Abstract
SCAN is a protein domain frequently found at the N termini of proteins encoded by mammalian tandem zinc finger (ZF) genes, whose structure is known to be similar to that of retroviral gag capsid domains and whose multimerization has been proposed as a model for retroviral assembly. We report that the SCAN domain is derived from the C-terminal portion of the gag capsid (CA) protein from the Gmr1-like family of Gypsy/Ty3-like retrotransposons. On the basis of sequence alignments and phylogenetic distributions, we show that the ancestral host SCAN domain (ESCAN for extended SCAN) was exapted from a full-length CA gene from a Gmr1-like retrotransposon at or near the root of the tetrapod animal branch. A truncated variant of ESCAN that corresponds to the annotated SCAN domain arose shortly thereafter and appears to be the only form extant in mammals. The Anolis lizard has a large number of tandem ZF genes with N-terminal ESCAN or SCAN domains. We predict DNA binding sites for all Anolis ESCAN-ZF and SCAN-ZF proteins and demonstrate several highly significant matches to Anolis Gmr1-like sequences, suggesting that at least some of these proteins target retroelements. SCAN is known to mediate protein dimerization, and the CA protein multimerizes to form the core retroviral and retrotransposon capsid structure. We speculate that the SCAN domain originally functioned to target host ZF proteins to retroelement capsids.
INTRODUCTION
Gmr1-like elements are a class of Ty3/Gypsy long terminal repeat (LTR) retrotransposons similar to most other Ty3/Gypsy elements but with a different protein domain order within the pol gene; the domain order of Gmr1-like elements is Ty1/copia-like, PRO-INT-RT-RNH (protease [PRO]-integrase [INT]-reverse transcriptase [RT]-RNase H [RNH]), rather than the typical Gypsy order PRO-RT-RNH-INT (15). Gmr1-like elements have an origin before the common ancestor of deuterostomes (vertebrates along with sea urchins, etc.) (15). In this study, we investigate the relationships of the capsid structural proteins encoded by Gmr1-like elements and the SCAN domain present in amniotes (mammals, birds, and reptiles). It has previously been noticed and briefly remarked upon that in lower vertebrates, sequences matching a SCAN domain profile reside in retrovirus-like polyproteins (36). In addition, protein structural similarity between the SCAN domain and the HIV C-terminal capsid has been observed. The SCAN domain has been shown to multimerize by a domain-swapping mechanism in which two monomers swap their major homology regions (MHRs), and it has been hypothesized that domain swapping in this fashion plays a role in retroviral assembly (23, 26). We speculate that the protein-protein interaction function of the SCAN domain was borrowed from the capsid domain of a Gmr1-like element, which multimerizes in vivo to form the core retrotransposon capsid structure.
The SCAN domain is a conserved motif of approximately 80 amino acids found at the N termini of many Cys2His2 (C2H2)-type zinc finger (ZF) proteins and is leucine rich and dominated by α-helical structure (32, 45, 46). The SCAN domain is known to be involved in protein-protein interactions and is capable of dimerization leading to the creation of homo- and heterodimer protein complexes (35, 38, 45). SCAN domains are a common feature of the tandem C2H2 zinc finger gene complements of many mammals and are found almost exclusively associated with C2H2 zinc finger genes. A SCAN domain is present in the proteins encoded by about 50 human ZF genes, about 1 in 10 of all human ZF genes (12, 27, 44). SCAN domains are often found alongside KRAB (Krüppel-associated box; 5) domains, in which case the SCAN, KRAB, and C2H2-ZF domains are present in that order and are usually found on separate exons. This association is not universal, however, and many SCAN-ZF genes exist with no associated KRAB domain (reviewed in reference 10).
