Abstract
Geminiviruses are plant-infecting viruses with small circular single-stranded DNA genomes. These viruses utilize nuclear shuttle proteins (NSPs) and movement proteins (MPs) for trafficking of infectious DNA through the nuclear pore complex and plasmodesmata, respectively. Here, a biochemical approach was used to identify host factors interacting with the NSP and MP of the geminivirus Bean dwarf mosaic virus (BDMV). Based on these studies, we identified and characterized a host nucleoprotein, histone H3, which interacts with both the NSP and MP. The specific nature of the interaction of histone H3 with these viral proteins was established by gel overlay and in vitro and in vivo coimmunoprecipitation (co-IP) assays. The NSP and MP interaction domains were mapped to the N-terminal region of histone H3. These experiments also revealed a direct interaction between the BDMV NSP and MP, as well as interactions between histone H3 and the capsid proteins of various geminiviruses. Transient-expression assays revealed the colocalization of histone H3 and NSP in the nucleus and nucleolus and of histone H3 and MP in the cell periphery and plasmodesmata. Finally, using in vivo co-IP assays with a Myc-tagged histone H3, a complex composed of histone H3, NSP, MP, and viral DNA was recovered. Taken together, these findings implicate the host factor histone H3 in the process by which an infectious geminiviral DNA complex forms within the nucleus for export to the cell periphery and cell-to-cell movement through plasmodesmata.
INTRODUCTION
Viruses are obligate parasites that utilize host cellular machinery to mediate their replication and infection processes. Plant-infecting viruses encode movement proteins (MPs) that facilitate the cell-to-cell trafficking of their infectious nucleic acids along endogenous pathways, such as plasmodesmata, during the process of systemic infection (11, 30, 47). Although it is widely recognized that the interactions between virus-encoded MPs and host proteins are essential for a viable systemic infection, much remains to be elucidated in terms of the actual pathways involved. Indeed, although a number of host proteins involved in the viral systemic infection process have been identified (47), no consensus pathway has emerged. This likely reflects the diversity of plant viruses and the different cellular pathways utilized.
Plant viruses with DNA genomes, such as members of the family Geminiviridae, must utilize cellular components to cross both the nuclear pore and plasmodesmata (PD) for cell-to-cell trafficking of an infectious form of the virus. Geminiviruses possess small circular single-stranded DNA (ssDNA) genomes encapsidated within twinned icosahedral virions, and they replicate in the nucleus via double-stranded DNA (dsDNA) forms (13, 45). To cross nuclear and plasmodesmal boundaries, the bipartite geminiviruses of the genus Begomovirus, such as Bean dwarf mosaic virus (BDMV) (27), encode two proteins from the DNA-B component (35, 45, 52). The product of the BV1 gene is a nuclear shuttle protein (NSP), whereas that of the BC1 gene is an MP that mediates cell-to-cell trafficking of the infectious DNA through PD (9, 24, 27, 35, 45). The NSP and MP bind DNA on the basis of size and form, rather than sequence, and this provides specificity for viral DNA forms as well as maintenance of viral genome size (12, 14, 27, 40, 43, 45). In the current model for cell-to-cell movement of bipartite begomoviruses, the NSP binds to and mediates export of nascent viral DNA from the nucleus. At the nuclear periphery or in the cytoplasm, an NSP-DNA complex interacts with MP, resulting in the delivery of the viral DNA to PD for cell-to-cell movement (9, 24, 45). A number of possibilities exist to explain the form in which viral DNA traffics through PD: (i) an MP-DNA complex, with NSP released to reenter the nucleus; (ii) an NSP-DNA complex, with trafficking facilitated by an MP-PD interaction; and (iii) an MP-NSP-DNA complex (14, 24, 35, 43).
Numerous questions remain regarding the mechanism(s) by which NSP and MP facilitate geminivirus movement, including the precise nature of the viral DNA form(s) that passes through PD (i.e., ssDNA or dsDNA and in what form[s]), the nature of the interactions between NSP and MP and with the viral DNA and host proteins, and the composition of the movement-competent complex. For some bipartite begomoviruses, such as BDMV, the movement complex is not the virion, as capsid protein (CP) mutants are highly infectious (12, 27, 39, 52).
A number of NSP-interacting proteins have been identified in Arabidopsis, including an acetyltransferase (3, 4, 33), a GTPase (5), leucine-rich repeat receptor-like kinase proteins, and a proline-rich extension-like receptor protein kinase (7, 8). These findings suggest that NSP undergoes posttranslational modification, although the significance of this in terms of viral movement remains to be established. Posttranslational modification also may be important for MP function, as a tobacco plasmodesma-associated protein kinase phosphorylated the BDMV MP (26), and mutation of phosphorylation sites in the C terminus of the MP of Abutilon mosaic virus (AbMV) (another bipartite begomovirus) impacted symptom development and viral DNA accumulation (20). The function of the AbMV MP may also be mediated by an interaction with a plastid-targeted 70-kDa heat shock protein (23).
In this study, a biochemical approach was used to identify host factors interacting with the NSP and MP of BDMV. Based on these studies, we identified and characterized a host nucleoprotein, histone H3, which interacts with both the NSP and MP. Additionally, we demonstrate that the BDMV NSP and MP directly interact with each other. Finally, using in vivo coimmunoprecipitation (co-IP) assays and a Myc-tagged histone H3, a putative movement complex composed of histone H3, NSP, MP, and viral DNA was recovered. These findings are discussed in terms of the process by which an infectious form of viral DNA is generated and relayed from the nucleus to the cell periphery and PD.
MATERIALS AND METHODS
Protein purification, probe synthesis, and overlay assays.
Total proteins were extracted from 10 g of leaf tissue (upper 4 to 6 leaves collected from plants at the 10- to 16-leaf stage) and separated into four enriched subcellular fractions (P1, P0, P30, and S30), as described previously (6). The 35S-labeled probes were prepared by cloning selected genes into the pSP64 vector (Promega, Madison, WI) and carrying out in vitro translation in the presence of [35S]methionine with the TnT SP6 Quick coupled transcription/translation system (Promega, Madison, WI), according to the manufacturer's instructions. Protein overlay assays were performed as described previously (53). Protein blots were overlaid with 35S-NSP or 35S-MP for 45 min at room temperature, and NSP/MP-host protein complexes were detected by autoradiography.
Identification of BDMV NSP- and MP-interacting proteins.
An ∼17-kDa protein band containing a host protein(s) interacting with the BDMV NSP/MP was excised from an SDS-polyacrylamide gel and trypsin-digested, and the resultant peptides were analyzed with an ABI 4700 matrix-assisted laser desorption ionization–time of flight tandem mass spectrometer (MALDI-TOF/TOF MS) (Proteomics Core Facility, Genome Center, University of California—Davis).
Cloning and sequencing of the histone H3 genes from tomato and Nicotiana benthamiana.
Full-length histone H3 cDNAs were generated from mRNAs extracted from tomato (Solanum lycopersicum) and Nicotiana benthamiana leaf tissue by reverse transcription-PCR (RT-PCR) as previously described (48). For the tomato histone H3 gene, two degenerate primers (P7F/P8R and P9F/P10R) were used, whereas for that of N. benthamiana, one primer pair (P11F/P12R) was used. Primer sequences are presented in Table S1 in the supplemental material. Sequence alignments and comparisons were performed with Vector NTI 10 software.
Protein expression and purification.
Recombinant His6-tagged proteins were generated using pRSET vectors (Invitrogen Corp., Carlsbad, CA). The full-length tomato histone H3 gene and N-terminal and C-terminal deletion mutants were PCR amplified, cloned into pRSET-A, and sequenced to determine their integrity. The full-length BDMV NSP, MP, and CP genes and NSP deletion mutants were similarly cloned into the pRSET-A vector. The same approach was also used to clone the Tobacco mosaic virus (TMV) MP and CP genes, the Cucumber mosaic virus (CMV) 3a gene, and the Beet mild curly top virus (BMCTV) CP gene. The Bean common mosaic necrosis virus (BCMNV) HC-Pro and Tomato yellow leaf curl virus (TYLCV) CP genes were expressed as previously described (42). For protein expression, recombinant plasmids were transformed into Escherichia coli strain BL21(DE3) (Invitrogen). Proteins were extracted from inclusion bodies (35, 42) and then purified with Ni-nitrilotriacetic acid (Ni-NTA) columns according to the manufacturer's instructions (Qiagen, Valencia, CA). Purified proteins were renatured by dialysis against native buffer (10 mM Tris-HCl [pH 8.0], 100 mM EDTA, 10% glycerol, 1 mM dithiothreitol [DTT], and 1 mM phenylmethanesulfonylfluoride [PMSF]. Core histones from chicken erythrocytes (here referred to as core histones) were obtained commercially (Upstate Inc., Lake Placid, NY).
Western blot analysis.
Proteins were fractionated in SDS-polyacrylamide gels, electroblotted onto polyvinylidene fluoride (PVDF) membranes and probed with NSP, MP, or histone H3 antibodies. The anti-BDMV NSP polyclonal antibody was raised against an oligopeptide (N′-CLRNKRGSSFSQRRFY-C′) corresponding to a portion of the N terminus, whereas the MP antibody was raised against an oligopeptide (N′-CINSNCKAYQPKSLQ-C′) corresponding to a portion of the C terminus (Genemed Synthesis Inc., San Francisco, CA). The histone H3 monoclonal antibody (MAb) was obtained from Upstate Inc. The polyclonal NSP and MP antibodies were detected with goat anti-rabbit antibodies (Bio-Rad, Hercules, CA), whereas the histone H3 monoclonal antibody was detected with anti-mouse antibodies (Bio-Rad).
