Abstract
Genomic DNA synthesis is a universally conserved process for all herpesviruses, including human cytomegalovirus (HCMV). HCMV UL70 is believed to encode the primase of the DNA replication machinery, a function which requires localization in the nucleus, the site of viral DNA synthesis. No host factors that interact with UL70 have been reported. In this study, we provide the first direct evidence that UL70 specifically interacts with Snapin, a human protein that is predominantly localized in the cytoplasm and is associated with cellular vesicles. The interaction between UL70 and Snapin was identified in both the two-hybrid screen in yeast and coimmunoprecipitation in human cells. The nuclear import of UL70 was decreased in cells overexpressing Snapin and increased in cells in which the expression of Snapin was downregulated with anti-Snapin small interfering RNA (siRNA) molecules, respectively. Furthermore, viral DNA synthesis and progeny production were decreased in cells overexpressing Snapin and increased in the anti-Snapin siRNA-treated cells, respectively. In contrast, no significant difference in the nuclear level of UL70, viral DNA synthesis, and progeny production was found among the parental cells and cells that either expressed a control empty vector or were treated with control siRNA molecules that did not recognize any viral or cellular transcripts. Our results suggest that Snapin may play a key role in regulating the cellular localization of UL70 in HCMV, leading to modulation of viral DNA synthesis and progeny production.
INTRODUCTION
Human cytomegalovirus (HCMV), the leading viral cause of congenital abnormalities and mental retardation in newborns, is a member of the herpesvirus family, which includes herpes simplex virus 1 (HSV-1), Epstein-Barr virus (EBV), and Kaposi's sarcoma-associated herpesvirus (KSHV) (24). The hallmarks of HCMV pathogenesis include infection of a wide range of tissues and cells, such as neuronal cells, and the establishment of lytic and latent infection in many of these tissues (7, 30). In lytic productive infection, the HCMV gene products are expressed in a temporally regulated cascade consisting of three sequential phases described as immediate-early (IE), early (E), and late (L) phases (24). Genomic DNA replication, which occurs in the nucleus of infected cells (25), represents one of the processes most highly conserved among all herpesviruses and is the target for most of the current FDA-approved antiherpesvirus therapeutic agents (6). Understanding the process of viral DNA replication is critical for developing new strategies and novel compounds for the treatment and prevention of HCMV infection.
During lytic infection, HCMV synthesizes its genome from a single origin (oriLyt) of replication using a rolling-circle mode of DNA replication, and this process requires the activities of specific viral proteins (24, 25). Six HCMV-encoded proteins, which are also highly conserved in other herpesviruses, provide the core functions for viral DNA replication: the DNA polymerase (UL54), the associated polymerase processivity factor (UL44), the single-stranded DNA binding protein (UL57), the helicase (UL105), the primase (UL70), and the primase-associated factor (UL102) (24, 25).
Primase is a DNA-dependent RNA polymerase that synthesizes a short RNA primer for the Okazaki fragments made by DNA polymerase during discontinuous DNA replication (3, 45). In eukaryotic cells, primase is a heterodimer of a small and a large subunit and usually forms a complex with DNA polymerase alpha (3). HCMV primase and its herpesvirus homologues form a tight helicase-primase complex, which consists of UL105, UL70, and UL102 in HCMV (21, 35, 36, 46). This heterotrimeric complex possesses three main activities: primase, DNA-dependent NTPase, and 5′–3′ helicase. The helicase tracks along the lagging strand and unwinds the DNA in front of the replication fork, and the NTPase provides the energy needed for unwinding (24, 25). It is believed that the UL70 primase synthesizes short RNA primers for single-stranded DNA, which the DNA polymerase UL54 extends via deoxynucleoside triphosphate (dNTP) polymerization (44). Although the precise role of each subunit needs further investigation, it would be expected, by analogy with observations for other herpesviruses (e.g., HSV-1), that an assembled subcomplex containing the UL105 and UL70 subunits retains all three activities, while the UL102 subunit modulates these activities (5, 13, 20, 21). UL70 is believed to contain the complete primase active site in terms of phosphodiester bond formation, although it might not be able to initiate primer synthesis in the absence of UL105 (44).
During lytic infection, viral DNA replication compartments, which are the sites for both input viral genome deposition and gene transcription, form dot-like subnuclear structures located in or close to specific nuclear regions named nuclear domain 10 (ND10) sites (27). However, little is known about the pathway by which the HCMV replication machinery is assembled. Equally elusive is how the UL70-encoded essential primase is imported in the nuclear compartment where viral DNA replication occurs (33).
Extensive studies have been carried out to investigate human-HCMV interactions and the host responses to HCMV infection (24). For example, many HCMV proteins, including the DNA replication core proteins UL44, UL54, and UL57, which possess nuclear localization signal sequences (NLS), have been found to interact with human cellular importin to facilitate importation of the newly synthesized viral proteins into the nucleus through the nuclear pore complex (1, 2, 14, 28, 29, 34). Currently there is no report of interactions of UL70 or other herpesvirus primases with any host proteins. While it is well known that the human primase forms a complex with DNA polymerase alpha, few interactions of human or eukaryotic primase with other eukaryotic proteins, including p53, RAE1, and MRPL38, have been reported (22, 37, 39). The interaction of p53 with the primase can help the polymerase-primase complex elongate a primer with a mispaired 3′ end by the exonuclease activity of p53, while other functions of these interactions are still unknown (22).
In an effort to gain a better understanding of the viral DNA replication process, in this study we performed a yeast two-hybrid (YTH) screen to identify human proteins that potentially interact with UL70. Our results provide the first direct evidence that UL70 interacts with Snapin, a cellular vesicle-associated protein that is localized predominantly in the cytoplasm (4, 8, 40). The level of UL70 in the nucleus, the site of viral DNA synthesis, is reduced in cells overexpressing Snapin and is increased in cells in which Snapin expression is downregulated. Furthermore, the level of viral DNA synthesis and progeny production is decreased and increased in cells with upregulated and downregulated levels of Snapin, respectively. These results suggest that Snapin may play a key role in regulating the cellular localization of UL70, leading to modulation of viral DNA synthesis and lytic productive infection.
MATERIALS AND METHODS
Construction of plasmids.
The constructs generated in this study are listed in Table 1. To generate pGBKT7-UL70 for the yeast two-hybrid screen and pCMV-Myc-UL70 for expression in human cells, the coding sequence of HCMV UL70 was amplified by PCR from the DNA of TowneBAC (10), using the primers 70-F (5′-CATCATATGGGGTCGACGATGACGCTCGTTCTGTT–3′) and 70-R (5′-GGGGATCCGGTACCGTGGAAAGTGAGGCTCAGAC-3′), and then inserted into NdeI/BamHI-digested pGBKT7 (Clontech, Mountain View, CA) and SalI/KpnI-digested pCMV-Myc (Clontech), respectively. The constructs that contained the sequences encoding different UL70 deletion mutants were generated by PCR using pGBKT7-UL70 as the template, followed by insertion of the amplified PCR products into NdeI/BamHI-digested pGBKT7 for yeast two-hybrid analysis and SalI/KpnI-digested pCMV-Myc for expression in human cells. The resultant constructs were confirmed by restriction digestion profile and sequencing. The constructs containing HCMV UL25 and UL83 sequences have been previously described (41).
Table 1.