The KRAB domain was probably derived originally from the Meisetz (PRDM9) gene (6), and recently, several studies have demonstrated its importance in the silencing of exogenous and endogenous retroviral elements via its known interaction partners KAP1 and SETDB1 (30, 34). Specifically, KRAB is thought to act mainly through KRAB-ZF genes in which the zinc finger array provides DNA target recognition and the KRAB domain recruits a protein complex (including KAP1 and SETDB1) that leads to transcriptional repression through a closed chromatin state (14, 37). Experiments in mouse embryonic stem cells demonstrate that the removal of KAP1 or SETDB1, both presumed to abrogate KRAB-mediated transcriptional silencing, lead to a large increase in the transcription of endogenous and exogenous mouse retroviral elements (30, 34), and two ZF genes have been directly shown to repress retroviral transcription (7, 20, 47). These studies provide evidence supporting a compelling hypothesis to explain the function and evolution of KRAB-ZF genes and hint that the SCAN domain, often found associated with KRAB in the form of SCAN-KRAB-ZF genes, may also function in these processes. Many genomes contain a large number of SCAN-ZF genes without a KRAB domain (36), but it has been observed that even SCAN-KRAB-ZF genes can function as transcriptional silencers in a KAP1-independent fashion (22). In contrast to the KRAB domain, the origin, evolution, and ultimate function of the SCAN domain are still something of a mystery. In this study, we investigate the origins of the SCAN domain and explore its possible functions, its relationship to Gmr1-like retrotransposons and the history of its association with KRAB-ZF genes. We present evidence that the SCAN domain was derived from the capsid (CA) protein of Gmr1-like retroelements in the ancestor of amniotes and was exapted, or adaptively coopted for a novel function, by C2H2-type zinc finger genes. We also provide binding site prediction evidence that the SCAN domain may play a role in the targeting and transcriptional silencing of Gmr1-like retrotransposons in the Anolis carolinensis lizard.
MATERIALS AND METHODS
Identification of Gmr1-like elements.
To detect genomic copies of Gmr1-like retroelements, we searched for matches to the characteristic Gmr1-like capsid (CA), protease (PRO), integrase (INT), reverse transcriptase (RT), and RNase H (RNH) domains. Search profiles were a custom CA profile and Pfam profiles PF00077 and PF08284 (retroviral aspartyl proteases), PF00078 (reverse transcriptase), PF00075 (RNase H), and PF00665 (integrase, catalytic). The RPS-BLAST program was run on target genomes with each of the profiles as queries and a very permissive E value to allow for fragmentary domains expected from older, neutrally evolving retroelements. Profile matches with a heuristically chosen BLAST score of 22 or higher were retained and sorted in genome order. When adjacent matches to the same profile overlapped by 30% or more, only the higher scoring match was retained. A custom algorithm was applied to extract matches within 3 kb of each side of RT matches and in the expected Gmr1-like order (INT before RT). The algorithm constrained the aligned matches as follows: 1 or more CA, 0 or more PRO, 1 or more INT, 1 or more RT, and 0 or more RNH. Match clusters that contained RT matches with a score sum of 40 or more, CA matches with a score sum of 60 or more, and a total match score sum of 150 or more were retained as Gmr1-like retroelements. Tests with shuffled genomic sequence showed that the false-positive rate was below detection. Examples of Gmr1-like matches to various genomes are given in Data S1 in the supplemental material. For Anolis Gmr1-like elements, sequence encompassing each match cluster and 5 kb to each side were extracted from the genomic sequence and submitted to the online LTR_FINDER program to identify other long terminal repeat (LTR) retroelement features (48).
Mammalian SCAN domains.
We detected no Gmr1-like retrotransposons in any mammal, so we extracted mammalian genomic SCAN domain matches as samples of host SCAN domains (we define a host SCAN domain to be a genomic SCAN domain not associated with any retroelement). SCAN domains were collected from platypuses, opossums, elephants, mice, and humans to attempt to capture the full diversity of mammalian SCAN domains.
RSCAN domains.
We extracted Gmr1-like retroelement SCAN domain matches from clear Gmr1-like retroelements from all genomes with these matches. We call these Gmr1-like CA domains that have high similarity to the SCAN domain profile RSCAN (retrotransposon SCAN-like) domains. We assumed that the close order and juxtaposition of integrase and reverse transcriptase domain hits identified these as RSCAN domains. Nonmammalian tetrapods were challenging because it was clear that the Anolis lizard has both host SCAN and RSCAN domains; this challenge was resolved as follows: SCAN domain hits in close association with a tandem zinc finger (ZF) open reading frame (ORF) and in the right orientation were presumed to be host SCAN domains, whereas SCAN domain hits in close association with INT and RT domains were presumed to be RSCAN domains. Phylogenetic analysis was conducted using phyml (17) to build maximum likelihood protein trees from PRANK (29) and DIALIGN (40) protein alignments. Branch support was computed using the approximate likelihood ratio test (aLRT) method (2). Results from these phylogenies were congruent with previous identifications; putative host SCAN domains formed a clade and nested within the broader diversity of putative RSCAN domains. For both host SCAN domains and RSCAN domains, some species had a huge number of matches, making it computationally prohibitive to build maximum likelihood trees. From these species, we used a preliminary species-specific tree to choose sequences that represent the diversity of each species; subsequent tree analysis used these representative sequences. This process left us with a broad sampling of clear host SCAN domains from species ranging from the lizard and finch through representative mammals, and a broad sampling of RSCAN domains from fish, basal deuterostomes, and basal tetrapods.