Immunoprecipitation assays.
In vitro coimmunoprecipitation (co-IP) assays were performed with a protein G immunoprecipitation kit (Sigma, St. Louis, MO) according to the manufacturer's instructions. Briefly, mixtures of 5 μg of His6-tagged fusion protein (NSP, MP, or histone H3) or 25 μg of core histones were incubated with NSP or MP antibodies (5 μg) for 2 h at 4°C. The co-IP fractions were recovered by loading the mixtures onto protein G-Sepharose columns, incubating the columns for 1 h at 4°C, and then washing according to manufacturer's instructions. The co-IP fraction was eluted and analyzed by SDS-PAGE and Western blotting with NSP, MP, and histone H3 antibodies.
For in vivo co-IP experiments performed with histone H3-enhanced cyan fluorescent protein (ECFP) fusion protein, leaves of N. benthamiana plants at the 5- to 7-leaf stage were coinfiltrated with combinations of Agrobacterium tumefaciens strains carrying binary vectors expressing H3-ECFP or BDMV NSP or MP under the control of the 35S promoter. The control consisted of equivalent leaves coexpressing ECFP, NSP, and MP. Total leaf proteins were extracted at 2 days postinfiltration (dpi), and co-IP assays were performed using the Pierce cross-link immunoprecipitation kit (Pierce Biotechnology, Rockford, IL), according to the manufacturer's instructions. For these assays, 10 μg of a GFP monoclonal antibody (Clontech, Mountain View, CA) was coupled to protein A/G-agarose for 1 h at 23°C, cross-linked with disuccinimidyl suberate for 40 min, and loaded onto a spin column. Total leaf proteins were then incubated on this spin column for 2 h at 4°C. The column was washed, and the co-IP fraction was eluted according to the manufacturer's instructions. Eluted proteins were separated by SDS-PAGE and analyzed by Western blotting with the ECL Western blotting system (GE Healthcare Biosciences, Pittsburgh, PA) and GFP monoclonal antibodies and purified BDMV NSP and MP polyclonal antibodies.
In vivo co-IP experiments were also performed using histone H3 tagged at the N terminus with a Mycx4 peptide (Mycx4-H3). In these experiments, leaves of N. benthamiana plants were coinfiltrated with A. tumefaciens strains carrying binary vectors expressing Mycx4-H3, BDMV NSP, or BDMV MP, each under the control of the 35S promoter, as well as A. tumefaciens strains carrying binary vectors with the infectious multimeric clones of the full-length DNA-A and DNA-B components of BDMV (15) (this is referred to as the Mycx4-H3 treatment). For these co-IP studies, controls were healthy N. benthamiana leaves and leaves coinfiltrated with the same A. tumefaciens strains as described above, except that a strain with a binary vector expressing Mycx4 was used in place of the one with the Mycx4-H3 construct (this is referred to as the Mycx4 treatment). Furthermore, in these co-IP experiments, an A. tumefaciens strain carrying a binary vector expressing the BCMNV HC-Pro silencing suppressor was coinfiltrated to enhance levels of protein expression.
Total proteins were extracted at 3 dpi, and co-IP assays were performed using the Myc-Tag (9B11) mouse MAb (Sepharose Bead Conjugate) kit (Cell Signaling Technology, Inc., Danvers, MA) according to the manufacturer's instructions. An aliquot of total protein extract (1 mg) was incubated with the Myc-Tag (9B11) mouse MAb beads (80 μl) overnight at 4°C. The beads were then washed and the co-IP fraction eluted according to the manufacturer's instructions. Eluted proteins were separated by 12% SDS-PAGE and analyzed by Western blotting with the ECL Western blotting system (GE Healthcare Biosciences), Myc-Tag (71D10) rabbit MAb antibody (Cell Signaling Technology, Inc.), and purified BDMV NSP and MP polyclonal antibodies.
Detection of viral DNA contained in the Mycx4-H3 co-IP fraction.
DNA was extracted from the co-IP fraction with the DNeasy plant minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The presence of the BDMV DNA-A and DNA-B components was determined by PCR with the begomovirus degenerate primer pairs PAR1-c496/PAL1-c1978 (DNA-A) and PCRc-1/BL1v2040 (DNA-B) as previously described (41). PCR products were sequenced to confirm their identity.
Colocalization assays.
Expression vectors having the 35S promoter, a multiple-cloning site, full-length EGFP/CFP or monomeric red fluorescent protein (mRFP) open reading frames (ORFs), and the NOS terminator (25) were used for subcellular localization studies. The histone H3, NSP, MP, and CP genes were PCR amplified and cloned into these vectors. These recombinant plasmids, individually or in various combinations, were then transfected into protoplasts prepared from N. tabacum cv. Xanthi nc cell suspension cultures (15). Transfected protoplasts were examined at 18 to 48 h postelectroporation with a confocal laser scanning microscopy (CLSM) system (Leica TCS-SP2 AOBS) (EGFP excitation, 488 nm [emission collected at 500 to 530 nm]; mRFP excitation, 543 nm [emission collected at 570 to 600 nm]; DAPI [4′,6′-diamidino-2-phenylindole] excitation, 405 nm [emission collected at 440 to 470 nm]). Image processing was performed with Adobe Photoshop 6.0 software (Adobe, Mountain View, CA).
In planta subcellular localization experiments were also conducted using the above-described C-terminal fusion proteins in A. tumefaciens-mediated transient assays. Here, N. benthamiana leaves were coinfiltrated with combinations of A. tumefaciens strains carrying binary vectors expressing histone H3, NSP, and MP fusion proteins or the infectious multimeric BDMV DNA-A and DNA-B clones (15). Infiltrated areas of leaves were examined by CLSM at 1 to 2 dpi.
TRV vector and BDMV inoculation.
To attempt to silence the N. benthamiana histone H3 gene, the Tobacco rattle virus (TRV) vector system was employed as described previously (28). Full-length and truncated forms of the N. benthamiana histone H3 gene were PCR amplified and cloned into pTRV2. A 335-bp phytoene desaturase (PDS) gene cDNA fragment, corresponding to nucleotides (nt) 898 to 1231 of the N. benthamiana PDS gene, was also PCR amplified and cloned into pTRV2 (28). The pTRV1 and pTRV2 plasmids were introduced into A. tumefaciens strain GV2260, C58, or EHA 105 by electroporation. Agrobacterium cultures (optical density [OD] of ∼0.8 at 600 nm) containing binary vectors with TRV constructs were mixed at a 1:1 ratio and infiltrated into the lower leaf surfaces of leaves of N. benthamiana plants at the 4-leaf stage. Plants were then transferred back to a growth chamber (25°C and with a 16-h light/8-h dark regimen). Eight days after TRV inoculation, plants were agroinoculated with BDMV by coinfiltrating leaves with A. tumefaciens strains carrying binary vectors with the multimeric infectious BDMV DNA-A or BDMV DNA-B clones. These experiments were performed at least 5 times with at least four plants per experiment.
RESULTS
Identification of a host factor that interacts with the BDMV NSP and MP.
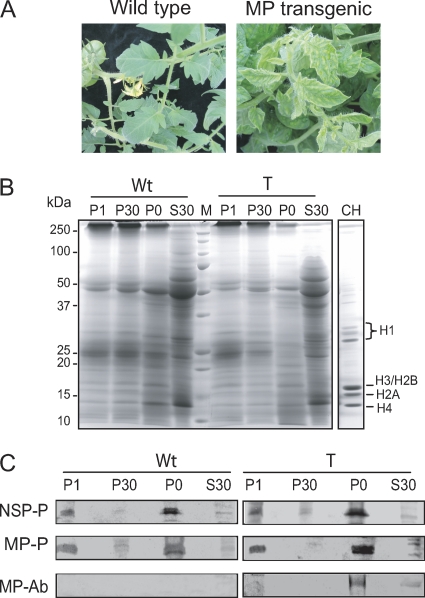
Gel overlay assays were carried out to detect host factors that interact with BDMV NSP and MP. For these experiments, two plant species were employed: N. benthamiana and tomato. A transgenic tomato line (MP-9B line [16]) was used in these studies because expression of the MP results in a phenotype that mimics that of virus-infected plants (Fig. 1A). Protein extracts were prepared from leaves collected from wild-type and transgenic tomato plants and from wild-type and BDMV-infected N. benthamiana plants. Four enriched subcellular fractions (P0, P1, P30, and S30) were generated by differential centrifugation: the P0 and P1 fractions were enriched in cell wall, nuclear, and chloroplast proteins; P30 was enriched in membrane and mitochondrial proteins; and S30 contained soluble proteins. The profiles of these isolated protein fractions from wild-type and transgenic tomato are shown in Fig. 1B.
Fig. 1.
Gel overlay assays detect interactions between Bean dwarf mosaic virus (BDMV) movement proteins (NSP and MP) and plant proteins. (A) Wild-type and BDMV MP transgenic tomato plants. Expression of MP resulted in disease symptom development. (B) Protein profiles obtained for wild-type (Wt) and MP-expressing (T) tomato leaves. Proteins were fractionated (P1, P30, P0, and S30), separated by SDS-PAGE, and visualized by Coomassie brilliant blue staining. CH, core histones from chicken erythrocytes. (C) Gel overlays of host protein fractions probed with 35S-labeled in vitro-translated NSP (NSP-P) or MP (MP-P) and Western blot analysis of equivalent fractions probed with MP antiserum (MP-Ab).
