods that we developed. The posterior probabilities of two such ancestors were identical and significantly higher than those of all other candidates. The cause of this ambiguity is the presence of allelic variants for the gene segment VH2-5. In addition, because the mutation frequency of 2F5 is relatively high, at 13%, and because such a large expanse of the CDR3 of 2F5 appears to be N-encoding nucleotides, each nucleotide in the ancestor that is attributed to an N region has an 87% posterior probability, which is low compared to the probabilities for template nucleotides.
The two alleles that are equally likely are VH2-5*1 and VH2-5*5. These alleles differ from each other in two places: VH2-5*1 encodes N at amino acid position 56, where VH2-5*5 encodes D, and VH2-5*1 encodes S at amino acid position 62, where VH2-5*5 encodes G. VH2-5*1 agrees with 2F5 at position 62, while VH2-5*5 agrees with 2F5 at position 54.
To estimate the population prevalence of VH2-5 alleles, we analyzed 10,625 rearranged human heavy-chain genes from NCBI GenBank. Four hundred sixty-two of these genes used VH2-5, and of these, 50% used VH2-5*1 and 15% used VH2-5*5. Among all VH2-5 alleles, 54% encode D at position 56, while 46% encode N at this position (Table 1). Thus, from these estimates, we expect to find at least one allele encoding D56 in nearly 80% of the population.
2F5 HCDR2 unmutated allelic variants bind gp41 peptide antigen with differing affinities.
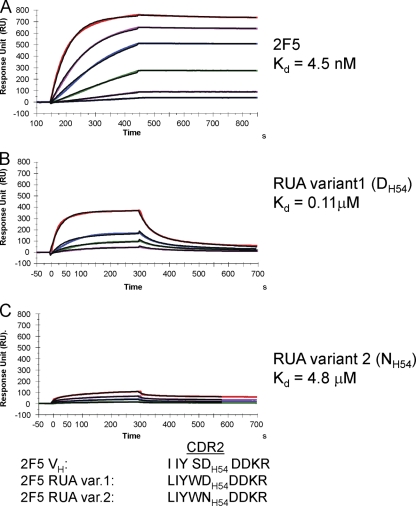
Based on HCDR2 substitutions in the two unmutated ancestors (UAs) (Fig. 1), we made two predictions about antigen reactivity of 2F5 UA antibodies: first, that both 2F5 UAs will have lower antigen affinities than the mature 2F5 antibody, and second, that with the HCDR2 DH54-to-NH54 mutation, the antigen binding of UA variant 2 (NH54) will be further weakened. The carboxylic acid of 2F5 HCDR2 residue DH54 forms a salt bridge with the side chain amine of gp41 665K (Fig. 1), one of the residues that forms the tripeptide core (664DKW) of the epitope (27) and is essential for 2F5 binding (25, 29, 48). Thus, compared to 2F5, we observed a markedly weaker binding Kd (dissociation constant) for the 2F5 UA variant 1 (Kd = 110 nM) (Fig. 2). In the case of UA variant 2, in which the DH54 is altered to NH54, structural analysis suggested that the critical H bonding with gp41 665K would be disrupted and, together with the potential interchain bonding between NH54 and the adjacent residue H56, would result in further weakening of UA variant 2 binding to the gp41 MPER containing the 2F5 epitope (Fig. 1). As predicted, while UA variant 1 bound to the 2F5 gp41 MPER epitope peptide with a Kd of 0.1 μM, a 40-fold-weaker binding Kd (4.8 μM) was observed for UA variant 2 (Fig. 2). Thus, the DH54-to-NH54 mutation in HCDR2 had a more adverse effect on the binding of UA variant 2 to gp41 peptide antigen.
Fig. 2.