Plasmid constructs used in the study
| Plasmid | Description (positions [bp]) | Reference/source |
|---|---|---|
| pGBKT7 | Cloning vector for protein expression fused with GAL4 DNA-binding domain in yeast | Clontech |
| pACT2 | Cloning vector for protein expression fused with GAL4 activation domain in yeast | Clontech |
| pCMV-HA | Cloning vector for protein expression fused with hemagglutinin (HA) tag in mammalian cell | Clontech |
| pCMV-Myc | Cloning vector for protein expression fused with c-Myc tag in mammalian cell | Clontech |
| pGBKT7-UL70 | pGBKT7 containing HCMV UL70 full-length sequence (1–2841) | This study |
| pGBKT7-UL70 (1–866aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–2600) | This study |
| pGBKT7-UL70 (1–800aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–2400) | This study |
| pGBKT7-UL70 (1–733aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–2200) | This study |
| pGBKT7-UL70 (1–666aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–2000) | This study |
| pGBKT7-UL70 (1–600aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–1800) | This study |
| pGBKT7-UL70 (1–533aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–1600) | This study |
| pGBKT7-UL70 (1–466aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–1400) | This study |
| pGBKT7-UL70 (1–400aa) | pGBKT7 containing UL70 C-terminally truncated sequence (1–1200) | This study |
| pGBKT7-UL70 (67–946aa) | pGBKT7 containing UL70 N-terminally truncated sequence (199–2841) | This study |
| pGBKT7-UL70 (134–946aa) | pGBKT7 containing UL70 N-terminally truncated sequence (400–2841) | This study |
| pGBKT7-UL70 (67–733aa) | pGBKT7 containing UL70 truncated sequence (199–2200) | This study |
| pGBKT7-UL70 (67–866aa) | pGBKT7 containing UL70 truncated sequence (199–2600) | This study |
| pCMV-Myc-UL70 | pCMV-Myc containing HCMV UL70 full-length sequence (1–2841) | This study |
| pCMV-Myc-UL70 (1–866aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–2600) | This study |
| pCMV-Myc-UL70 (1–800aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–2400) | This study |
| pCMV-Myc-UL70 (1–733aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–2200) | This study |
| pCMV-Myc-UL70 (1–666aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–2000) | This study |
| pCMV-Myc-UL70 (1–600aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–1800) | This study |
| pCMV-Myc-UL70 (1–533aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–1600) | This study |
| pCMV-Myc-UL70 (1–466aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–1400) | This study |
| pCMV-Myc-UL70 (1–400aa) | pCMV-Myc containing UL70 C-terminally truncated sequence (1–1200) | This study |
| pCMV-Myc-UL70 (67–946aa) | pCMV-Myc containing UL70 N-terminally truncated sequence (199–2841) | This study |
| pCMV-Myc-UL70 (134–946aa) | pCMV-Myc containing UL70 N-terminally truncated sequence (400–2841) | This study |
| pCMV-Myc-UL70 (67–733aa) | pCMV-Myc containing UL70 truncated sequence (199–2200) | This study |
| pCMV-Myc-UL70 (67–866aa) | pCMV-Myc containing UL70 truncated sequence (199–2600) | This study |
| pCMV-Snapin | pCMV containing full-length human Snapin-encoding sequence (1–411) | This study |
| pCMV-HA-Snapin | pCMV-HA containing full-length human Snapin-encoding sequence (1–411) | This study |
YTH analysis.
Saccharomyces cerevisiae strain AH109, control vectors pGADT7, pGADT7-T, and pGBKT7-p53, and a human fetal brain cDNA library were obtained from Clontech (Mountain View, CA). The yeast strain containing pGBKT7-UL70 was transformed with the library, and positive clones were selected on synthetic dropout (SD) medium lacking four nutrients, tryptophan, leucine, adenine, and histidine (SD-minus Trp/Leu/Ade/His [QDO]), and tested for β-galactosidase activity by a colony-lift filter assay according to the manufacturer's instructions and following the procedures described previously (41). DNA constructs that contained the sequence coding for the interaction partners of UL70 (designated pACT2-cDNA) from positive colonies were extracted from yeast and rescued by transformation of competent DH5α Escherichia coli. Each pACT2-cDNA plasmid rescued from E. coli was used to cotransform the yeast with pGBKT7-UL70 or empty bait vector pGBKT7 as a control. The transformed cells were plated on QDO and also subjected to a β-galactosidase activity test. Blue yeast colonies were selected, and the human gene sequences in the pACT2-cDNA constructs from these selected colonies were determined with a sequencing primer (5′-AATACCACTACAATGGAT-3′).
Antibodies, viruses, and cells.
The antibodies against human actin and Snapin were purchased from Sigma, Inc. (St. Louis, MO) and Santa Cruz Biotech, Inc. (Santa Cruz, CA), respectively. The monoclonal antibodies that react with the HCMV proteins UL44, IE1, and UL99 have been described previously (43, 49). Human foreskin fibroblasts (HFFs), astrocytoma U373MG cells, HeLa cells, and 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum as described previously (17, 41). HCMV (strain Towne and TowneBAC) was propagated in HFFs and U373MG cells (10).
To generate the U373-S and U373-C cell lines, DNAs of the constructs pCMV-Snapin and the empty vector pCMV were cotransfected into U373MG cells with DNA of the LXSN vector (17). At 48 to 72 h postinfection, neomycin was added to the culture medium at a final concentration of 600 μg/ml. Cells were subsequently selected in the presence of neomycin for 2 weeks, and neomycin-resistant cells were cloned (18, 23). The levels of Snapin in individual cell clones were determined by Western analysis.
Coimmunoprecipitation and Western blot analysis.
Cells were cotransfected with the DNAs of pCMV-Myc-UL70 and its derivative UL70 mutant constructs and pCMV-HA-Snapin with the aid of Lipofectamine (Invitrogen, Carlsbad, CA). At 48 h posttransfection, cell lysates were harvested and coimmunoprecipitation experiments were performed using the ProFound mammalian hemagglutinin (HA) tag immunoprecipitation/coimmuniprecipitation (IP/co-IP) and c-Myc tag IP/co-IP kits, following the manufacturer's protocol (Pierce, Rockford, IL) (41).
To carry out Western analysis, the denatured polypeptides from cell lysates or co-IP were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrically to nitrocellulose membranes. Membranes were blocked in 5% dried milk in phosphate-buffered saline (PBS) plus 0.2% Tween 20. Detection of tagged proteins was performed by incubation with 1:5,000-diluted mouse anti-Myc or 1:500-diluted mouse anti-HA antibodies (Santa Cruz Biotech, Santa Cruz, CA) and reacted in an enzyme-linked immunoassay with 1:2,000-diluted horse anti-mouse IgG antibody conjugated with horseradish peroxidase (Vector Laboratories, Burlingame, CA). The membranes were subsequently stained with a chemiluminescent substrate with the aid of ECL Western blotting detection reagent kits (GE Healthcare) and quantitated either by densitometry of the films exposed to the samples or with a Storm840 phosphorimager. The experiments were carried out in duplicate and repeated three times. Quantitation was performed in the linear range of protein detection.
Transfection of siRNA into cells.
Cells (n = 1 × 105) were seeded in 12-well plates and transfected with chemically synthetic anti-Snapin siRNA (S-siRNA) and control siRNA (C-siRNA) molecules (Santa Cruz Biotech, Santa Cruz, CA). For each well, 3 μl 20 mM siRNA and 3 μl Lipofectamine (Invitrogen) were diluted in 50 μl and 12 μl Opti-MEM (Invitrogen), respectively. After 5 min at room temperature, both solutions were combined. After 20 min, 400 μl prewarmed Opti-MEM was added to each transfection reaction mixture, which was subsequently added to the cells. At 10 h posttransfection, siRNA-containing medium was removed. Cells were washed and incubated with complete DMEM supplemented with 10% fetal bovine serum, and the transfection was repeated once. Forty-eight hours after the second round of transfection, cells were either infected or prepared for analysis by immunoblotting.
Immunofluorescence microscopy analysis.