Gel overlay assays were next performed on these fractions with an in vitro-translated 35S-labeled BDMV NSP or MP probe. Preliminary experiments established that although both probes interacted with a number of host proteins, a predominant interacting protein appeared in the ∼17-kDa region. For the NSP, this ∼17-kDa host protein was detected in the P1 and P0 fractions from both wild-type and transgenic tomato plants (Fig. 1C). Equivalent results were obtained using fractionated proteins from wild-type and BDMV-infected N. benthamiana (data not shown). The MP probe also interacted with a protein in the ∼17-kDa regions of the P1 and P0 fractions. Note also that the BDMV MP was detected in the P0 fractions obtained from MP-transgenic tomato plants (Fig. 1C). Similar results were obtained in three independent overlays, with new extracts prepared for each experiment.
Histone H3 identified as the interacting protein.
The ∼17-kDa protein band with the NSP- and MP-interacting protein(s) was excised from an SDS-polyacrylamide gel and trypsin digested, and the resultant peptides were analyzed by MALDI-TOF mass spectrometry. The most abundant peptide had the sequence STELLIR, and a database search identified the protein as histone H3. A second, less abundant peptide had the sequence LVLPGELAK, and this was identified as histone H2B. Note that histones H3 and H2B are both core histones (i.e., they are part of the nucleosome [34, 49, 55]). Furthermore, these proteins have a molecular mass (∼15 kDa) which is in the range of that of the NSP- and MP-interacting protein(s) (Fig. 1B and C) (49).
To confirm the interaction between BDMV NSP and MP and histone H3, the most abundant interacting protein, full-length histone H3 cDNAs were amplified from tomato and N. benthamiana by RT-PCR with degenerate primers designed based on sequences in GenBank (see Table S1 in the supplemental material). The cloned histone H3 genes from tomato and N. benthamiana were each 411 nt and encoded predicted proteins of 136 amino acids, which were 97% identical (see Fig. S1 in the supplemental material) (GenBank accession numbers EF661028.1 [tomato] and EF661029.1 [N. benthamiana]).
Recombinant tomato histone H3 (R-H3) protein was expressed in E. coli as a His6 fusion protein from the pRSET expression vector, and this R-H3 protein was ∼21 kDa. To confirm the functionality of the R-H3 protein, gel mobility shift assays were performed. In these experiments, R-H3 was added to a total genomic DNA extract prepared from leaves of N. benthamiana plants infected with the geminivirus Honeysuckle yellow vein virus, and the viral DNA forms were revealed by Southern blot hybridization analysis. Here, R-H3 bound ssDNA and dsDNA forms; no binding was observed in a control in which R-H3 was not added (data not shown). These results indicate the functional nature of the E. coli-expressed R-H3 and the capacity of the protein to bind various forms of geminivirus DNA.
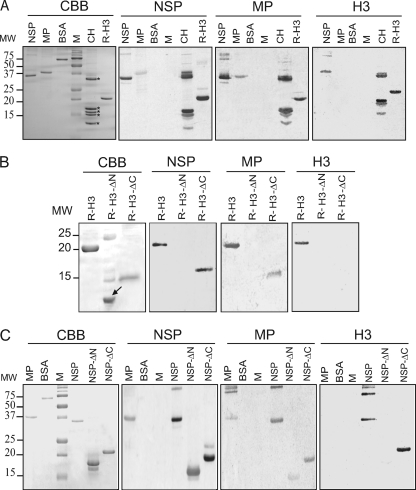
For overlay assays, recombinant NSP, MP, and H3 were separated by PAGE. As controls, bovine serum albumin (BSA) and core histones were included. Proteins were transferred onto PVDF membranes and probed with 35S-labeled in vitro-translated NSP, MP, or histone H3. As illustrated in Fig. 2A, the NSP probe interacted strongly with itself, MP (weakly), the core histones (H3 and also H2 and H1), and R-H3 but not with BSA. Similarly, the MP probe interacted with itself, NSP, the core histones, and R-H3, and no interaction was observed with BSA. The histone H3 probe strongly interacted with itself, NSP, and the core histones. As expected, the histone H3 probe did not interact with BSA, but it also did not interact with MP, and the latter result was obtained in multiple independent experiments. As the in-gel form of the MP was expressed in E. coli, the possibility exists that this protein requires posttranslational modification for efficient binding to NSP and H3. Taken together, these results indicate that histone H3 is the ∼17-kDa NSP- and MP-interacting protein detected in the initial overlay assays, although we cannot rule out that other interacting proteins (e.g., histone H2B) also were present.
Fig. 2.
Gel overlay assays establish the interaction between Bean dwarf mosaic virus (BDMV) movement proteins (NSP and MP) and histone H3 and identify domains involved in the protein-protein interaction. (A) Purified recombinant BDMV NSP, MP, and R-H3 were separated by SDS-PAGE and visualized by Coomassie brilliant blue (CBB) staining. Proteins were transferred to PVDF membranes and probed with 35S-labeled in vitro-translated NSP, MP, or H3. BSA and core histones (CH) (indicated by asterisks) from chicken erythrocytes were included as negative and positive controls, respectively. (B) Gel overlay assay to identify interacting domains of histone H3 and BDMV movement proteins. Mutant H3 proteins with N-terminal (residues 1 to 68 deleted; R-H3-ΔN; the arrow indicates the truncated histone H3 protein) or C-terminal (residues 69 to 136 deleted; R-H3-ΔC) deletions were separated by SDS-PAGE and visualized by CBB staining. Protein interactions were tested as for panel A. (C) Gel overlay assay to identify interacting domains of BDMV NSP and histone H3. Mutant NSP proteins with N-terminal (residues 1 to 128 deleted; NSP-ΔN) or C-terminal (residues 129 to 256 deleted; NSP-ΔC) deletions were separated by SDS-PAGE and visualized by CBB staining. Protein-protein interactions were tested as for panel A. BDMV MP and BSA were employed as positive and negative controls, respectively.
To assess the specificity of this interaction, we next performed overlay assays with MPs from selected plant-infecting RNA viruses. These assays revealed no interaction between histone H3 and BCMNV HC-Pro, CMV 3a, or TMV MP (see Fig. S2 in the supplemental material). Here, it is important to emphasize that like histone H3 and BDMV NSP, CMV 3a and TMV MP are basic proteins. Thus, the fact that the histone H3 did not interact with the MPs of these RNA viruses indicates specificity in the interaction of histone H3 with NSP and MP rather than a nonspecific interaction between basic proteins. As an additional control for these experiments, overlay assays were performed using NSP as a probe; here, no interaction was detected with the MPs of these three RNA viruses (data not shown).
Molecular determinants of the H3 interaction with BDMV NSP and MP.
To further investigate the nature of the interaction between NSP or MP and histone H3, truncated forms of histone H3 were engineered and used in gel overlay assays. As shown in Fig. 2B, R-H3-ΔC (lacking residues 69 to 136) interacted with both NSP and MP. In contrast, R-H3-ΔN (lacking residues 1 to 68) failed to interact with either NSP or MP. It is noteworthy that neither truncated form interacted with the histone H3 probe. Thus, the domain(s) involved in NSP/MP interaction appears to reside in the N-terminal region of histone H3.
To determine the region of the BDMV NSP required for histone H3 interaction, overlay assays were next performed with truncated forms of this protein. As shown in Fig. 2C, NSP-ΔC (lacking residues 129 to 256) still interacted with itself, MP, and R-H3. Equivalent overlay assays performed with NSP-ΔN (lacking residues 1 to 128) revealed that this mutant form still interacted with itself and MP, albeit weakly, but not with R-H3. These studies established that the NSP N terminus is necessary for histone H3 interaction and is also involved in NSP-MP binding.
Histone H3 also interacts with CPs of different geminiviruses.
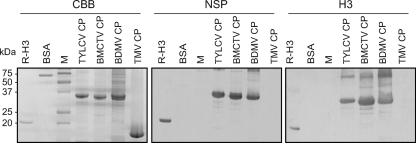
The CP is the functional equivalent of NSP in the monopartite begomoviruses (44) and is considered the progenitor of the NSP of the bipartite begomoviruses (19, 45). Consistent with this evolutionary relationship, the CPs of bipartite begomoviruses can complement certain NSP mutants (39), as well as mask recognition of NSP by host defense responses (58). Overlay assays were next performed to assess the capacity for interactions between histone H3 and the CPs of the monopartite begomovirus TYLCV, the monopartite curtovirus BMCTV, and the bipartite begomovirus BDMV. The BDMV NSP and TMV CP served as controls. Strong interactions were detected between R-H3 and NSP (positive control). In addition, strong interactions were detected between R-H3 and the CPs of the three geminiviruses, whereas no interaction was observed between R-H3 and TMV CP (Fig. 3). Interestingly, in parallel overlay assays performed with BDMV MP as the probe, MP interacted with the three geminiviral CPs but not with the TMV CP (data not shown).
Fig. 3.
The Bean dwarf mosaic virus (BDMV) NSP and histone H3 proteins interact with capsid proteins (CPs) from a range of geminiviruses but not with the CP of an RNA virus. Purified recombinant CPs of Tomato yellow leaf curl virus (TYLCV), Beet mild curly top virus (BMCTV), BDMV, and TMV were separated by SDS-PAGE and visualized by Coomassie brilliant blue (CBB) staining. Proteins were transferred to PVDF membranes and probed with 35S-labeled in vitro-translated NSP or H3. Recombinant histone H3 (RH-3) and BSA were included as positive and negative controls, respectively.