2F5 UAs bind to 2F5 nominal epitope peptide with weaker affinity than 2F5 MAb. The gp41 MPER peptide (QQEKNEQELLELDKWASLWN), which includes the 2F5 nominal epitope peptide, was anchored (200 to 300 RU) on a streptavidin (SA) chip via a biotin tag attached to the N terminus of the peptide. Each MAb was injected at various concentrations: 0.67, 1.7, 3.3, 6.7, 13.3, and 26.7 nM (2F5) (A); 3.3, 6.7, 13.3, and 33.3 nM (2F5 UA variant 1) (B); and 66.7, 165.7, 333.3, and 500 nM (2F5 UA variant 2) (C). UA variant 1 with HCDR2 DH54 bound to gp41 MPER peptide with a 40-fold-higher binding Kd than that of UA variant 2 (NH54). Rate constants, ka and kd, for 2F5 were 1.97 × 105 M−1 s−1 and 8.8 × 10−4 s−1; those for UA variant 1 were 3.0 × 105 M−1 s−1 and 3.26 × 10−2 s−1; those for UA variant 2 were 8.4 × 103 M−1 s−1 and 4.1 × 10−2 s−1, respectively. Data are representative of at least two independent experiments.
2F5 UAs show differences in fine epitope specificity from the mature 2F5 antibody.
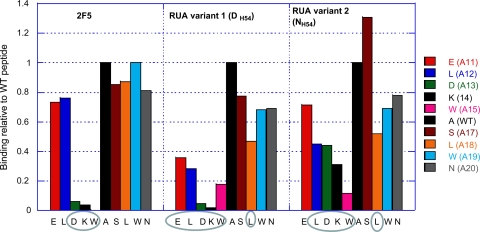
Alanine scanning mutagenesis of the 2F5 nominal epitope residues showed that both UAs had differences in sensitivity to alanine substitutions compared to the mature 2F5 (Fig. 3). As previously reported (27), the specificity of 2F5 was restricted to the tripeptide 664DKW residues, and restriction to those same residues was observed with UA variant 1 but less so with variant 2. However, both UAs were less tolerant to residues outside the 2F5 DKW core, with the UA's specificity extended further to include 662ELDKW residues and the C-terminal residue 669L (Fig. 3). Residue 669L is not involved in 2F5 binding and has been previously reported to be a critical residue for the nonneutralizing gp41 MPER MAb 13H11 (26, 33). Some notable differences in the UA variant 2 fine specificity include the relatively weaker involvement of residues 664D and 665K. The almost-complete abrogation of binding to the 665K mutant suggested that, like 2F5 MAb, UA variant 1 also relies on bonding with 665K, perhaps with similar contacts utilizing a salt bridge (Fig. 1). On the other hand, the HCDR2 alteration of DH54 to NH54 allowed UA variant 2 to better tolerate the 665K mutation (Fig. 3), which suggests that the salt bridge contacts were very likely disrupted and resulted in the observed weaker-affinity binding of UA variant 2 to the MPER epitope peptide (Fig. 2). Overall, the alanine scanning mutagenesis studies showed that the two UAs have specificities distinct from that of the mature 2F5 antibody, which has a more restricted specificity, allowing it to focus on the gp41 epitope core tripeptide residues DKW. However, this relatively broader binding epitope of the two 2F5 UAs was not associated with their ability to neutralize any of three HIV-1 pseudoviruses, HIV-1 MN.B, SF162.B, and BG1168.B, which were all sensitive to the mutated 2F5 MAb (Table 1).
Fig. 3.
2F5 UAs show broader specificity than does the mature 2F5 antibody. The effect of alanine substitution of each of the indicated residues of the 2F5 nominal epitope peptide on the binding of 2F5, UA variant 1, or UA variant 2 is plotted as a ratio of the binding of mutant to wild-type (WT) peptide. Critical residues showing >50% reduction in steady binding responses as measured by SPR analysis are circled.
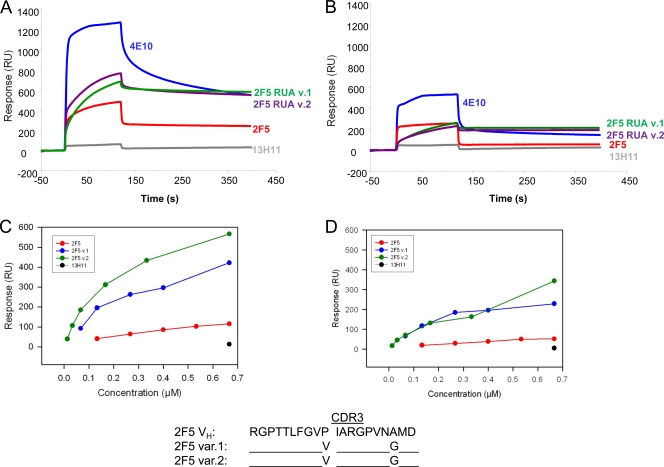
2F5 UAs bind gp41-inter protein with weaker affinities than MAb 2F5.