Cells that were grown on glass coverslips were cotransfected with DNAs of the different constructs with the aid of Lipofectamine (Invitrogen) (11). At 48 h posttransfection, cells were either mock infected or infected with HCMV at a multiplicity of infection (MOI) of 1 to 5. At different time points postinfection, cells were fixed with 4% formaldehyde and blocked with 0.1% bovine serum albumin (BSA) and 0.1% sodium azide in PBS and stained with primary antibodies, followed by incubation with the secondary antibody Alexa Fluor 555 anti-mouse IgG or Alexa Fluor 488 anti-rabbit IgG (Invitrogen). Nuclear staining was performed with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Images were captured with a Nikon Eclipse TE300 microscope using a Spot RT Slider camera and imaging software (Diagnostic Instruments, Inc., Sterling Heights, MI) (11). The digital images were subsequently merged using the SimplePCI software program (Compix Inc., Sewickley, PA).
Viral infection and assays for viral gene expression and growth.
Cells (n = 1 × 106) were either mock infected or infected with HCMV at a multiplicity of infection (MOI) of 0.5 to 5 in an inoculum of 1.5 ml of DMEM supplemented with 1% fetal bovine serum. The inoculum was replaced with DMEM supplemented with 10% fetal bovine serum after 2 h of incubation with cells. Protein extracts were prepared from the infected cells at 12 to 72 h postinfection and analyzed by Western blot experiments as described previously (43, 49).
To determine the level of viral growth, cells (n = 1 × 105) were infected with HCMV at an MOI of 1. The cells and medium were harvested at 1, 2, 3, 4, 5, 6, and 7 days postinfection, and viral stocks were prepared by adding an equal volume of 10% (vol/vol) skim milk, followed by sonication. The titers of the viral stocks were determined by infecting 1 × 105 human foreskin fibroblasts and counting the number of plaques 10 to 14 days after infection. The values obtained were averages from triplicate experiments.
Nuclear and cytoplasmic extract preparation.
Cells that were either mock infected or infected with HCMV were harvested at different time points postinfection and resuspended in buffer A (10 mM HEPES [pH 7.4], 10 mM KCl, 1 mM dithiothreitol, 0.6% NP-40) at 4°C for 10 min. After centrifugation at 1,000 × g for 5 min, the supernatant was collected as the cytoplasmic fraction and the nuclear fraction was prepared by resuspending the remaining pellet in buffer B (20 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM dithiothreitol). Equal amounts of nuclear and cytoplasmic extracts were used for immunoblotting of UL70, actin, and histone H1.
Assaying the level of intracellular HCMV genome.
Quantitative real-time PCR (qPCR) analysis of viral DNA was carried out as described elsewhere (15). Briefly, DNA was isolated from mock-infected or HCMV-infected cells using a DNeasy tissue kit (Qiagen) according to the manufacturer's instructions. The levels of intracellular viral DNAs were quantified with a primer pair (P53 [5′-GTCAGCGTTCGTGTTTCCCA-3′] and P33 [5′-GGGACACAACACCGTAAAGC-3′]) and a probe for amplifying the HCMV UL83 sequence. The TaqMan probe (5′-6-carboxy-fluorescein [FAM]CCCGCAACCCGCAACCCTTCATG [TAMRA]-3′) was purchased from Applied Biosystems Inc. (Foster City, CA) and was labeled at the 5′ end with FAM and at the 3′ end with the fluorescent quencher TAMAR. The number of viral genomes was normalized to the number of cellular copies of β-actin with a previously described set of primers and probe (15). Unknown sample values were determined on the basis of a standard curve of known copy numbers of UL83 (TowneBAC) and β-actin (pβ-actin). Amplification was performed in a 50-μl reaction mixture (which contained 21 μl of DNA extract, 25 μl of 2× TaqMan Universal PCR master mix [Applied Biosystems Inc.], 1 μl of each primer at 10 μM, and 2 μl of the fluorogenic probe at 5 μM) using either the iCycler real-time PCR detection system (Bio-Rad, Hercules, CA) or an ABI 7500 device (Applied Biosystems Inc., Foster City, CA). Thermal cycling conditions were as follows: 50°C for 2 min, 95°C for 15 min, and 45 cycles of 95°C for 30 s and 60°C for 1 min. The PCR results were derived from three independent experiments.
RESULTS
Identification of potential interaction of Snapin with UL70 by yeast two-hybrid analysis.
We previously determined the genomic sequence of an HCMV Towne strain that was cloned into a bacterial artificial chromosome (BAC) vector (19) and was maintained as a single-copy bacterial artificial chromosome (BAC)-based plasmid in E. coli (10). When introduced into human cells, the viral DNA sequence of this construct, TowneBAC, produces infectious progeny and retains wild-type growth characteristics in vitro. These results suggested that the genome sequence of TowneBAC encodes a collection of functional viral genes responsible for viral replication and infection (10, 12). Construct pGBKT7-UL70, in which the DNA sequence encoding the UL70 open reading frame (ORF) was amplified by PCR using the DNA of TowneBAC as the template and inserted into the pGBKT7 yeast expression vector, was generated. The inserted UL70 sequence was further confirmed by sequencing analysis (data not shown).
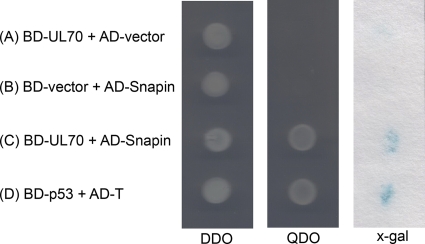
The GAL4-based yeast two-hybrid system was employed to identify cellular partners that potentially interact with HCMV UL70. To identify cellular factors that potentially interact with UL70, the yeast two-hybrid screen was performed by transformation of S. cerevisiae AH109 containing pGBKT7-UL70 with a cDNA library derived from human fetal brain. Those yeast cells that grew on synthetic dropout (SD) medium lacking four nutrients, tryptophan, leucine, adenine, and histidine (SD-minus Trp/Leu/Ade/His [QDO]) and yielded blue signals in a colony-lift filter test for β-galactosidase activity were identified and defined as primary positive clones. One of the positive constructs, pACT2-SNAPIN, which contained the full-length coding sequence of Snapin, was consistently found to interact with UL70 in our yeast two-hybrid screens (Fig. 1C). Of nearly 1.5 × 107 independent cDNA clones tested, 13 yeast colonies yielded positive results, 3 of which contained plasmid constructs with the full-length coding sequence of Snapin. Snapin is a ubiquitous and predominantly cytoplasmic protein known to associate with several soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), which are central components of the cellular vesicle and fusion machinery (4, 8, 40). The vectors did not appear to be responsible for the observed interactions since no positive signals were found in the interactions between BD-UL70 and AD-vector or between BD-vector and AD-Snapin (Fig. 1A and B). BD-p53 contains the sequence coding for the murine p53 protein fused to the binding domain (BD) of GAL4, while AD-T contains the sequence coding for the simian virus 40 (SV40) large T-antigen fused to the activation domain (AD) of GAL4. Murine p53 and SV40 T-antigen interacted with each other and served as a positive control (Fig. 1D).
Fig. 1.
Identification of the UL70-Snapin interaction by yeast two-hybrid analysis. Yeast strain AH109 was cotransformed with the combination of one BD and one AD plasmid as indicated (A to D). Transformed yeast cells containing both plasmids were first grown on SD-minus Trp/Leu plates (DDO) to maintain the two plasmids, and then colonies were replica plated on SD-minus Trp/Leu/Ade/His plates (QDO) and also subjected to a β-galactosidase activity test (x-gal) by filter lift staining. No interactions were identified between BD-UL70 and the control empty vector AD-vector (A) or between the control empty vector BD-vector and AD-Snapin (B). Positive interactions were identified between BD-UL70 and AD-Snapin (C), as well as BD-p53 and AD-T (D), which served as a positive control.