Co-IP assays confirm that histone H3 interacts with NSP and MP, both in vitro and in vivo.
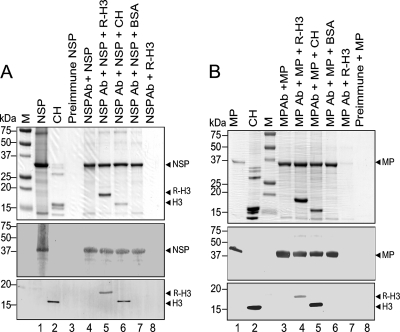
Co-IP assays were next performed to further confirm the interaction between histone H3 and the BDMV NSP and MP. Purified recombinant NSP was mixed with purified R-H3 or core histones, and NSP polyclonal antiserum was added. This mixture was loaded onto a protein G-Sepharose column and incubated for 1 h. After washing, bound proteins were eluted, fractionated in a 12% SDS-polyacrylamide gel, and transferred to PVDF membranes. Membranes with bound proteins were analyzed by Western blot analysis with NSP or histone H3 antiserum (Fig. 4A). Controls included NSP incubated with preimmune serum, NSP incubated with NSP antiserum, and NSP incubated with BSA and NSP antiserum.
Fig. 4.
In vitro coimmunoprecipitation assays demonstrate an interaction between histone H3 and the Bean dwarf mosaic virus (BDMV) movement proteins NSP (A) and MP (B). Recombinant BDMV NSP and MP were expressed in and purified from E. coli (lanes 1). Recombinant tomato histone H3 (R-H3) was similarly expressed and purified. The indicated combinations of proteins and antibodies were incubated and then passed over a protein G-Sepharose affinity column. Coimmunoprecipitated proteins were eluted, separated by SDS-PAGE, and visualized by Coomassie brilliant blue staining (upper panels). Proteins were transferred to PVDF membranes and Western blot analyses performed with NSP and histone H3 antibodies or MP and histone H3 antibodies (middle and lower panels, respectively). Ab, antibodies; BSA, bovine serum albumin; CH, chicken core histones; M, marker proteins.
As expected, NSP was not recovered when incubated with preimmune serum (Fig. 4A, lane 3), whereas it was recovered when incubated with NSP antiserum (Fig. 4A, lane 4). When NSP was incubated with R-H3 and NSP antisera, both NSP and R-H3 were recovered (Fig. 4A, lane 5). Similarly, the histone H3 of the core histones was recovered when incubated with NSP and NSP antisera (Fig. 4A, lane 6). The specificity of these interactions was confirmed by using BSA or by omitting NSP (Fig. 4A, lanes 7 and 8, respectively). Similar experiments were performed with MP. Here, MP was not recovered when incubated with preimmune serum (Fig. 4B, lane 8), whereas it was recovered when incubated with MP antiserum (Fig. 4B, lane 3). When MP was incubated with R-H3 and MP antisera, both MP and R-H3 were recovered (Fig. 4B, lane 4). Similarly, when the core histones were incubated with MP and MP antisera, the histone H3 was recovered (Fig. 4B, lane 5). The specificity of these interactions was again confirmed by using BSA or by omitting MP (Fig. 4B, lanes 6 and 7, respectively). Taken together, these in vitro co-IP experiments provide strong supporting evidence for a specific interaction between histone H3 and the BDMV NSP and MP.
To further confirm the interaction between histone H3 and the BDMV NSP and MP, in vivo co-IP experiments were performed. Here, N. benthamiana leaves were coinfiltrated with A. tumefaciens strains carrying binary vectors expressing the H3-ECFP fusion protein, NSP, and MP. Total proteins were extracted from tissues at 2 dpi. Western blot analysis performed on these total protein extracts with the GFP antibody revealed that H3-ECFP was highly expressed in this system (see Fig. S3A, lane 1, in the supplemental material), and co-IP assays revealed efficient recovery of H3-ECFP using the GFP monoclonal antibody spin column (see Fig. S3A, lane 2). Western blot analyses performed on these total protein extracts with MP and NSP antibodies indicated that both proteins were expressed at low levels (see Fig. S3A, lanes 5 and 9, respectively), which may reflect gene silencing or protein instability (21). However, results of Western blot analyses of proteins recovered in the co-IP fraction revealed both the presence and enrichment of MP and NSP (see Fig. S3A, lanes 6 and 10, respectively). Three independent experiments yielded equivalent results.
To establish that the histone H3 portion of the H3-ECFP fusion protein was responsible for the recovery of MP and NSP in the above-described co-IP assays, N. benthamiana leaves were coinfiltrated with A. tumefaciens strains carrying binary vectors expressing ECFP, MP, and NSP, and equivalent co-IP assays were performed. Western blot assays performed with the GFP antiserum revealed a strong ECFP signal in the total protein extracts prepared from infiltrated leaves (see Fig. S3B, lane 1, in the supplemental material); in addition, a strong ECFP signal also was detected in the co-IP fraction (see Fig. S3B, lane 2). Equivalent Western blot assays performed with MP and NSP antisera on total protein extracts revealed the presence of both proteins (see Fig. S3B, lanes 3 and 5, respectively); however, neither viral protein was detected in the co-IP fraction (see Fig. S3B, lanes 4 and 6, respectively). Similar results were obtained in three independent experiments. These results indicate that the recovery of NSP and MP in the co-IP assays performed with the H3-ECFP fusion protein was due to the presence of histone H3. Taken together, these in vivo co-IP studies confirmed that histone H3 interacts with MP and NSP in planta.
Co-IP studies establish the presence of an H3-NSP-MP-viral DNA complex.
As demonstrated above, co-IP assays performed with H3-ECFP allowed for the recovery of NSP and MP. However, when the BDMV DNA-A and DNA-B components were included in these co-IP experiments, viral DNA was not detected in the co-IP fraction. This may have been due to interference from the relatively large ECFP tag (∼27 kDa); thus, we next performed co-IP experiments with a Myc-tagged histone H3 protein. Here, a Mycx4 peptide tag (∼5 kDa) was fused to the N terminus of histone H3 to generate Mycx4-H3. Control experiments performed with total (input) proteins and the co-IP fraction from healthy N. benthamiana leaves revealed that there were no endogenous proteins that interacted with the Myc monoclonal antibody (see Fig. S5A, lanes 1 and 2, in the supplemental material). In an additional series of control experiments, total (input) proteins and co-IP fractions were obtained from N. benthamiana leaves coinfiltrated with combinations of A. tumefaciens strains carrying binary vectors expressing Mycx4, HC-Pro, and the infectious multimeric BDMV DNA-A and DNA-B clones. Whereas no proteins interacting with the Myc antibody were detected in total (input) protein extracts from any of the treatments (see Fig. S5, lanes 1, 3, 5, and 7), two protein bands (∼30 and ∼60 kDa) were detected in the co-IP fractions from the treatments with Mycx4 expression (see Fig. S5, lanes 6 and 8). Given that these bands were detected by the Myc monoclonal antibody, they presumably represent components of a protein complex containing the Mycx4 peptide. Interestingly, these interacting proteins were present in the co-IP fraction at much higher levels in leaves infected with BDMV DNA-A and DNA-B (see Fig. S5 [compare lanes 6 and 8]).
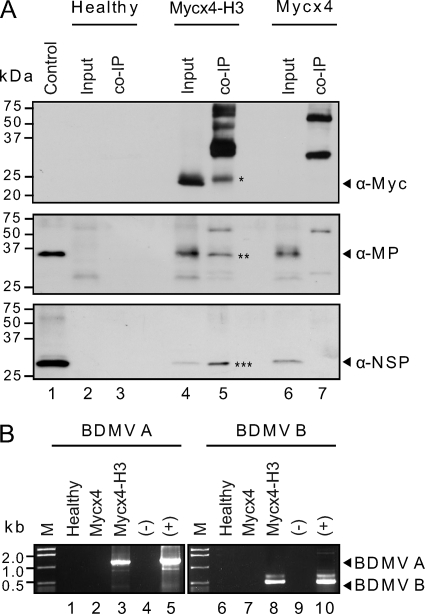
Experiments were next performed with N. benthamiana leaves coinfiltrated with A. tumefaciens strains carrying binary vectors expressing Mycx4-H3, NSP, MP, HC-Pro, and BDMV DNA-A and DNA-B (Mycx4-H3 treatment). Controls were healthy N. benthamiana leaves and leaves coinfiltrated with the same A. tumefaciens strains as in the Mycx4-H3 treatment except that the Mycx4-H3 strain was replaced with a strain expressing Mycx4 (Mycx4 treatment). Total (input) proteins and co-IP fractions were obtained, proteins separated by PAGE (see Fig. S4 in the supplemental material), and Western blot analyses performed with Myc, NSP, and MP antibodies. Results for the controls were similar to those described above: no proteins interacting with the Myc antibody were detected in total (input) proteins or the co-IP fraction from healthy N. benthamiana leaves (Fig. 5A, lanes 2 and 3, upper panel), whereas no interacting proteins were detected in total (input) proteins from the Mycx4 treatment (Fig. 5A, lane 6, upper panel), but ∼30- and 60-kDa interacting proteins were detected in the co-IP fraction for this treatment (Fig. 5A, lane 7, upper panel). Furthermore, Western blot analyses with NSP and MP antisera revealed the presence of NSP and MP in the total (input) protein extract of the Mycx4 treatment (Fig. 5A, lane 6, middle and lower panels, respectively) but not in the co-IP fraction (Fig. 5A, lane 7, middle and lower panels, respectively). On the other hand, equivalent Western blot analyses of total (input) proteins and the co-IP fraction from the Mycx4-H3 treatment confirmed the interaction between H3, NSP, and MP. Here, Mycx4-H3, NSP, and MP were detected in the total (input) proteins (Fig. 5A, lane 4, upper, middle, and lower panels, respectively), as well as in the co-IP fraction recovered with the Myc monoclonal antibody (Fig. 5A, lane 5, upper, middle, and lower panels, respectively). Note also the presence of the additional protein bands detected in the co-IP fraction of the Mycx4-H3 treatment when probed with the Myc monoclonal antibody (Fig. 5A, lane 5, upper panel); these were similar to those recovered in the co-IP fraction of the Mycx4 treatment (Fig. 5A, lane 7, upper panel).