From online BLAST searches, we identified a single Xenopus tropicalis sequence predicted to encode a SCAN-ZF protein (XP_002942163.1), but the SCAN-like sequence cluster deep inside the RSCAN clade is adjacent in the genome to a weak retroviral integrase match, and the SCAN domain is in an unprecedented internal location in the SCAN-ZF prediction (data not shown). We interpret this as an erroneous prediction that fuses a SCAN domain from a retroviral fragment to a nearby ZF exon, which is plausible given the large numbers of both Gmr1-like retrotransposons and tandem ZF sequences in the frog genome. In any case, on the basis of its tree position, this sequence is unlikely to be the predecessor of other host SCAN domains.
Prediction of putative SCAN-ZF genes.
To identify candidate SCAN- and KRAB-containing genes, we used genomic RPS-BLAST searches with Pfam profiles PF02023 (SCAN), PF01352 (KRAB), and a custom ZF profile constructed from a composite of human Cys2His2 (C2H2) ZF domains repeated 60 times to identify blocks of tandem ZF domains. All of the above domains are detected with high sensitivities, so we used highly restrictive E-value cutoffs to avoid false-positive results. Genomic locations with C2H2 ZF tandem arrays located near and in the correct orientation with respect to KRAB or SCAN domains were considered putative KRAB-ZF, SCAN-ZF, or SCAN-KRAB-ZF genes as appropriate.
Prediction of SCAN-ZF binding profiles.
In order to predict the binding affinity of putative SCAN-ZF genes and test their affinity for retroelement sequences, a binding profile for each was constructed as follows, following a simplified version of a previously reported dynamic programming method (31). Each putative SCAN-ZF protein was trimmed to only contiguous C2H2 zinc fingers. A support vector machine (SVM) model developed to evaluate the binding of zinc fingers to DNA targets was employed with its polynomial kernel using SVMlight software to predict the four best 4-nucleotide (nt) binding sites for the first zinc finger, one ending with each of the four nucleotides (24, 33). These 4-nt sites were then extended to 7 nucleotides (representing all 256 3-nt extensions to the first set of possible binding sites), and each was tested to find the optimal predicted binding site for the first two zinc fingers. This process was repeated for each additional zinc finger until a full predicted binding site of 3N + 1 nucleotides was created for each array of N zinc finger domains. The full binding site calculated in this fashion is the optimal predicted binding site, given the sequence inputs and the model.
This binding site was then transformed into a binding profile by simulating 3(3N + 1) sequences, each differing from the predicted binding site by one nucleotide. The binding model was applied to each to generate a new score so that at each position of the predicted binding target, there exist four SVM scores: scoremax representing the SVM score with the optimal nucleotide, and three additional scores representing the three other possible nucleotides. Following Myers et al. (31), we interpret these SVM scores as log-scaled binding probabilities such that:
represents the difference in score (ΔS) between nucleotides a and b when Pa is the binding probability of the full profile including nucleotide a and Pb is the binding probability of the full profile including nucleotide b. We assume that the maximum binding probability (Pmax) is 0.1 at all positions, and this allows the calculation of the three other binding probabilities. We then calculate the enrichment in binding probability for each nucleotide over the mean binding probability and transform this to a final position-specific scoring matrix (PSSM) score using the following heuristic equation:
where Scorea is the score for nucleotide a and P̄ is the mean binding probability.
Repeating this process for each position in the predicted binding profile leads to a full PSSM which can be used to search DNA sequences for significant matches. See Data S3 in the supplemental material for the Anolis putative SCAN-ZF genes used and Data S4 for the Anolis PSSMs calculated by this method.
Identification of Gmr1-like targets of SCAN-ZF binding profiles.
The predicted binding profile of each SCAN-ZF, determined as described above, was used as a PSSM and searched against each Gmr1-like element in the Anolis lizard without allowing gaps. The highest scoring match for each SCAN-ZF/Gmr1-like pair was recorded as well as its P value, calculated using the TFMPvalue software, which allows efficient and accurate computation of PSSM P values given a PSSM and nucleotide frequencies (43). Bonferroni's correction was applied to this P value to account for testing multiple sites within each Gmr1-like element, giving a corrected P value for each SCAN-ZF/Gmr1-like pair. These corrected P values were analyzed using the QVALUE framework which uses P values to estimate a false discovery rate (FDR) (39). See Data S5 in the supplemental material for a list of Anolis Gmr1-like elements scanned in this fashion, and see Data S6 in the supplemental material for information on significant hits produced.
RESULTS
Gypsy SCAN domain.