Identification of the interaction of Snapin with UL70 in human cells by coimmunoprecipitation.
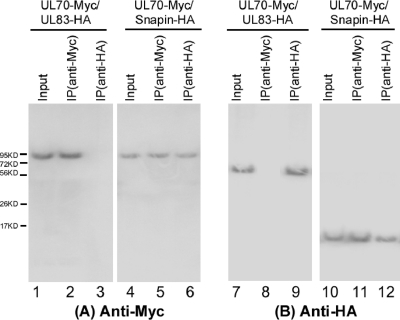
A coimmunoprecipitation (co-IP) assay was employed to validate the results from the yeast two-hybrid approach and to determine whether the identified interaction between UL70 and Snapin occurs in human cells. In these experiments, the sequences coding for UL70 and Snapin were cloned into the mammalian expression vectors pCMV-Myc and pCMV-HA to generate the constructs pCMV-Myc-UL70 and pCMV-HA-Snapin, in which each ORF was expressed as a fusion protein with an amino-terminal Myc or HA epitope tag, respectively. Human astrocytoma U373MG cells were transfected with the mammalian expression constructs and were harvested at 48 h. Protein samples were separated electrophoretically on SDS-containing gels, transferred electrically to membranes, and reacted with anti-HA and anti-Myc antibodies. HCMV UL70 and Snapin were detected as proteins of about 100 and 15 kDa, respectively (Fig. 2), consistent with their coding sequences of 947 and 137 amino acids, as predicted from the HCMV TowneBAC and human genomic sequences (4, 8, 10, 40).
Fig. 2.
Coimmunoprecipitation of viral proteins and Snapin. Human U373MG cells were cotransfected with a combination of two plasmids expressing HA- and Myc-tagged proteins. The input protein samples (80 μg) (Input) (lanes 1, 4, 7, and 10) and samples (15 μg) that were either immunoprecipitated with anti-Myc [IP (anti-Myc)] (lanes 2, 5, 8, and 11) or anti-HA antibodies [IP (anti-HA)] (lanes 3, 6, 9, and 12) were separated on SDS-containing polyacrylamide gels and assayed with Western blot analysis using anti-Myc (A) or anti-HA (B) antibodies, respectively.
Our co-IP experiments showed that UL70 was found to be associated with Snapin in U373MG cells transfected with constructs expressing the Myc-tagged UL70 and HA-tagged Snapin proteins (Fig. 2). Protein lysates from the transfected cells were first immunoprecipitated with either anti-Myc or anti-HA and then immunoblotted with antibodies against the Myc and HA epitope tags. Myc-tagged UL70 was coprecipitated with HA-tagged Snapin (Fig. 2, lanes 6 and 11). In contrast, in control experiments, we observed no significant binding or coprecipitation between Myc-tagged UL70 and HA-tagged UL83 of HCMV, which is not known to interact with UL70 and serves as a negative control (lanes 1 to 3 and 7 to 9) (41). These results confirmed the specificity of the co-IP assay and suggest that the UL70-Snapin interaction may occur in human cells during HCMV infection. Similar results were also found in HeLa cells and HCMV-infected U373MG cells transfected with the constructs expressing the Myc-tagged UL70 and HA-tagged Snapin proteins (data not shown).
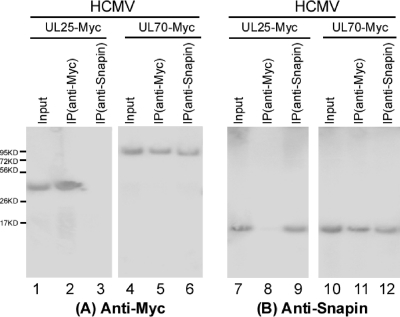
Snapin is a cytoplasmic protein expressed at a substantial level in neuronal cells (4, 8, 40). To further confirm the association between UL70 and endogenous Snapin in human cells and in the presence of HCMV infection, U373MG cells were transfected with mammalian expression constructs expressing Myc-tagged UL70 as well as the HCMV UL25 protein and then infected with HCMV. UL25 was not known to interact with Snapin and was used as a negative control in the experiments. Protein lysates from the cells were first immunoprecipitated with either anti-Myc or anti-Snapin and then immunoblotted with antibodies against the Myc epitope tag and Snapin (Fig. 3). Myc-tagged UL70 was coprecipitated with endogenous Snapin (Fig. 3, lanes 6 and 11). In contrast, in control experiments, we observed no significant binding or coprecipitation between Myc-tagged UL25 and Snapin (lanes 1 to 3 and 7 to 9). Similar results were also observed in uninfected cells that were transfected with pCMV-Myc-UL70 (data not shown). These results confirm the specificity of the co-IP assay and further suggest that UL70 may interact with Snapin in human cells in the absence and presence of HCMV infection.
Fig. 3.
Coimmunoprecipitation of transiently expressed viral proteins and endogenous cellular Snapin. Human U373MG cells were transfected with a plasmid expressing the Myc-tagged viral protein UL70 or UL25 and then infected with HCMV (MOI = 1) at 48 h posttransfection. Cellular lysates were prepared at 48 to 72 h postinfection. The input protein samples (50 μg) (Input) (lanes 1, 4, 7, and 10) and samples (10 μg) that were either immunoprecipitated with anti-Myc [IP (anti-Myc)] (lanes 2, 5, 8, and 11) or anti-Snapin [IP (anti-Snapin)] antibodies (lanes 3, 6, 9, and 12) were separated on SDS-containing polyacrylamide gels and assayed with Western blot analysis using anti-Myc (A) or anti-Snapin antibodies (B), respectively.
Important roles of the amino-terminal domain of UL70 in the interaction with Snapin.
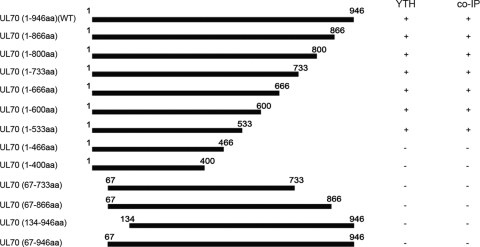
To further investigate the association of UL70 and Snapin and to map the domains of UL70 required for this association, we constructed a series of UL70 truncation mutants (Fig. 4). Yeast two-hybrid analyses were carried out to investigate the binary interactions of these UL70 mutants and Snapin. Furthermore, a series of mammalian expression constructs (Table 1), which encoded the Myc-tagged UL70 truncation mutants, were generated (Fig. 4). These constructs were then coexpressed with the HA-tagged Snapin in U373MG cells, and the interactions between HA-tagged Snapin and different Myc-tagged UL70 mutants were examined in coimmunoprecipitation experiments. The results, summarized in Fig. 4, indicate that the minimal UL70 mutant that binds to Snapin contains amino acids 1 to 533 and that a deletion of the amino-terminal sequence from amino acid 1 to 67 of UL70 abolishes the binding. Thus, the amino-terminal sequence of UL70 is essential for its interaction with Snapin.
Fig. 4.
Schematic diagram of UL70 and its deletion mutants that interact with Snapin as identified by the two-hybrid screen in yeast (YTH) and coimmunoprecipitation (co-IP) in U373MG cells. The interactions that were positive and negative in the two-hybrid screen or co-IP were marked as “+” and “−,” respectively.
Cellular localization of UL70 and Snapin in human cells.