Fig. 5.
In vivo coimmunoprecipitation (co-IP) assays confirm the interactions between histone H3 and the Bean dwarf mosaic virus (BDMV) movement proteins (NSP and MP) and reveal a complex composed of histone H3, NSP, MP, and viral DNA. (A) The Mycx4-H3, NSP, MP, and BCMNV HC-Pro suppressor and BDMV DNA-A and DNA-B components were coexpressed in leaves of N. benthamiana via A. tumefaciens-mediated transient expression. Controls were healthy N. benthamiana leaves and leaves expressing Mycx4, NSP, MP, HC-Pro, and BDMV DNA-A and DNA-B. Total proteins were extracted from healthy and infiltrated leaves (3 dpi), and co-IP was performed with the Myc-Tag (9B11) mouse MAb (Sepharose bead conjugate) kit. Input (total) and eluted (co-IP fraction) proteins were separated by SDS-PAGE, and proteins were transferred to PVDF membranes. Western blot analyses were performed with Myc, MP, and NSP antibodies. Asterisks indicate the immunoprecipitated Mycx4-H3 (*), MP (**), and NSP (***) proteins. (B) Detection of BDMV DNA-A and DNA-B components in co-IP fractions by PCR with DNA-A and DNA-B primers. DNA extracts were prepared from co-IP fractions from healthy N. benthamiana leaves and leaves infiltrated with Mycx4, NSP, MP, HC-Pro, and BDMV DNA-A and DNA-B (Mycx4) or with Mycx4-H3, NSP, MP, HC-Pro, and BDMV DNA-A and DNA-B (Mycx4-H3). These extracts were used in the PCR with degenerate primers for the begomovirus DNA-A component (BDMV A) or DNA-B component (BDMV B). Internal controls were DNA extracts prepared from healthy (−) and BDMV-infected (+) N. benthamiana leaves. The expected sizes of the DNA-A (1.1 kb) and DNA-B (0.5 kb) fragments are indicated by arrowheads.
We next tested for the presence of viral DNA in the co-IP fractions from healthy N. benthamiana leaves and the Mycx4 and Mycx4-H3 treatments using PCR and the degenerate DNA-A or DNA-B primers (41). When DNA extracts prepared from co-IP fractions from healthy leaves were used in the PCR, no DNA fragments were amplified (Fig. 5B, lanes 1 and 6). Similarly, when DNA extracts prepared from co-IP fractions from leaves of the Mycx4 treatment were used in the PCR, no DNA fragments were amplified (Fig. 5B, lanes 2 and 7). However, when DNA extracts prepared from co-IP fractions from leaves expressing Mycx4-H3, NSP, MP, and HC-Pro and infected with BDMV DNA-A and DNA-B (Mycx4-H3 treatment) were used in the PCR, DNA fragments of the expected size were amplified with the DNA-A and DNA-B primers (Fig. 5B, lanes 3 and 8). Sequence analyses confirmed that these were BDMV DNA-A and DNA-B fragments. Similar results were obtained from three independent experiments. Collectively, these findings support the hypothesis that during the process of BDMV infection, a complex comprised of histone H3, NSP, MP, and viral DNA is formed.
Colocalization of histone H3, NSP, CP, and MP.
To further investigate the interactions between histone H3 and the BDMV NSP, MP, and CP, we next conducted subcellular localization studies with N. tabacum protoplasts. To this end, we generated transient-expression vectors to express NSP, MP, CP, and histone H3 fusion proteins having EGFP or monomeric red fluorescent protein (mRFP) at the C termini. In preliminary experiments, expression of the fusion proteins from these vectors in transfected protoplasts was confirmed by CLSM.
In control experiments, EGFP/mRFP fluorescence was observed over the cytoplasm and nucleoplasm but not over the nucleolus (see Fig. S6A in the supplemental material). Both H3-EGFP and H3-mRFP were localized over the nucleoplasm of transfected protoplasts, with only a weak signal observed over the nucleolus (see Fig. S6B). In these assays, DAPI staining confirmed the location of the nucleus (see Fig. S6C). Fluorescence associated with NSP-EGFP was also detected over the nucleoplasm, but in contrast to the pattern of fluorescence associated with histone H3 fusions, a strong signal also was detected over the nucleolus (see Fig. S6D). Similar results were obtained with CP-EGFP (data not shown). In protoplasts expressing MP-EGFP, fluorescence was detected at the nuclear and cell peripheries (see Fig. S6E). In other cases, the MP-EGFP fluorescence displayed a punctate pattern at the cell periphery (see Fig. S6F).
Coexpression of NSP-EGFP and histone H3-mRFP fusion proteins revealed their colocalization within the nucleus, with the signal localized predominantly over the nucleolus (see Fig. S6G to I in the supplemental material). Whereas some colocalization of the proteins was observed over the nucleoplasm, a particularly strong area of colocalization was a ring-like structure in or around the nucleolus (see Fig. S6I). Similar results were obtained when CP-EGFP and H3-mRFP fusions were coexpressed (see Fig. S6J to L). Although relatively little colocalization was observed when MP-EGFP and H3-mRFP were coexpressed in protoplasts, discrete spots of yellow fluorescence, indicating colocalization, were consistently observed at the nuclear periphery (see Fig. S6M to O). For all these combinations, similar results were obtained when the reverse combinations were used (e.g., H3-EGFP and NSP-mRFP/MP-mRFP/pCP-mRFP) (data not shown). Furthermore, these findings represent multiple independent experiments involving many hundreds of protoplasts per treatment.
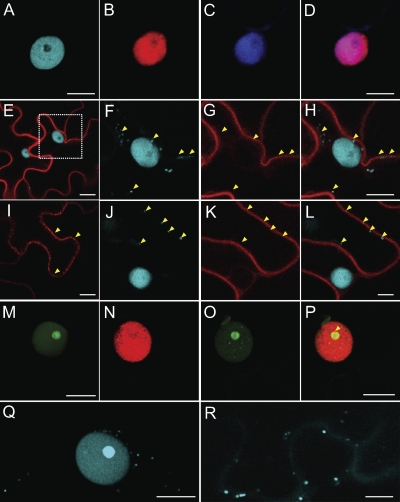
Colocalization studies were next performed by infiltrating N. benthamiana leaves with combinations of A. tumefaciens strains carrying binary vectors expressing the histone H3, NSP, and MP fusion proteins. As in our protoplast experiments, H3-ECFP and H3-mRFP localized over the nucleoplasm, with little signal over the nucleolus (Fig. 6A and B); DAPI staining confirmed the location of the nucleus (Fig. 6C and D). Also, similar to results with the protoplast experiments, MP-mRFP fluorescence was localized to the nuclear and cellular peripheries (data not shown). However, unlike the protoplast results, MP-mRFP fluorescence also was observed in punctate structures located over the cell wall (Fig. 6I). Evidence that these structures represented plasmodesmal pitfields came from coexpression experiments in which A. tumefaciens strains carrying binary vectors expressing BDMV MP-mRFP and the PD marker, TMV MP-GFP, were coinfiltrated into N. benthamiana leaves. Here, the BDMV MP-mRFP and TMV MP-GFP showed strong colocalization to the cell periphery and these punctate structures in the cell wall (see Fig. S7 in the supplemental material). Thus, these results suggest that these punctuate structures are PD, and they support results of previous studies indicating that the MP is targeted to PD (21, 35). When H3-ECFP and MP-mRFP strains were coinfiltrated into N. benthamiana leaves, MP-mRFP was localized at the nuclear and cellular peripheries (Fig. 6E, G, I, and K). Importantly, in these MP-mRFP/histone H3-ECFP coexpression experiments, H3-ECFP underwent a relocalization from the nucleus to sites within the cytoplasm and cell periphery (Fig. 6F and H). Furthermore, when MP-mRFP was localized to plasmodesmal pitfields (Fig. 6I and K), H3-ECFP was similarly colocalized (Fig. 6J and L).
Fig. 6.