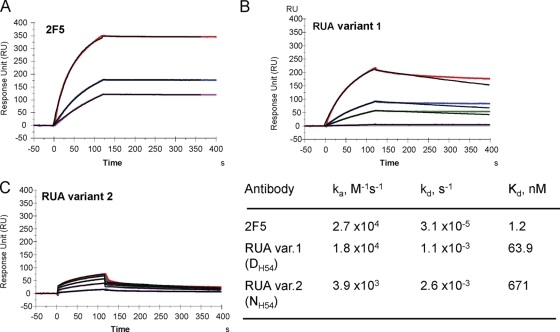
The broadly neutralizing gp41 MAbs 2F5 and 4E10 have been proposed to target the transiently exposed prehairpin intermediate state of gp41 that is conformationally distinct from the prefusion Env trimers on HIV-1 virions (12, 13). Thus, the effect of HCDR2 substitutions on antigen binding was further examined by comparing the binding of the antibodies to a GCN4-trimeric gp41-inter protein, a mimic of the fusion intermediate state of gp41 (13). While nonneutralizing gp41 antibodies do not bind to gp41-inter protein (12, 26), 2F5 and 4E10 bind strongly to gp41 fusion intermediate protein with almost irreversible off-rates (2, 13) (Fig. 4) (Kd = 3 × 10−5 s−1). Both UAs did bind to gp41-inter protein, but with weaker affinities, largely due to off-rates that were 2 orders of magnitude higher than those of 2F5 binding (Fig. 4). Thus, the extremely low off-rate that is required for 2F5 to form a stable complex with the transient prehairpin intermediate state is acquired in the mature 2F5 antibody. However, UA variant 1 with its likely ability to bond with the 665K residue also showed relatively stronger binding to the gp41 fusion intermediate conformation, with an overall binding Kd an order of magnitude lower than that of UA variant 2 (Fig. 4).
Fig. 4.
Binding of 2F5 and 2F5 UAs to trimeric gp41-inter protein. Each of the antibodies, 2F5 (A), UA variant 1 (B), or UA variant 2 (C), was captured to about 600 to 1,000 RU on individual flow cells immobilized with anti-human Fc antibody. 92UG gp41-inter protein (13) was injected over each of the captured antibodies at concentrations of 38.5, 76.9, 192.3, 384.6, and 769 nM (UA variant 1) or 769, 1,538.5, 2,307, 3,077, and 3,846 nM (UA variant 2). The rate constants and Kd values shown in the table were derived from a global curve analysis using the 1:1 Langmuir equation and are representative of two experiments.
2F5 UAs have hydrophobic HCDR3 loops and bind to lipids.
The 2F5 HCDR3 is long and highly hydrophobic, and while its apex does not make direct contact with antigen (27), the hydrophobicity is required for membrane lipid binding and HIV-1 neutralization (2, 18, 28, 37). The weak and reversible binding interactions with viral membrane allow the antibody to capture the transient prehairpin intermediate structure (2). The two 2F5 UA variants bound to anionic phospholipid-containing liposomes equally well and gave binding responses higher than those of 2F5 MAb binding (Fig. 5). These observations are consistent with the overall hydrophobicity of UA HCDR3 compared to that of 2F5 HCDR3 (Fig. 5). Thus, the 2F5 bNAb may have been preferentially selected due to its HCDR3 hydrophobicity and the associated favorable antibody binding energies when encountering the gp41 prehairpin intermediate on the viral surface.
Fig. 5.
2F5 UAs bind to anionic phospholipids. (A and B) Synthetic liposomes with either cardiolipin (CL) (A) or phosphatidylserine (PS) (B) were prepared at a percent molar ratio of 25:75 with phosphatidylcholine (PC) (PC/CL or PC/PS ratio) as described earlier (1, 2, 11). Each of the liposomes was captured to about 500 RU on an L1 sensor chip, and the indicated antibodies at 100 μg/ml were injected over the lipid captured surfaces. No binding was observed on a control PC liposome captured surface (not shown). The control was the nonneutralizing gp41 MPER antibody 13H11, which does not bind to phospholipids (1, 26). (C and D) Binding in resonance units (RU) of each antibody at various concentrations to cardiolipin (C)- or PS (D)-containing liposomes shows higher binding responses of both 2F5 UA variants than of 2F5 MAb. No binding was observed for the 13H11 MAb at the highest concentration of 100 μg/ml (C and D). 2F5 HCDR3 sequences show two substitutions in 2F5 UA HCDR3, V to P and A to G. The residue P100E is at the apex of the 2F5 HCDR3 loop (27).