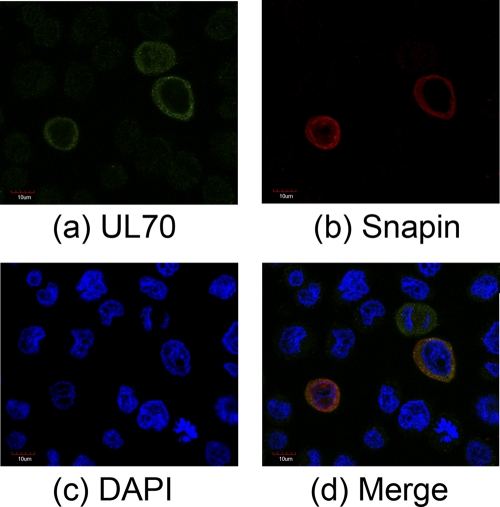
If UL70 is associated with (or binds to) Snapin in cells, it is expected that these proteins would localize within the same cellular compartments. To determine whether this is the case, cells were transfected with constructs expressing HA-tagged Snapin and Myc-tagged UL70, and the cellular localization of the expressed proteins was studied using immunofluorescence microscopy. Snapin has been previously found to be localized predominantly in the cytoplasm and is associated with intracellular vesicles (4, 8, 40). In our experiments, both Myc-tagged UL70 and HA-tagged Snapin were found to be localized predominately in the cytoplasm in UL373MG cells (Fig. 5). Cytoplasmic localization for both proteins was also observed in HeLa cells (data not shown). Cells that had not been exposed to antiserum recognizing Snapin, HA, or Myc epitopes showed no obvious fluorescence, indicating that the staining seen in cells is not due to a nonspecific cross-reaction of the secondary antibody with a viral or cellular protein. These observations are consistent with our results of the UL70-Snapin interactions from the two-hybrid screens in yeast and the co-IP experiments in human cells. Furthermore, these results provide direct evidence to suggest that UL70 is localized primarily in the cytoplasmic compartment in uninfected cells.
Fig. 5.
Localization of UL70 and Snapin expressed in human cells. Cells were transfected with constructs containing the sequences of the Myc-tagged UL70 or HA-tagged Snapin, fixed at 48 h posttransfection, stained with antibodies, and visualized using a microscope. The images of Myc-tagged UL70 (a), HA-tagged Snapin (b), and the nuclei stained with DAPI (c) were used to generate the composite images (d).
Effects of up- and downregulation of expression of Snapin on cellular localization of UL70.
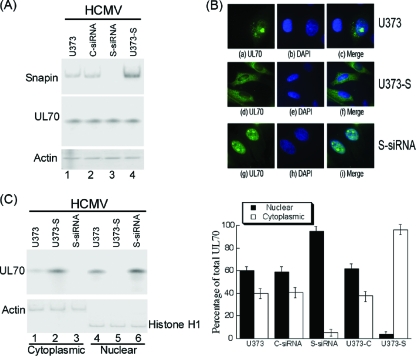
UL70 is required to be in the nuclei, where viral DNA synthesis occurs during HCMV infection. Thus, our findings that UL70 interacts with the cytoplasmic Snapin protein and is localized primarily in the cytoplasm in uninfected cells raise the possibility that Snapin may sequester UL70 and affect its cellular localization through binding. It is conceivable that overexpression of Snapin, a predominantly cytoplasmic protein, may lead to an accumulation of UL70 in the cytoplasm. To study the effect of overexpression of Snapin on the cellular localization of UL70, we constructed a stable U373MG cell line, U373-S, which constitutively expressed Snapin, and a control cell line, U373-C, which contained the empty expression vector. Western blotting of HCMV-infected cell lysates indicated an ∼5-fold-elevated level of Snapin in U373-S cells compared to that in the parental U373MG and control U373-C cells (Fig. 6 A). The constructed cell lines and the parental U373MG cells were indistinguishable in terms of their growth and viability for 3 months (data not shown), suggesting that the overexpression of Snapin did not result in significant cytotoxicity.
Fig. 6.
Effect of up- and downregulation of the expression of Snapin on the cellular distribution of UL70. (A) Western blot analysis of the levels of Snapin and Myc-tagged UL70 in the parental U373MG cells (U373) (lane 1), the Snapin-expressing U373-S cells (U373-S) (lane 4), or U373MG cells that were transfected with either anti-Snapin siRNA (S-siRNA) (lane 3) or control siRNA (C-siRNA) (lane 2). The expression of cellular actin was used as the internal control. Cells were transfected with pCMV-Myc-UL70 in the presence and absence of siRNAs. Forty-eight hours after transfection, cells were infected with HCMV at an MOI of 1. Protein samples were prepared at 48 to 72 h postinfection. (B) Immunofluorescence microscopy of the cellular localization of UL70 in the parental U373MG cells (U373), the anti-Snapin siRNA-treated cells (S-siRNA), or the Snapin-expressing cells (U373-S). Cells were infected with HCMV (MOI = 5) at 48 h posttransfection, fixed at 48 to 72 h postinfection, stained with antibodies, and visualized. The images of Myc-tagged UL70 (green) (a, d, and g) and the nuclei stained with DAPI (blue) (b, e, and h) were used to generate the composite images (c, f, and i). (C) Effect of Snapin on the distribution of UL70 in nuclear and cytoplasmic fractions. Different cells (e.g., parental U373MG cells, U373-S, and S-siRNA-treated cells) were transfected with pCMV-Myc-UL70 in the absence and presence of siRNAs. At 48 h posttransfection, cells were infected with HCMV (MOI = 1). At 48 to 72 h postinfection, cells were harvested and separated into nuclear and cytoplasmic fractions. Equivalent amounts of each fraction were analyzed by immunoblotting with anti-Myc. The purity of the nuclear and cytoplasmic fractions was assayed by immunoblotting with anti-histone H1 and anti-actin, respectively. The analyses were repeated three times. The standard deviation is indicated by the error bar (C, right panel).
To determine the localization of UL70 in HCMV-infected cells, the constructed cell lines were transfected with pCMV-Myc-UL70 and then infected with HCMV. An MOI of 1 to 5 was used to ensure that most cells were infected. Indeed, IE1 expression was detected in a majority of the cells when cells were strained with an anti-IE1 antibody (data not shown). HCMV infection appeared to promote nuclear import of UL70 in U373MG cells since a substantial amount of UL70 signal was found in the nuclei at 48 to 72 h postinfection compared to results for the uninfected cells (compare Fig. 6B, a to c, to Fig. 5a to c). In the presence of HCMV infection, the localization of UL70 in the Snapin-expressing U373-S cells was significantly different from that in the parental U373MG cells and the control U373-C cells. At 48 h postinfection, UL70 was found exclusively in the cytoplasm of U373-S cells (Fig. 6B, d to f). In contrast, both nuclear and cytoplasmic expression of UL70 was found in the U373MG (Fig. 6B, a to c) and U373-C (data not shown) cells. These results suggest that overexpression of Snapin leads to a reduction of nuclear import of UL70 and an accumulation of UL70 in the cytoplasm.
Since overexpression of Snapin leads to the cytoplasmic accumulation of UL70, it is conceivable that downregulation of Snapin expression may promote nuclear import of UL70. To determine whether this is the case, U373MG cells were cotransfected with pCMV-Myc-UL70 and siRNA molecules that were designed either to recognize the Snapin mRNA (S-siRNA) or not to recognize any viral or cellular transcripts (control siRNA [C-siRNA]) and then either mock infected or infected with HCMV at 48 h posttransfection. Downregulation of Snapin expression mediated by siRNA has been shown to have no effect on cell viability (4, 40). Western blotting of transfected cell lysates indicated that the level of the Snapin protein was reduced by more than 80% in cells transfected with S-siRNA compared to that in cells transfected with control C-siRNA (Fig. 6A, lanes 2 and 3). In the presence of HCMV infection, the localization of UL70 in the S-siRNA-treated cells was predominantly nuclear, while both nuclear and cytoplasmic expression of UL70 was found in the parental U373MG cells and the control-siRNA treated cells (Fig. 6B, g to i, and data not shown). These results suggest that downregulation of the expression of Snapin reduces the cytoplasmic accumulation of UL70 and increases its nuclear localization.