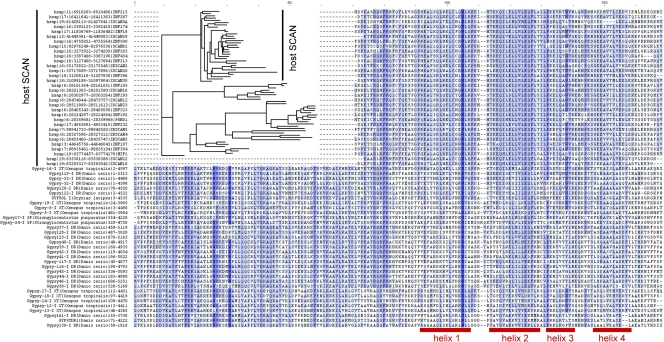
In the course of identifying SCAN-ZF genes in various vertebrate genomes, we noticed that the zebrafish genome has a large number of SCAN-related genomic matches that are not associated with zinc finger (ZF) sequences. Further investigation revealed that many of these matches are located within annotated long terminal repeat (LTR) retrotransposon sequences. An association of SCAN domain matches with putative retrotransposon sequences was previously noticed but not further characterized (36). We characterized this association by testing for Pfam SCAN profile matches in the complete set of RepBase 15.05 deuterostome sequences (which includes consensus sequences for all known retrotransposons and endogenous retroviruses) and found strong matches to LTR retrotransposons of the Gypsy/Ty3 group from zebrafish, Xenopus, and sea urchins. We extracted the complete open reading frames (ORFs) containing these SCAN matches and characterized their domain content. Many of the ORFs include clear matches to LTR retroelement protein domains in the order SCAN-PRO-INT-RT-RNH (SCAN-protease-integrase-reverse transcriptase-RNase H). This domain order is unusual and is characteristic of Gmr1-like Gypsy elements (11, 15). The SCAN domain is invariably a short distance upstream of the protease domain and an RNA binding CCHC motif is often located between the SCAN and protease domains (see Fig. S1 in the supplemental material). This position suggested that the SCAN domain is part of the Gmr1-like gag capsid (CA) protein (11). A multiple alignment of the ORFs indicated that the SCAN match is the C-terminal segment of a longer block of high conservation (Fig. 1; see Fig. S1 in the supplemental material), which we call the RSCAN domain (retrotransposon SCAN-like CA domain). The crystal structure of one SCAN domain is known to have a protein fold very similar to the C-terminal segment of the HIV retroviral CA domain, though no protein sequence similarity was detected (23, 26). We conclude that the RSCAN domain corresponds to the CA protein of Gmr1-like Gypsy retrotransposons and that the SCAN domain is derived from the C-terminal segment of this CA protein. On the basis of the protein fold similarity, other retrotransposon and retroviral CA sequences presumably share a ancestor but are highly divergent in sequence.
Fig. 1.
SCAN and Gmr1-like CA alignment. Alignment of SCAN domains from Homo sapiens with Gmr1-like capsid sequences from various species. The SCAN domain shows strong similarity to the C-terminal portion of current Gmr1-like CA sequences, and human SCAN domains form a clade with respect to Gmr1-like CA sequences. The four helices in the known structure of the SCAN domain (23) are indicated below the sequence alignment.
To test whether the RepBase retroelements with an RSCAN domain generally correspond to the Gmr1-like class, we built a tree from RT protein sequence extracted from each RepBase element annotated as a deuterostome Gypsy (25). We mapped onto this tree those elements with high-scoring RSCAN matches and those with a domain order INT-RT (rather than the typical Gypsy order RT-INT). The INT-RT domain order and RSCAN matches were perfectly congruent and monophyletic on the RT protein tree (see Fig. S2 in the supplemental material). We also characterized the phylogenetic distribution of RSCAN retroelements in complete genome sequences using profile searches and a profile match alignment method. Clear matches to LTR retroelements with high-scoring RSCAN domains were identified in Porifera (sponge Amphimedon queenslandica), Placozoa (Trichoplax adherens), Arachnida (Ixodes scapularis), Mollusca (Lottia gigantea), Echinodermata (Strongylocentrotus purpuratus), Hemichordata (Saccoglossus kowalevskii), Urochordata (Ciona intestinalis), Cephalochordata (Branchiostoma floridae), Hyperoartia (the Petromyzon marinus lamprey), several Actinopterygii (ray-finned fishes), Amphibia (Xenopus tropicalis), and Reptilia (the Anolis carolinensis lizard), but not in birds or mammals. In almost all cases in which high-scoring INT and RT domains were also present, the domain order was RSCAN-INT-RT, further indicating that these elements correspond to the widespread Gmr1-like Gypsy group. Gmr1-like elements were not found in the Caenorhabditis elegans or Drosophila melanogaster genomes, suggesting loss somewhere on those lineages. Genomic positions of representative matches are given in Data S1 in the supplemental material.