Subcellular localization of Bean dwarf mosaic virus (BDMV) movement proteins (NSP and MP) and histone H3 in epidermal cells of N. benthamiana leaves following delivery and expression via Agrobacterium tumefaciens-mediated transient expression. (A) The nucleus of an epidermal cell in leaves infiltrated with H3-ECFP, exhibiting strong fluorescent signal over the nucleoplasm, but not over the nucleolus. (B) As in panel A but with H3-mRFP. (C) A representative nucleus revealed by DAPI staining. (D) Merged images of panels B and C, confirming localization of H3-mRFP over the nucleoplasm. (E) Coexpression of MP-mRFP and H3-ECFP. (F) Magnification of boxed area in panel E, showing egress of H3-ECFP from the nucleus into the cytoplasm in the presence of MP-mRFP; arrowheads identify regions of H3-ECFP accumulation. (G) Localization of MP-mRFP to the nuclear and cellular peripheries (arrowheads). (H) Merged images of panels F and G; arrowheads identify regions of H3-ECFP accumulation outside the nucleus. (I) MP-mRFP localized to punctate structures (arrowheads) either along or over the cell wall and that were subsequently identified as plasmodesmata (see Fig. S7 in the supplemental material). (J) H3-ECFP localized over the nucleus and punctate structures either along or over the cell wall (arrowheads). (K) MP-mRFP localized to cytoplasmic strands (arrowheads) extending from the nuclear to cell periphery. (L) Merged images of panels J and K; arrowheads indicate regions of H3-ECFP accumulation along the cell periphery. (M) NSP-EGFP strongly localized over the nucleolus. (N) H3-mRFP localized over the nucleus and nucleolus in coexpression experiments with NSP-EGFP. (O) Accumulation of NSP-EGFP within the nucleolus and foci in the nucleoplasm in coexpression experiments with H3-mRFP. (P) Merged images of panels N and O; note the colocalization of NSP-EGFP and H3-mRFP over a ring-like structure in or around the nucleolus (arrowhead). (Q) Accumulation of H3-ECFP within the nucleoli of cells infected with BDMV DNA-A and DNA-B. Note also the presence of an H3-ECFP signal located within the cytoplasm. (R) As in panel Q but at a later stage in the BDMV infection process; note the continued egress of H3-ECFP into the cytoplasm and cell periphery, including over PD-like punctate bodies in the cell wall. Scale bars = 10 μm. All CLSM images were collected 48 h after infiltration, except those in panels M to P, which were taken 24 h postinfiltration.
Parallel experiments performed with NSP-EGFP confirmed that it accumulated to high levels in the nucleolus (Fig. 6M). Coexpression experiments with NSP-EGFP and H3-mRFP revealed relocalization of H3-mRFP from the nucleoplasm to the nucleolus (Fig. 6N) and of NSP-EGFP from the nucleolus to discrete bodies within the nucleoplasm (Fig. 6O). Perhaps more notable was the colocalization of NSP-EGFP and H3-mRFP to a ring-like structure in or around the nucleolus (Fig. 6P); this pattern of colocalization was similar to that observed in the protoplast experiments (see Fig. S6I in the supplemental material). The relocalization of the NSP from the nucleolus to the nucleoplasm and the colocalization of histone H3 and NSP to the ring-like structure around the nucleolus support the notion that, in vivo, histone H3 interacts with the BDMV NSP.
Further insight into the involvement of histone H3 in the BDMV infection process was gained by performing experiments in which leaves were coinfiltrated with A. tumefaciens strains carrying binary vectors expressing H3-ECFP and the infectious multimeric BDMV DNA-A and DNA-B clones. Here, in contrast to the situation where H3-ECFP accumulated in the nucleoplasm when expressed alone (Fig. 6A), H3-ECFP accumulated to high levels in the nucleolus in the presence of BDMV infection (Fig. 6Q). Subsequently, the H3-ECFP signal egressed from the nucleus and accumulated in discrete bodies within the cytoplasm (Fig. 6Q) and over the cell wall (Fig. 6R); these bodies have been previously shown to be PD (see Fig. S7 in the supplemental material). These results provide evidence that histone H3 interacts with BDMV NSP and MP during the viral infection process.
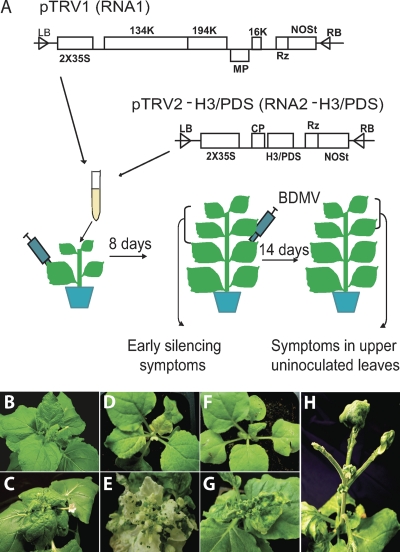
TRV-mediated expression of H3 induces abnormal plant development.
To further investigate the role of histone H3 in the BDMV infection process, the Tobacco rattle virus (TRV) silencing system (28, 29) was used to express various forms of the endogenous histone H3 gene in N. benthamiana in an attempt to silence the histone H3 gene. A subset of these plants was then inoculated with BDMV to assess the effect on the viral infection process (Fig. 7A). The phenotypes of wild-type and BDMV-infected plants are illustrated in Fig. 7B and C. In control experiments, TRV carrying the PDS gene was introduced via agroinoculation, and in these plants, the expected PDS silencing developed (Fig. 7D) (28). When a subset of these plants undergoing PDS silencing were agroinoculated with BDMV (8 days after inoculation with TRV-PDS), typical symptoms of BDMV infection developed at 7 to 10 days after inoculation and viral DNA was readily detected in symptomatic leaves by PCR (Fig. 7E). Thus, silencing of the PDS gene had no effect on the BDMV systemic infection process.
Fig. 7.
Attempts to silence the N. benthamiana histone H3 gene via the TRV vector elicit the development of an abnormal development phenotype. (A) Schematic of the TRV silencing vector used to express forms of the histone H3 or PDS gene in N. benthamiana. Plants were first infiltrated with Agrobacterium tumefaciens strains carrying binary vectors expressing pTRV1 and pTRV2-H3/PDS; 8 days later, an upper newly emerged leaf was infiltrated with A. tumefaciens strains carrying binary vectors with the infectious multimeric components of Bean dwarf mosaic virus (BDMV) DNA-A and DNA-B. At 14 days after BDMV inoculation (dpi), newly emerged leaves were examined for symptom development, and DNA and RNA were extracted for RT-PCR and Southern blot hybridization analyses, respectively (see Fig. S8 in the supplemental material). (B) Uninoculated control plant. (C) Stunted growth and leaf epinasty symptoms in a plant systemically infected with BDMV. (D) Plant infected with TRV-PDS, showing the phenotype associated with an inhibition of carotenoid biosynthesis (8 days dpi). (E) BDMV-infected plants from panel D at 14 dpi, showing both symptom development and silencing of the PDS gene. (F) Plant infected with TRV-H3 (full length) at 8 dpi; similar results were observed with plants infiltrated with TRV carrying various truncated forms of histone H3. (G) BDMV-infected plants from panel F at 14 dpi, showing symptom development and additional perturbation of apical development. (H) Plant infected with TRV-H3 (full length) at 21 dpi, exhibiting abnormal leaf development; similar results were observed with plants infiltrated with TRV carrying various truncated forms of histone H3.
An equivalent series of experiments was then performed using TRV carrying various forms of the N. benthamiana histone H3 gene. As illustrated in Fig. 7F and G, infection of plants with these TRV constructs failed to retard BDMV systemic infection, based upon the severity and time of appearance of the disease symptoms and the detection of viral DNA in symptomatic leaves. Unexpectedly, control plants agroinoculated with TRV-histone H3 constructs alone developed striking developmental abnormalities, including the failure of leaves to develop blades (Fig. 7H). To ascertain whether this abnormal phenotype was a consequence of histone H3 silencing, RT-PCR assays were performed. Whereas plants silenced for the PDS gene with the TRV system had a substantial reduction in the level of PDS gene expression, plants infected with TRV-histone H3 constructs showed equivalent or even enhanced levels of histone H3 gene expression relative to those in wild-type plants or in plants silenced for PDS gene expression (see Fig. S8 in the supplemental material). Thus, regardless of the histone H3 construct used, the TRV silencing system was unable to induce silencing of the N. benthamiana histone H3 gene. Moreover, plants in which histone H3 silencing was attempted developed an abnormal developmental phenotype, indicating that this led to a perturbation of normal plant development. Together, these results precluded a direct test of the role played by histone H3 in BDMV infection.
DISCUSSION
In the current model for cell-to-cell movement of bipartite begomoviruses, NSP and MP coordinate the trafficking of infectious viral DNA out of the nucleus, via the nuclear pore complex, and then cell to cell through PD (9, 14, 24, 30, 45, 46). What are not well understood are the nature of the DNA form(s) that moves from cell to cell and the means by which NSP and MP coordinate this process. In this study, we present evidence for interactions among the BDMV NSP and MP and the host factor histone H3. We further demonstrate the existence of a complex composed of histone H3, NSP, MP, and viral DNA, thereby suggesting that histone H3 may play a role in geminivirus cell-to-cell movement through the formation of a movement-competent complex.
The interaction of NSP and MP with histone H3 was established by gel overlay assays, and evidence for the specificity of these interactions was provided by the failure to detect an interaction between histone H3 and MPs of three RNA viruses and the CP of TMV. Gel overlay assays were also used to map the NSP and MP interaction domains to the N-terminal region of histone H3, a domain known to be involved in protein-protein interactions. In vitro and in vivo co-IP experiments confirmed the interactions among NSP, MP, and histone H3, and in vivo co-IP experiments performed with Myc-tagged histone H3 allowed for the recovery of a complex composed of histone H3, NSP, MP, and viral DNA. Transient-expression experiments revealed colocalization of histone H3 and NSP (and CP) in the nucleolus and redirection of histone H3 from the nucleoplasm into the nucleolus in the presence of NSP and CP, as well as during viral infection (Fig. 6Q). Perhaps more strikingly, in the presence of MP, the H3 histone nucleoprotein was redirected and localized to the cytoplasm and punctate bodies in the cell wall, which were identified as PD (Fig. 6R). Thus, the results of these transient-expression experiments supported the results of the gel overlay assays and the co-IP experiments and were fully consistent with the known subcellular localization of the BDMV NSP and MP (reference 44 and this study). Finally, gel overlay assays also revealed a direct interaction between the NSP and MP, which is likely to be essential in mediating the export of the histone H3-associated movement complex from the nucleus.