DISCUSSION
In this study, we described two HCDR2 allelic variants of the VH2-5 inferred unmutated ancestor germ line of the 2F5 bNAb and showed that both UA antibodies bound to gp41 peptide and protein antigens and are thus capable of recognizing either linear or conformational gp41 epitopes. However, their binding affinities for the gp41-inter protein are an order of magnitude weaker than those of the mature 2F5 antibody. The use of NH54 rather than DH54 in HCDR2 resulted in further weakening of antigen reactivity for UA variant 2. These results showed that there is considerable disparity in antigen binding affinities of the two inferred 2F5 HCDR2 allelic variant germ line antibodies.
The mature 2F5 antibody relies on the positioning of two HCDR2 residues, DH54 and DH56, such that bonding and salt bridge connection can be made with a critical residue (665K) within the tripeptide core of the 2F5 epitope (27) (Fig. 1). An explanation of the large differences in antigen binding of the 2F5 UAs could be that the HCDR2 differences in the UAs adversely affect the local structure such that the important bonding of 665K of gp41 is disrupted. This critical bond is likely to be more adversely compromised in UA variant 2, which has the D-to-N mutation at position 54 of the HCDR2 loop. The added potential for HCDR2 NH54 to H-bond with DH56 would further preclude establishing interactions with gp41 665K. In UA variant 1, the HCDR2 DH54 residue is retained and thus could potentially bond with gp41 665K. However, the HCDR2 SH53-to-WH53 mutation in both UAs will introduce a bulky side chain that can generate steric clashes with CDR H1 backbone and the side chain of FH32, in addition to disruption of bonding with CDR H1 G33 (Fig. 1). Thus, these considerations suggest that the HCDR2 substitutions in the 2F5 UAs are likely to have large adverse effects on the local structure where the core residues (DKW) of gp41 are positioned and, as described here, subsequently result in weaker antigen binding affinities.
Neither of the two 2F5 UAs showed neutralization activity against pseudotyped viruses (Table 1). The weaker binding affinity of UA variant 2 for the fusion intermediate state of gp41 (Fig. 3) can explain its lack of neutralization of pseudoviruses that are sensitive to the mature antibody 2F5 (Table 1). UA variant 1 has a Kd and off-rate for gp41-inter binding that are comparable to those of Z13e1 (12), a relatively less potent neutralizing MAb than 2F5. Both UA variants, however, showed lower on- and off-rates for lipid binding than did mature 2F5, and such slow kinetics might not be favorable for capturing the intermediate conformation of gp41. The fast kinetics for 2F5 lipid binding is optimal for preconcentration and for positioning close to the gp41 MPER and for subsequent high-affinity binding to the transient prehairpin intermediate gp41 (2). Thus, while the observed HCDR3 hydrophobicity in the UAs is retained in the 2F5 antibody (Fig. 4), the enhanced gp41 antigen binding affinity of 2F5 MAb (Fig. 3) and its ability to neutralize HIV-1 are acquired in the matured 2F5.
The potency and breadth of HIV-1 neutralization of 2F5 indicate that the gp41 membrane proximal external region is an important target for the induction of humoral responses, both during natural infection and in response to a vaccine. However, Xiao et al. reported that one UA of 2F5 (equivalent to our UA variant 1) did not bind to a gp140 oligomer in an enzyme-linked immunosorbent assay (ELISA) and postulated that the UA lacked reactivity with gp41 (46). Our data showed that both 2F5 inferred germ line allelic variants are capable of binding not only to the linear nominal 2F5 epitope but also to a gp41 conformation that mimics the fusion intermediate state, thus implying that the human B cell repertoire has naïve germ line B cell receptors that recognize transient antigenic structures on HIV-1 Env. One potential explanation for the differences between the study by Xiao et al. and our present work is that we used SPR for detection of direct binding to gp41-inter protein while they used ELISA for measuring the binding to a recombinant gp140 Env oligomer, which has been previously shown to bind suboptimally to 2F5 (13).