To confirm the effects of Snapin expression on the localization of UL70, we determined the distribution of UL70 by cell fractionation and Western blot analysis (Fig. 6C). Different cells (e.g., U373-C and U373-S cells and cells treated with C-siRNA or S-siRNA) were first transfected with pCMV-Myc-UL70 and then infected with HCMV and subjected to differential centrifugation into nuclear and cytoplasmic fractions. The purity of the fractions was confirmed by immunoblotting for histone H1 (nuclear marker) and actin (cytoplasmic marker) (Fig. 6C). In parental U373MG cells that were transfected with pCMV-Myc-UL70 and infected with HCMV, 60% of UL70 was found in the nuclei while 40% was in the cytoplasm (Fig. 6C). Similar results were also found in control cells that either contained the empty expression vector (U373-C cells) or were treated with siRNA molecules designed not to recognize any viral or cellular transcripts (C-siRNA) (Fig. 6C, right panel). However, downregulation of the expression of Snapin resulted in an increase (more than 95%) in the level of UL70 in the nuclear fractions in the S-siRNA-treated cells. Less than 5% of UL70 was found in the nuclear fractions in cells overexpressing Snapin (i.e., U373-S cells) (Fig. 6C). These observations confirm our results from the immunofluorescence microscopy experiments and suggest that Snapin plays an important role in the cellular localization of UL70.
Effect of up- and downregulation of the expression of Snapin on HCMV DNA synthesis and lytic infection.
It is expected that a change of the UL70 level in the nucleus as a result of altered expression of Snapin may affect the level of HCMV DNA synthesis and lytic infection. This is because UL70 encodes the viral primase essential for HCMV DNA replication, which occurs in the nuclei (24). To determine the effect of altered expression of Snapin on HCMV infection, we examined the profiles of viral gene expression by assaying the production of viral proteins of different kinetic classes during replication in cells where the expression of Snapin had been altered.
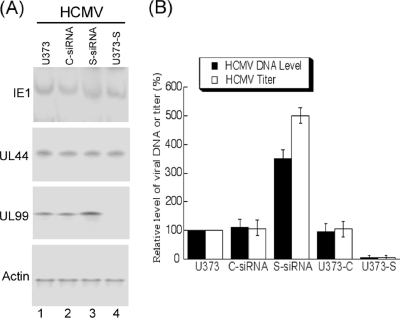
Different cells (e.g., U373-C and U373-S cells and cells treated with C-siRNA or S-siRNA) were infected with HCMV and then harvested at several time points postinfection. Protein levels were assayed by Western blot analysis. At 48 to 72 h postinfection, a reduction of more than 80% in the expression of Snapin was found in cells treated with S-siRNA molecules, while the level of Snapin in U373-S cells was about 5-fold higher than that in U373MG cells (Fig. 6A and data not shown). The up- and downregulation of the expression of Snapin did not appear to affect the protein levels of either the immediately-early viral gene product IE1 or the early gene product UL44 (Fig. 7 A). However, a decrease in the protein level of UL99 was detected in the Snapin-expressing U373-S cells, while an increase in the level of UL99 was found in cells treated with S-siRNA molecules (Fig. 7A). UL99 is the product of a viral late gene, whose expression is dependent upon viral DNA synthesis (24). We speculated as to whether the change in UL99 levels in these cells might be an effect of modulation of viral DNA synthesis as a result of a change in the nuclear UL70 level due to altered expression of Snapin.
Fig. 7.
Effect of Snapin on HCMV gene expression (A) and viral genomic synthesis and production (B). (A) Western blot analysis of the levels of HCMV IE1, UL44, and UL99 in the parental U373MG cells (U373) (lane 1), the Snapin-expressing U373-S cells (lane 4), or U373MG cells that were transfected with either anti-Snapin siRNA (S-siRNA) (lane 3) or control siRNA (C-siRNA) (lane 2). The expression of cellular actin was used as the internal control. At 48 h posttransfection, cells were infected with HCMV at MOI of 1. Protein samples were prepared at 48 to 72 h postinfection. (B) To assay the level of intracellular viral DNA, cells were harvested at 72 h postinfection. To assay the level of viral production, total infection cultures were collected at 5 days postinfection and viral titers were determined. The values of the relative levels of HCMV DNA and titers, which are the means from triplicate experiments, represent the ratios of the levels of viral DNA or titers in different cells to those in the parental U373MG cells (U373), respectively. The analyses were repeated three times, and the standard deviation is indicated by the error bar.
Two sets of experiments were carried out in these cells to determine whether this is the case. In the first set of experiments, total intracellular DNAs were isolated and the levels of viral DNA synthesis were assayed. The number of viral genomes present in each sample was determined by quantitative real-time PCR (qPCR), normalizing the copy number of the viral UL83 sequence to the copy number of the cellular β-actin gene sequence. At 72 h postinfection, a reduction of more than 90% in the number of copies of UL83/β-actin was observed in the Snapin-expressing U373-S cells while an increase of about 3.5-fold was found in cells treated with anti-Snapin S-siRNA molecules (Fig. 7B). In contrast, we observed no significant difference in the number of copies of UL83/β-actin among the parental U373MG cells, the empty expression vector-containing U373-C cells, and the control C-siRNA-treated cells (Fig. 7B). These results suggest that up- and downregulation of Snapin expression reduces and increases HCMV DNA synthesis, respectively.
In the second set of experiments, viral progeny production in these cells was determined. Virus stocks were prepared from the infected cultures (cells and culture medium together) at 1-day intervals through 7 days after infection, and their titers were determined. At 5 days after infection, a reduction of at least 20-fold in viral yield was observed in the Snapin-expressing U373-S cells, while an increase of about 5-fold was found in cells treated with S-siRNA molecules (Fig. 7B). In contrast, we observed no significant difference in viral yield among the parental U373MG cells, the empty expression vector-containing U373-C cells, and the control C-siRNA-treated cells (Fig. 7B). These results suggest that altered expression of Snapin modulates viral lytic replication and production.
DISCUSSION
Genomic DNA replication is a universal and essential process for herpesvirus infection (24, 31). Each component of the DNA replication machinery, including the primase-helicase complex, which in HCMV consists of UL70, UL102, and UL105, is highly conserved among all herpesviruses (5, 13, 26, 42, 47). Understanding the potential interactions of these virally encoded proteins, as well as their interactions with human proteins, is critical for elucidating the mechanism of herpesvirus genomic DNA synthesis and for developing novel strategies for the treatment and prevention of herpesvirus infections. In this study, we have provided the first direct evidence that the HCMV UL70 protein specifically interacts with human Snapin. This interaction was identified by our results from the two-hybrid screen in yeast and coimmunoprecipitation in human cells. We also showed that the level of UL70 in the nucleus, the site of viral DNA synthesis, is reduced in cells overexpressing Snapin with an expression vector and is increased in cells in which Snapin expression is downregulated with siRNA molecules specifically recognizing the Snapin mRNA transcript. Furthermore, the level of viral DNA synthesis and progeny production was decreased and increased in cells with upregulated and downregulated levels of Snapin, respectively. In contrast, no significant differences in the levels of UL70 in the nuclei, viral DNA synthesis, and HCMV particle production were found in the parental cells and those that expressed the empty expression vector or were treated with control siRNAs that did not recognize any viral or cellular transcripts. These results suggest that Snapin functions to bind to UL70 and reduce its nuclear import and availability for viral DNA synthesis, leading to a decrease in viral DNA replication and particle production.