The finding that histone H3 interacts with geminiviral MPs and is part of a movement complex is perhaps not surprising given that these single-stranded DNA viruses replicate in the nucleus. Evidence for the interaction of histones with viral nucleic acids has been previously reported, including the formation of minichromosomes for a range of animal and plant viruses having a nuclear component in their life cycle. These include the bipartite begomovirus AbMV (37, 38), dsDNA viruses such as the plant-infecting pararetrovirus Cauliflower mosaic virus (36), the papillomavirus Simian virus 40 (54), and the herpesvirus Herpes simplex virus type 1 (18), and influenza virus, a segmented negative-stranded RNA virus (2). Geminiviruses replicate in the nucleus via a dsDNA replicative form (13, 50), and these forms can be assembled into minichromosomes via the formation of nucleosomes with histone proteins (37, 38). In addition, an interaction between the geminivirus replication-associated protein and histone H3 was detected in yeast two-hybrid assays. This was hypothesized to facilitate the replication and/or transcription of the viral genome, possibly via the displacement or alteration of nucleosomes (22). Thus, a role for histones in geminiviral replication and transcription has been previously suggested; however, our results provide evidence that histones are also involved in viral movement.
The interactions of NSP, MP, histone H3, and viral DNA are consistent with the formation of a movement-competent nucleoprotein complex. The role of histone H3 in this process may be in the initial packaging and compaction of viral DNA into a form that can be trafficked through the nuclear pore complex. This is consistent with the role of histones, especially H3, in the formation of nucleosomes and the packaging and compaction of host chromosomal DNA into chromatin and heterochromatin in the nucleus (34, 49). Nucleosome formation involves coiling of approximately 150 bp of DNA around a histone octamer core, with anywhere from ∼0 to 90 bp of DNA in between nucleosomes (31, 34, 49, 55). This results in generation of a DNA fiber of ∼10 nm in diameter, which has a “beads-on-a-string” appearance. Direct electron microscopy (EM) observation and mapping studies of AbMV minichromosomes have revealed 11 to 12 nucleosomes per ∼2,700-bp circular dsDNA molecule and a number of gaps or exposed stretches of DNA (37, 38). Thus, the geminivirus minichromosome would appear to have regions of DNA available for additional protein binding.
It is well established that the NSP is a DNA binding protein and that it binds ssDNA and dsDNA in a size- and form-specific manner (14, 43). Furthermore, NSP can cooperatively bind DNA, resulting in the generation of compact nucleoprotein structures (43). Thus, the finding that NSP strongly interacted with itself in gel overlay assays (Fig. 2A) is consistent with its cooperative DNA binding properties. In the formation of a movement complex, NSP may bind to exposed regions of DNA between the nucleosomes of the geminivirus minichromosome and/or to histone H3 to activate cooperative binding and DNA compaction. This could lead to the formation of a highly compact NSP-histone H3-DNA complex (<10 nm in diameter). The finding that NSP interacts with the N terminus of histone H3 suggests that this interaction may occur via the N-terminal tail of H3, a positively charged domain that protrudes from the nucleosome core and plays a role in protein-protein interactions (1, 34, 51). The influenza virus matrix protein interacts with histone tails in the formation of a nucleoprotein complex that is exported from the nucleus (10, 57). Thus, the NSP may bind to histone tails protruding from a histone-bound viral DNA form, possibly a dsDNA minichromosome, and trigger the formation of a compact infectious viral DNA form suitable for nuclear export (12). Furthermore, the observation that BDMV NSP and histone H3 were colocalized in or around the nucleolus provides evidence that these proteins interact, in vivo, and indicates that the NSP-histone H3-viral DNA complex may well form in the nucleolus, an organelle targeted by the BDMV NSP (44).
Nucleosomes typically are formed with dsDNA, but histones can also interact with ssDNA, and the R-H3 bound geminiviral dsDNA and ssDNA forms in gel shift assays. However, because nucleosomes form with geminiviral dsDNA in minichromosomes and NSP preferentially bound dsDNA in vitro (43), the form involved in the movement complex may well be dsDNA. Furthermore, ssDNA may be less available for formation of such a complex, as it is both encapsidated by CP to form virions and converted to dsDNA in the nucleus. However, the involvement of an ssDNA form in the formation of a movement complex cannot be discounted. The interaction of NSP with histone H3 may also be influenced by other host factors and processes involved in chromatin remodeling (e.g., acetylation, carbonylation, methylation, phosphorylation, sumoylation, and ubiquitination [34]). For example, AtNSI, an Arabidopsis acetyltransferase, acetylates histones H2A and H3 and interacts with the NSP and CP of the bipartite begomovirus Cabbage leaf curl virus (33). Additionally, several protein kinases interact with and phosphorylate NSP (7, 8, 32), and this could directly or indirectly alter interaction with histone H3 (17, 34, 51). Insight into the DNA form(s) in the movement complex and the role of posttranslational modifications and other host factors will come from biochemical analyses of the complex.
Our overlay assays revealed direct interactions between MP/NSP and histone H3, as well as between MP and NSP. A direct interaction between NSP and MP has long been postulated based on results of genetic analyses and protoplast studies in which NSP was redirected to the cell periphery in the presence of MP (9, 14, 24, 45, 46). However, our gel overlay assay results now provide direct evidence that these proteins interact. Support for a direct interaction between MP and histone H3 in planta came from results of our in vivo colocalization studies, in which histone H3 was redirected to the cell periphery and PD-like punctate structures in the presence of MP (Fig. 6I to L). Evidence that these punctuate structures were PD came from the colocalization of the BDMV MP with the TMV MP, a known PD marker. Together, these results suggest that histone H3 is exported from the nucleus and delivered to PD through an interaction with MP, which is targeted to the nuclear and cellular peripheries (44, 56). This likely occurs via the histone H3-NSP-DNA complex (Fig. 6N to P) and may involve MP interacting with both the NSP and histone H3 components of the complex during the process of nuclear export and delivery to PD. Consistent with this notion was our discovery that during the BDMV infection process, histone H3 first is directed into the nucleolus and then is subsequently exported into the cytoplasm and to PD-like structures in the cell wall (Fig. 6Q and R).
The nature of the presumed BDMV DNA complex that passes through PD is unknown. At least in some hosts, the complex is not the virion, nor does it require the CP, because BDMV CP mutants are highly infectious and effectively spread cell to cell and long distance (12, 52, 58). On the other hand, the movement complex most likely contains MP, as this viral protein is required for BDMV cell-to-cell movement, moves through PD, and binds to and mediates cell-to-cell movement of ssDNA and dsDNA (12, 35). Furthermore, the AbMV MP has been immunolocalized to PD (21). The binding of MP to histone H3 and NSP may further compact or modify the histone H3-NSP-viral DNA complex, thereby making it competent for movement through PD. In this scenario, both histone H3 and NSP would be trafficked cell to cell with the complex and could possibly mediate subsequent nuclear import of the viral DNA in adjacent uninfected cells. Alternatively, MP may displace NSP or histone H3 from the complex to form a new type of complex that is competent for cell-to-cell spread through PD. The latter scenario may better accommodate the DNA binding properties of the BDMV MP and its role in imposing a size constraint on the viral genome (12).
To directly investigate the role played by histone H3 in BDMV infection, we attempted to silence the N. benthamiana histone H3 gene with the TRV vector system. We were unable to trigger efficient silencing of H3 (see Fig. S8 in the supplemental material), perhaps because of the large family of histone genes in plants (e.g., 45 core histone genes in Arabidopsis) (55) and/or the essential role that these proteins play in plant growth and development. Thus, our inability to silence the histone H3 gene with the TRV system and the resulting abnormal developmental phenotype associated with these plants (Fig. 7H) are fully consistent with the influence of histones and histone modification on almost all aspects of plant growth and development (34, 49, 55). Hence, silencing of the histone H3 gene would likely have produced plants with major development defects, and such plants would have been unsuitable for viral infection assays.
Supplementary Material
ACKNOWLEDGMENTS
The TRV vector was kindly provided by S. P. Dinesh-Kumar. We thank Tatsuya Kon for assistance with the R-H3 in vitro DNA binding experiments.
This research was supported by U.S. Department of Agriculture National Research Initiative Competitive Grant 2002-01418 (to R.L.G. and W.J.L.). Y.Z. was supported in part by a Graduate Fellowship from the University of California—Davis.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 7 September 2011.