An important implication of this study is in unmutated ancestor-based immunogen design. In addition to the differences in the binding strengths from that of the mutated 2F5, both UAs showed fine specificity that extended beyond the core DKW epitope of 2F5 (Fig. 3). Differences in fine specificity as a result of sequence diversity due to somatic hypermutation rather than combinatorial joining had been previously described for antihemagglutinin antibodies (6). The extended footprint and enhanced polyreactivity of the 2F5 UAs suggest that immunogen constructs may be designed for enhanced UA reactivity in order to optimally trigger naïve B cells. Our data suggest that appropriate substitution of residues flanking the core DKW might result in enhanced affinity for the UAs, and such immunogens could potentially be more effective in inducing 2F5-like HIV-1 antibodies.
A second important implication of this study is that a significant proportion of the population is likely to be predisposed toward having naïve B cell receptors (BCR) (i.e., UA variant 1) with an order-of-magnitude-higher threshold for activation by the gp41 MPER neutralizing epitope. However, it cannot be ruled out that the μM antigen binding affinity observed for UA variant 2 is below the threshold of BCR activation. In the absence of competition from high-affinity ligands, low-affinity interactions with μM Kd can effectively trigger BCR (9, 35, 36). However, in the case of B cells carrying the NH54 allele, such low-affinity interactions with subdominant gp41 epitopes might be outcompeted by B cells with higher-affinity receptors that target the dominant epitopes on HIV-1 envelope. In contrast, carriers of DH54 alleles, having naïve B cells with relatively higher gp41 antigen affinity, are likely to have a fairer chance in such a competition. Thus, in addition to genes that regulate tolerance control and/or B cell survival (17, 42), antibody responses to an HIV-1 envelope neutralizing epitope may be also influenced in part by genetically determined VH alleles. Thus, a vaccine designed using either 2F5 allelic variant UA as a template to define gp41 MPER constructs that bind to each allelic variant with enhanced affinity may provide a more avid humoral response against the 2F5 MPER epitope.
ACKNOWLEDGMENTS
This research was conducted as part of the Collaboration for AIDS Vaccine Discovery (CAVD) with support from the Bill & Melinda Gates Foundation to B.F.H. (38643) and the Center for HIV/AIDS Vaccine Immunology (CHAVI) AI067854 from the NIAID, NIH.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Alam S. M., et al. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178: 4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alam S. M., et al. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 106: 20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alam S. M., et al. 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berman J. E., Alt F. W. 1990. Human heavy chain variable region gene diversity, organization, and expression. Int. Rev. Immunol. 5: 203–214 [DOI] [PubMed] [Google Scholar]
- 5. Cardoso R. M., et al. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22: 163–173 [DOI] [PubMed] [Google Scholar]
- 6. Clarke S. H., et al. 1985. Inter- and intraclonal diversity in the antibody response to influenza hemagglutinin. J. Exp. Med. 161: 687–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook G. P., Tomlinson I. M. 1995. The human immunoglobulin VH repertoire. Immunol. Today 16: 237–242 [DOI] [PubMed] [Google Scholar]
- 8. Cook G. P., et al. 1994. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat. Genet. 7: 162–168 [DOI] [PubMed] [Google Scholar]
- 9. Dal Porto J. M., Haberman A. M., Kelsoe G., Shlomchik M. J. 2002. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J. Exp. Med. 195: 1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dennison S. M., et al. 2011. Nonneutralizing HIV-1 gp41 envelope cluster II human monoclonal antibodies show polyreactivity for binding to phospholipids and protein autoantigens. J. Virol. 85: 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dennison S. M., et al. 2009. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J. Virol. 83: 10211–10223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frey G., et al. 2010. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat. Struct. Mol. Biol. 17: 1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frey G., et al. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 105: 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedman D. F., Kraj P., Silberstein L. E. 1995. VH4.21 expression in the normal human B-cell repertoire. Ann. N. Y. Acad. Sci. 764: 285–292 [DOI] [PubMed] [Google Scholar]
- 15. Gray E. S., et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83: 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haynes B. F., et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308: 1906–1908 [DOI] [PubMed] [Google Scholar]
- 17. Haynes B. F., Moody M. A., Verkoczy L., Kelsoe G., Alam S. M. 2005. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum. Antibodies 14: 59–67 [PMC free article] [PubMed] [Google Scholar]