Snapin, a predominantly cytoplasmic protein originally found in neuronal cells, is ubiquitously expressed in other types of cells, such as adipocytes (4, 8, 40). In neuronal cells, it is a component of the SNARE complex of proteins that is required for synaptic vesicle docking and fusion (8). In other types of cells, Snapin is also found to interact with components of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which is a ubiquitously expressed multisubunit protein complex required for normal biogenesis of specialized organelles of the endosomal-lysosomal system, such as melanosomes and platelet-dense granules (32, 38). It has not been reported that Snapin interacts with any viral proteins. How Snapin interacts with UL70 is currently unknown. Our results suggest that the binding of Snapin sequesters UL70 in the cytoplasm and reduces its nuclear import, leading to an inhibition of HCMV DNA synthesis and particle production. It will be interesting to determine whether such inhibition, mediated by Snapin interaction with UL70, constructs an antiviral defense for the benefit of the host or represents a means of viral temperance (10) for the benefit of HCMV, which is well known for its ability for highly controlled replication during persistent and latent infections.
Our findings with respect to the Snapin-UL70 interaction may have general implications regarding the pathways for localization and assembly of the components of DNA replication machinery of herpesviruses. Each component of the tripartite helicase-primase complex is highly conserved in all herpesviruses (16, 24, 31). For example, this complex is encoded by UL52 (helicase), UL5 (primase), and UL8 (primase-associated factor) in HSV-1, by BBLF (helicase), BSLF1 (primase), and BBLF2/3 (primase-associated factor) in EBV, and by ORF44 (helicase), ORF56 (primase), and ORF40/41 (primase-associated factor) in KSHV (5, 13, 26, 42, 47). Proper localization of each component of the helicase-primase complex in the nucleus, where viral DNA synthesis occurs, is essential for viral infection, and its process is believed to be conserved among all herpesviruses (24). For HSV-1, when expressed individually in human cells, UL5, UL8, and UL52 were localized in the cytoplasm (5, 9, 20). However, when all three were expressed in the same cells, these proteins were localized in the nuclei. Similar results were also observed in the localization of BBLF, BSLF1, and BBLF2/3 of EBV (13). In KSHV, when expressed individually in cells, ORF56 (primase), ORF44, and ORF40/41 were localized in the cytoplasm (42). However, these three proteins were localized in the nuclei only when they were coexpressed with other core components of the viral DNA replication machinery, including the DNA polymerase, the polymerase processivity factor, and single-strand DNA binding protein (42). UL70 has been shown to interact with UL102 and UL105 (21). However, little is currently known about how UL70 (and its counterparts in other viruses) interacts with UL102 and UL105 (and their counterparts in other viruses) to achieve proper localization in the nuclei. No cellular factors that interact with these proteins have been reported. In this study, we provide the first direct evidence that UL70, when expressed alone in human cells, is exclusively localized in the cytoplasm. This is consistent with the cytoplasmic localization pattern of its counterpart in HSV, EBV, and of KSHV when these proteins were expressed alone in the absence of other viral proteins (9, 13, 48). Furthermore, we showed for the first time that UL70 specifically interacts with Snapin, which is localized predominantly in the cytoplasm. Snapin may play a key role in the cellular localization of UL70 by binding to and sequestering this protein in the cytoplasmic compartment. Whether Snapin also interacts with the counterparts of UL70 in other viruses is currently unknown. Our results indicate that Snapin binds to the amino-terminal domain of UL70, which is highly conserved among the primases encoded by herpesviruses (46). It will be interesting to determine whether Snapin also interacts with the amino-terminal domains of these primase proteins.
In addition to that of Snapin, the coding sequences of several human proteins have also been found to be present in the yeast colonies that are positive in the YTH screen experiments. Further studies, including additional and independent experiments (e.g., coimmunoprecipitation), are needed to confirm the interactions of these proteins with UL70. Confirmation and future investigation of these interactions will provide insight into how host proteins affect UL70 function during HCMV infection.
Using several different types of cells, including U373MG, 293T, and HeLa cells, we showed that UL70 potentially interacts with Snapin in co-IP experiments. Furthermore, the roles of Snapin in UL70 localization and HCMV infection were investigated using U373MG cells. U373MG cells, which are derived from neuronal tissues infected naturally with HCMV in vivo, are highly permissive to HCMV replication and have been widely used to study HCMV infection (24). It may be important to test the role of Snapin in fibroblasts that are fully permissive and commonly used to study HCMV infection. However, it is technically difficult to carry out similar experiments in fibroblasts, since the poor efficiency of transfection in these cells resulted in a low level of UL70 and anti-Snapin siRNA. Moreover, fibroblasts are primary cells and therefore have a limited number of passage. It is difficult to generate a fibroblast cell line that constitutively and stably expresses high levels of Snapin after a substantial number of passages. In contrast, U373MG cells are immortalized, and therefore we can easily generate U373MG cell lines that constitutively express stable and high levels of Snapin. Further experiments in fibroblasts and other cells permissive to HCMV infection should elucidate the role of Snapin in these cells.
Our results indicate that overexpression of Snapin reduces the nuclear import of UL70 and increases its accumulation in the cytoplasm. The mechanism of how UL70 is retained in the cytoplasm is currently unknown. It is conceivable that Snapin sequesters UL70 in specific cytoplasmic compartments so that UL70 is not available for interaction with other viral or cellular protein carriers for nuclear transport. Meanwhile, downregulation of the expression of Snapin increases nuclear import of UL70 and decreases its cytoplasmic accumulation. These observations suggest that the proper cellular localization of UL70 in either the cytoplasm or nucleus is in a delicate balance and may be dictated by the cellular expression level of Snapin. There are no known nuclear localization signals (NLS) in UL70, although we cannot completely exclude the possibility of any potential unknown and hidden NLS. Currently, the mechanism of how UL70 is imported into the nucleus is unknown. It is conceivable that a specific cellular carrier(s) for nuclear transport competes with Snapin for binding of UL70. When Snapin expression is reduced, binding of UL70 to the cellular nuclear carrier(s) is favored, leading to increased nuclear import of UL70. In contrast, Snapin, when overexpressed, interacts with and sequesters UL70, leading to its accumulation in the cytoplasm. This hypothesis is supported by our observations that up- and downregulation of Snapin promotes cytoplasmic and nuclear accumulation of UL70, respectively. It will be interesting to determine whether the cellular localization of the homologues of UL70 in other viruses also employ similar cellular pathways. These and future studies of the interactions of the herpesvirus primase proteins with other viral and cellular proteins will provide significant insight into the mechanism of proper localization and assembly of the helicase-primase complex during lytic infection of herpesviruses.
ACKNOWLEDGMENTS
We are indebted to Paul Rider and Xiaohong Jiang for technical assistance and invaluable suggestions.
A.S. was a recipient of a China Graduate Student Scholarship from the Chinese Ministry of Education (NSFC, no. 30528022). Y.-C.C. and E.Y. were partially supported by a Block Grant Predoctoral Fellowship (University of California—Berkeley). This research has been supported by grants from the National Basic Research Program of China (973 program, no. 2011CB504800), National Natural Science Foundation of China (no. 31100128 and 81030031), and NIH (AI050468 and DE014145).