REFERENCES
- 1. Arents G., Burlingame R. W., Wang B. W., Love W. E., Moudrianakis E. N. 1991. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. U. S. A. 88: 10148–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bui M., Wills E. G., Helenius A., Whittaker G. R. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74: 1781–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carvalho M. F., Lazarowitz S. G. 2004. Interaction of the movement protein NSP and the Arabidopsis acetyltransferase AtNSI is necessary for cabbage leaf curl geminivirus infection and pathogenicity. J. Virol. 78: 11161–11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carvalho M. F., Turgeon R., Lazarowitz S. G. 2006. The geminivirus nuclear shuttle protein NSP inhibits the activity of AtNSI, a vascular-expressed Arabidopsis acetyltransferase regulated with the sink-to-source transition. Plant Physiol. 140: 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carvalho C. M., et al. 2008. A novel nucleocytoplasmic traffic GTPase identified as a functional target of the bipartite geminivirus nuclear shuttle protein. Plant J. 55: 869–880 [DOI] [PubMed] [Google Scholar]
- 6. Donald R. G. K., Zhou H., Jackson A. O. 1993. Serological analysis of Barley stripe mosaic virus-encoded proteins in infected barley. Virology 195: 659–668 [DOI] [PubMed] [Google Scholar]
- 7. Florentino L. H., et al. 2006. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J. Virol. 80: 6648–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fontes E. P., Santos A. A., Luz D. F., Waclawovsky A. J., Chory J. 2004. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 18: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gafni Y., Epel B. L. 2002. The role of host and viral proteins in intra- and inter-cellular trafficking of geminiviruses. Physiol. Mol. Plant Pathol. 60: 231–241 [Google Scholar]
- 10. Garcia-Robles I., Akarsu H., Muller C. W., Ruigrok R. W., Baudin F. 2005. Interaction of influenza virus proteins with nucleosomes. Virology 332: 329–336 [DOI] [PubMed] [Google Scholar]
- 11. Gilbertson R. L., Lucas W. J. 1996. How do viruses traffic on the vascular highway. Trends Plant Sci. 1: 260–268 [Google Scholar]
- 12. Gilbertson R. L., Sudarshana M., Jiang H., Rojas M. R., Lucas W. J. 2003. Limitations on geminivirus genome size imposed by plasmodesmata and virus-encoded movement protein: insights into DNA trafficking. Plant Cell 15: 2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanley-Bowdoin L., Settlage S. B., Orozco B. M., Nagar S., Robertson D. 1999. Geminiviruses: models for plant DNA replication, transcription and cell cycle regulation. Crit. Rev. Plant Sci. 18: 71–106 [PubMed] [Google Scholar]
- 14. Hehnle S., Wege C., Jeske H. 2004. Interaction of DNA with movement proteins of geminiviruses revisited. J. Virol. 78: 7698–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hou Y.-M., Paplomatas E. J., Gilbertson R. L. 1998. Host adaptation and replication properties of two bipartite geminiviruses and their pseudorecombinants. Mol. Plant Microbe Interact. 11: 208–217 [Google Scholar]
- 16. Hou Y.-M., Sanders R., Ursin V. M., Gilbertson R. L. 2000. Transgenic plants expressing geminivirus movement proteins: abnormal phenotypes and delayed infection by Tomato mottle virus in transgenic tomatoes expressing the Bean dwarf mosaic virus BV1 or BC1 proteins. Mol. Plant Microbe Interact. 13: 297–308 [DOI] [PubMed] [Google Scholar]
- 17. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- 18. Kent J. R., et al. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78: 10178–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kikuno R., Toh H., Hayashida H., Miyata T. 1984. Sequence similarity between putative gene products of geminivirus DNAs. Nature 308: 562. [DOI] [PubMed] [Google Scholar]
- 20. Kleinow T., et al. 2009. Three C-terminal phosphorylation sites in the Abutilon mosaic virus movement protein affect symptom development and viral DNA accumulation. Virology 390: 89–101 [DOI] [PubMed] [Google Scholar]
- 21. Kleinow T., et al. 2009. Expression dynamics and ultrastructural localization of epitope-tagged Abutilon mosaic virus nuclear shuttle and movement proteins in Nicotiana benthamiana cells. Virology 391: 212–220 [DOI] [PubMed] [Google Scholar]
- 22. Kong L.-J., Hanley-Bowdoin L. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krenz B., Windeisen V., Wege C., Jeske H., Kleinow T. 2010. A plastid-targeted heat shock cognate 70 kDa protein interacts with the Abutilon mosaic virus movement protein. Virology 401: 6–17 [DOI] [PubMed] [Google Scholar]
- 24. Lazarowitz S. G., Beachy R. N. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J. Y., et al. 2003. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299: 392–396 [DOI] [PubMed] [Google Scholar]
- 26. Lee J.-Y., et al. 2005. Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levy A., Tzfira T. 2010. Bean dwarf mosaic virus: a model system for the study of viral movement. Mol. Plant Pathol. 11: 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y., Schiff M., Dinesh-Kumar S. P. 2002. Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- 29. Lu R., Martin-Hernandez A. M., Peart J. R., Malcuit I., Baulcombe D. C. 2003. Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- 30. Lucas W. J. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- 31. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- 32. Mariano A. C., et al. 2004. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 318: 24–31 [DOI] [PubMed] [Google Scholar]
- 33. McGarry R. C., et al. 2003. A novel Arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. Plant Cell 15: 1605–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nelissen H., Boccardi T. M., Himanen K., van Lijsebettens M. 2007. Impact of core histone modifications on transcriptional regulation and plant growth. Crit. Rev. Plant Sci. 26: 243–263 [Google Scholar]
- 35. Noueiry A. O., Lucas W. J., Gilbertson R. L. 1994. Proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76: 925–932 [DOI] [PubMed] [Google Scholar]
- 36. Olszewski N., Hagen G., Guilfoyle T. J. 1982. A transcriptionally active, covalently closed minichromosome of cauliflower mosaic virus DNA isolated from infected turnip leaves. Cell 29: 395–402 [DOI] [PubMed] [Google Scholar]
- 37. Pilartz M., Jeske H. 1992. Abutilon mosaic geminivirus double-standed DNA is packed into minichromosomes. Virology 189: 800–802 [DOI] [PubMed] [Google Scholar]
- 38. Pilartz M., Jeske H. 2003. Mapping of Abutilon mosaic geminivirus minichromosomes. J. Virol. 77: 10808–10818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin S., Ward B. M., Lazarowitz S. G. 1998. The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 72: 9247–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Radhakrishnan G. K., Splitter G. A., Usha R. 2008. DNA recognition properties of the cell-to-cell movement protein (MP) of soybean isolate of Mungbean yellow mosaic India virus (MYMIV-Sb). Virus Res. 131: 152–159 [DOI] [PubMed] [Google Scholar]
- 41. Rojas M. R., Gilbertson R. L., Russell D. R., Maxwell D. P. 1993. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 77: 340–347 [Google Scholar]
- 42. Rojas M. R., Zerbini F. M., Allison R. F., Gilbertson R. L., Lucas W. J. 1997. Capsid protein and helper component-proteinase function as potyvirus cell-to-cell movement proteins. Virology 237: 283–295 [DOI] [PubMed] [Google Scholar]
- 43. Rojas M. R., Noueiry A. O., Lucas W. J., Gilbertson R. L. 1998. Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form-and size-specific manner. Cell 95: 105–113 [DOI] [PubMed] [Google Scholar]
- 44. Rojas M. R., et al. 2001. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 291: 110–125 [DOI] [PubMed] [Google Scholar]
- 45. Rojas M. R., Hagen C., Lucas W. J., Gilbertson R. L. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43: 361–394 [DOI] [PubMed] [Google Scholar]
- 46. Sanderfoot A. A., Lazarowitz S. G. 1995. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scholthof H. B. 2005. Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10: 376–382 [DOI] [PubMed] [Google Scholar]
- 48. Seo Y.-S., et al. 2006. A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc. Natl. Acad. Sci. U. S. A. 103: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith J. G., Hill R. S., Baldwin J. P. 1995. Plant chromatin structure and post-translational modifications. Crit. Rev. Plant Sci. 14: 299–328 [Google Scholar]
- 50. Stenger D. C., Revington G. N., Stevenson M. C., Bisaro D. M. 1991. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-cycle replication of a plant viral DNA. Proc. Natl. Acad. Sci. U. S. A. 88: 8029–8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Strahl B. D., Allis C. D. 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- 52. Sudarshana M. R., Wang H. L., Lucas W. J., Gilbertson R. L. 1998. Dynamics of Bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol. Plant Microbe Interact. 11: 277–291 [Google Scholar]
- 53. Taoka K.-I., Ham B.-K., Xoconostle-Cázares B., Rojas M. R., Lucas W. J. 2007. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPPl interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell 19: 1866–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Varshavsky A. J., Bakayev V. V., Chumackov P. M., Georgiev G. P. 1976. Minichromosome of simian virus 40: presence of histone H1. Nucleic Acids Res. 3: 2101–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Verbsky M. L., Richards E. J. 2001. Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4: 494–500 [DOI] [PubMed] [Google Scholar]
- 56. Zhang S. C., Wege C., Jeske H. 2001. Movement proteins (BC1 and BV1) of Abutilon mosaic geminivirus are cotransported in and between cells of sink but not of source leaves as detected by green fluorescent protein tagging. Virology 290: 249–260 [DOI] [PubMed] [Google Scholar]
- 57. Zhirnov O. P., Klenk H. D. 1997. Histones as a target for influenza virus matrix protein M1. Virology 235: 302–310 [DOI] [PubMed] [Google Scholar]
- 58. Zhou Y. C., Garrido-Ramirez E. R., Sudarshana M. R., Yendluri S., Gilbertson R. L. 2007. The N-terminus of the begomovirus nuclear shuttle protein (BV1) determines virulence or avirulence in Phaseolus vulgaris. Mol. Plant Microbe Interact. 20: 1523–1534 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.