- 18. Julien J. P., et al. 2010. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J. Virol. 84: 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohsaka H., et al. 1996. The human immunoglobulin V(H) gene repertoire is genetically controlled and unaltered by chronic autoimmune stimulation. J. Clin. Invest. 98: 2794–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kraj P., Friedman D. F., Stevenson F., Silberstein L. E. 1995. Evidence for the overexpression of the VH4-34 (VH4.21) Ig gene segment in the normal adult human peripheral blood B cell repertoire. J. Immunol. 154: 6406–6420 [PubMed] [Google Scholar]
- 21. Kwong P. D., et al. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420: 678–682 [DOI] [PubMed] [Google Scholar]
- 22. Li H., Cui X., Pramanik S., Chimge N. O. 2002. Genetic diversity of the human immunoglobulin heavy chain VH region. Immunol. Rev. 190: 53–68 [DOI] [PubMed] [Google Scholar]
- 23. Liao H. X., et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353: 268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsuda F., et al. 1998. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 188: 2151–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muster T., et al. 1993. A conserved neutralizing epitope on gp41 of human deficiency virus type 1. J. Virol. 67: 6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicely N. I., et al. 2010. Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nat. Struct. Mol. Biol. 17: 1492–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ofek G., et al. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78: 10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ofek G., et al. 2010. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J. Virol. 84: 2955–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purtscher M., et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 10: 1651–1658 [DOI] [PubMed] [Google Scholar]
- 30. Rabbitts T. H., Matthyssens G., Hamlyn P. H. 1980. Contribution of immunoglobulin heavy-chain variable-region genes to antibody diversity. Nature 284: 238–243 [DOI] [PubMed] [Google Scholar]
- 31. Richman D. D., Wrin T., Little S. J., Petropoulos C. J. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100: 4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sather D. N., et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen X., et al. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc. Natl. Acad. Sci. U. S. A. 107: 5972–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen X., et al. 2009. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J. Virol. 83: 3617–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shih T. A., Meffre E., Roederer M., Nussenzweig M. C. 2002. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat. Immunol. 3: 570–575 [DOI] [PubMed] [Google Scholar]
- 36. Shih T. A., Roederer M., Nussenzweig M. C. 2002. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat. Immunol. 3: 399–406 [DOI] [PubMed] [Google Scholar]
- 37. Song L., et al. 2009. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc. Natl. Acad. Sci. U. S. A. 106: 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stewart A. K., Huang C., Stollar B. D., Schwartz R. S. 1993. High-frequency representation of a single VH gene in the expressed human B cell repertoire. J. Exp. Med. 177: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tomlinson I. M., et al. 1995. A complete map of the human immunoglobulin VH locus. Ann. N. Y. Acad. Sci. 764: 43–46 [DOI] [PubMed] [Google Scholar]
- 40. Veijola R., et al. 1996. The immunoglobulin heavy-chain variable region in insulin-dependent diabetes mellitus: affected-sib-pair analysis and association studies. Am. J. Hum. Genet. 59: 462–470 [PMC free article] [PubMed] [Google Scholar]
- 41. Vencovsky J., Mageed R. A., Ollier W. E., Maini R. N. 1995. Monozygotic rheumatoid arthritis twin pairs express similar levels of conserved immunoglobulin V gene in polyclonal rheumatoid factors irrespective of disease status. Scand. J. Immunol. 42: 147–157 [DOI] [PubMed] [Google Scholar]
- 42. Verkoczy L., et al. 2010. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc. Natl. Acad. Sci. U. S. A. 107: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walker L. W., et al. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422: 307–312 [DOI] [PubMed] [Google Scholar]
- 45. Wood N. W., et al. 1995. Susceptibility to multiple sclerosis and the immunoglobulin heavy chain variable region. J. Neurol. 242: 677–682 [DOI] [PubMed] [Google Scholar]
- 46. Xiao X., et al. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390: 404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang P. M., et al. 1990. Possible deletion of a developmentally regulated heavy-chain variable region gene in autoimmune diseases. Proc. Natl. Acad. Sci. U. S. A. 87: 7907–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zwick M. B., et al. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 79: 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]