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Alvisi G., Jans D. A., Guo J., Pinna L. A., Ripalti A. 2005. A protein kinase CK2 site flanking the nuclear targeting signal enhances nuclear transport of human cytomegalovirus ppUL44. Traffic 6: 1002–1013 [DOI] [PubMed] [Google Scholar]
- 2. Alvisi G., et al. 2006. Human cytomegalovirus DNA polymerase catalytic subunit pUL54 possesses independently acting nuclear localization and ppUL44 binding motifs. Traffic 7: 1322–1332 [DOI] [PubMed] [Google Scholar]
- 3. Arezi B., Kuchta R. D. 2000. Eukaryotic DNA primase. Trends Biochem. Sci. 25: 572–576 [DOI] [PubMed] [Google Scholar]
- 4. Bao Y., Lopez J. A., James D. E., Hunziker W. 2008. Snapin interacts with the Exo70 subunit of the exocyst and modulates GLUT4 trafficking. J. Biol. Chem. 283: 324–331 [DOI] [PubMed] [Google Scholar]
- 5. Barnard E. C., Brown G., Stow N. D. 1997. Deletion mutants of the herpes simplex virus type 1 UL8 protein: effect on DNA synthesis and ability to interact with and influence the intracellular localization of the UL5 and UL52 proteins. Virology 237: 97–106 [DOI] [PubMed] [Google Scholar]
- 6. Biron K. K. 2006. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 71: 154–163 [DOI] [PubMed] [Google Scholar]
- 7. Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325: 417–470 [DOI] [PubMed] [Google Scholar]
- 8. Buxton P., et al. 2003. Identification and characterization of Snapin as a ubiquitously expressed SNARE-binding protein that interacts with SNAP23 in non-neuronal cells. Biochem. J. 375: 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calder J. M., Stow E. C., Stow N. D. 1992. On the cellular localization of the components of the herpes simplex virus type 1 helicase-primase complex and the viral origin-binding protein. J. Gen. Virol. 73: 531–538 [DOI] [PubMed] [Google Scholar]
- 10. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100: 14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunn W., Trang P., Khan U., Zhu J., Liu F. 2001. RNase P-mediated inhibition of cytomegalovirus protease expression and viral DNA encapsidation by oligonucleotide external guide sequences. Proc. Natl. Acad. Sci. U. S. A. 98: 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunn W., et al. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 7: 1684–1695 [DOI] [PubMed] [Google Scholar]
- 13. Gao Z., et al. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72: 8559–8567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giesen K., Radsak K., Bogner E. 2000. The potential terminase subunit of human cytomegalovirus, pUL56, is translocated into the nucleus by its own nuclear localization signal and interacts with importin alpha. J. Gen. Virol. 81: 2231–2244 [DOI] [PubMed] [Google Scholar]
- 15. Hanfler J., et al. 2003. Quantitation of cytomegalovirus (hCMV) DNA and beta-actin DNA by duplex real-time fluorescence PCR in solid organ (liver) transplant recipients. Med. Microbiol. Immunol. 192: 197–204 [DOI] [PubMed] [Google Scholar]
- 16. Kieff E., Rickinson A. B. 2007. Epstein-Barr virus and its replication, p. 2604–2654In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 17. Kilani A. F., et al. 2000. RNase P ribozymes selected in vitro to cleave a viral mRNA effectively inhibit its expression in cell culture. J. Biol. Chem. 275: 10611–10622 [DOI] [PubMed] [Google Scholar]
- 18. Liu F., Altman S. 1995. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev. 9: 471–480 [DOI] [PubMed] [Google Scholar]
- 19. Marchini A., Liu H., Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75: 1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsden H. S., et al. 1996. The herpes simplex virus type 1 UL8 protein influences the intracellular localization of the UL52 but not the ICP8 or POL replication proteins in virus-infected cells. J. Gen. Virol. 77(Pt. 9): 2241–2249 [DOI] [PubMed] [Google Scholar]
- 21. McMahon T. P., Anders D. G. 2002. Interactions between human cytomegalovirus helicase-primase proteins. Virus Res. 86: 39–52 [DOI] [PubMed] [Google Scholar]
- 22. Melle C., Nasheuer H. P. 2002. Physical and functional interactions of the tumor suppressor protein p53 and DNA polymerase alpha-primase. Nucleic Acids Res. 30: 1493–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller A. D., Rosman G. J. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7: 980–982, 984–986, 989–990 [PMC free article] [PubMed] [Google Scholar]
- 24. Mocarski E. S., Shenk T., Pass R. F. 2007. Cytomegalovirus, p. 2701–2772In Knipe D. M., et al. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Pari G. S. 2008. Nuts and bolts of human cytomegalovirus lytic DNA replication. Curr. Top. Microbiol. Immunol. 325: 153–166 [DOI] [PubMed] [Google Scholar]
- 26. Pari G. S., Anders D. G. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67: 6979–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Penfold M. E., Mocarski E. S. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239: 46–61 [DOI] [PubMed] [Google Scholar]
- 28. Pizzorno M. C., Mullen M. A., Chang Y. N., Hayward G. S. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65: 3839–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Plafker S. M., Gibson W. 1998. Cytomegalovirus assembly protein precursor and proteinase precursor contain two nuclear localization signals that mediate their own nuclear translocation and that of the major capsid protein. J. Virol. 72: 7722–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reeves M., Sinclair J. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325: 297–313 [DOI] [PubMed] [Google Scholar]
- 31. Roizman B., Knipe D. M., Whitley R. J. 2007. Herpes simplex viruses, p. 2503–2601In Knipe D. M., Howley P. M.(ed.), Fields virology, 5th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 32. Ruder C., et al. 2005. EBAG9 adds a new layer of control on large dense-core vesicle exocytosis via interaction with Snapin. Mol. Biol. Cell 16: 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salsman J., Zimmerman N., Chen T., Domagala M., Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 4: e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmolke S., Drescher P., Jahn G., Plachter B. 1995. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J. Virol. 69: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith J. A., Jairath S., Crute J. J., Pari G. S. 1996. Characterization of the human cytomegalovirus UL105 gene and identification of the putative helicase protein. Virology 220: 251–255 [DOI] [PubMed] [Google Scholar]
- 36. Smith J. A., Pari G. S. 1995. Human cytomegalovirus UL102 gene. J. Virol. 69: 1734–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Starcevic M., Dell'Angelica E. C. 2004. Identification of Snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279: 28393–28401 [DOI] [PubMed] [Google Scholar]
- 39. Stelzl U., et al. 2005. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- 40. Suzuki F., Morishima S., Tanaka T., Muramatsu I. 2007. Snapin, a new regulator of receptor signaling, augments alpha1A-adrenoceptor-operated calcium influx through TRPC6. J. Biol. Chem. 282: 29563–29573 [DOI] [PubMed] [Google Scholar]
- 41. To A., et al. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6: e17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tortorella D., Gewurz B. E., Furman M. H., Schust D. J., Ploegh H. L. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18: 861–926 [DOI] [PubMed] [Google Scholar]
- 43. Trang P., et al. 2000. Effective inhibition of human cytomegalovirus gene expression and replication by a ribozyme derived from the catalytic RNA subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Urban M., Joubert N., Hocek M., Alexander R. E., Kuchta R. D. 2009. Herpes simplex virus-1 DNA primase: a remarkably inaccurate yet selective polymerase. Biochemistry 48: 10866–10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waga S., Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67: 721–751 [DOI] [PubMed] [Google Scholar]
- 46. Woon H. G., Scott G. M., Yiu K. L., Miles D. H., Rawlinson W. D. 2008. Identification of putative functional motifs in viral proteins essential for human cytomegalovirus DNA replication. Virus Genes 37: 193–202 [DOI] [PubMed] [Google Scholar]
- 47. Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. 1988. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Gen. Virol. 62: 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu F. Y., et al. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75: 1487–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zou H., et al. 2003. Engineered RNase P ribozymes are efficient in cleaving a human cytomegalovirus mRNA in vitro and are effective in inhibiting viral gene expression and growth in human cells. J. Biol. Chem. 278: 37265–37274 [DOI] [PubMed] [Google Scholar]