Many other retrotransposons and endogenous retroviruses have a much weaker match to an RSCAN domain profile. These matches are located in an appropriate position for the gag CA region of their retroelements. Highly significant matches in RepBase include retroelements annotated as copia, BEL, and endogenous retrovirus (e.g., Copia-11_SB-I [Sorghum bicolor] [2E−07], BEL3-I_AG [Anopheles gambiae] ]1E−5[, and ERV31 [Monodelphis domestica] [1E−5]). We conclude that the CA proteins from all of these classes of retroelements have statistically significant, though distant, sequence similarity to the Gmr1-like Gypsy CA protein.
First appearance of a host SCAN domain.
Detecting the first appearance of a bona fide host-encoded SCAN domain is complicated by finding SCAN domain matches in a widely distributed retrotransposon. We solved this problem by first obtaining representative host SCAN sequences (i.e., SCAN domains not associated with retroelements) from species that clearly lack Gmr1-like retroelements and by obtaining representative RSCAN sequences from clear Gmr1-like retroelements from a variety of species. A protein tree of these representative sequences showed that all the host SCAN domains are monophyletic and have relatively little sequence diversity compared to RSCAN domains (Fig. 2). This pattern allowed us to tentatively assign all genomic SCAN domain matches of unclear origin to either the host SCAN or RSCAN type. Only the Anolis lizard had both types of genomic SCAN matches: a minority of the matches appeared on a sequence tree as RSCAN type, and the rest appeared as host SCAN type. Nearly all of those classified as host SCAN domains are closely upstream of tandem zinc finger ORFs. Representative sequences are provided in Data S2 in the supplemental material.
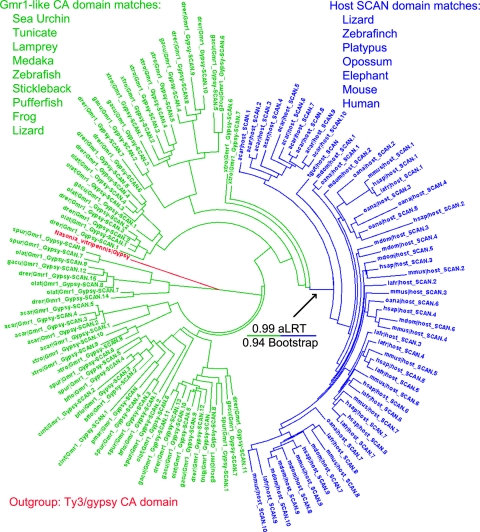
Fig. 2.
Outgrouped tree of SCAN and Gmr1-like CA. Phylogeny of SCAN domain matches (blue) and Gmr1-like element CA domains (green) chosen from a variety of species to represent the phylogenetic diversity of each. One Ty3/Gypsy element capsid sequence was chosen for its high score to the SCAN domain profile and is included as an outgroup and shown in red. Host genomic SCAN domains form a clade within the capsid sequences of Gmr1-like elements with excellent branch support (>0.99 by the approximate likelihood ratio test [aLRT] and 188/200 bootstraps). The tree was constructed using phyml 3.0 (17) and was visualized with Dendroscope (21). Notably, the Anolis carolinensis lizard is alone in having a large number of Gmr1-like elements and host SCAN domains. Sequences from the following species are shown: elephant, Loxodonta africana (lafr); frog, Xenopus tropicalis (xtro); pufferfish, Tetraodon nigroviridis (tnig); human, Homo sapiens (hsap); lamprey, Petromyzon marinus (pmar); lizard, Anolis carolinensis (acar); medaka, Oryzias latipes (olat); mouse, Mus musculus (mmus); opossum, Monodelphis domestica (mdom); platypus, Ornithorhyncus anatinus (oana); sea urchin, Strongylocentrotus purpuratus (spur); stickleback, Gasterosteus aculeatus (gacu); tunicate, Ciona intestinalis (cint); zebra finch, Taeniopygia guttata (tgut); zebrafish, Danio rerio (drer).
If the host SCAN domain arose once by exaptation of an RSCAN (Gmr1-like CA) sequence, then extant host SCAN sequences should appear on an outgroup-rooted tree as a clade coming from within the Gmr1 CA sequences. Since outgroup CA sequences (from non-Gmr1-like retrotransposons) are highly divergent from Gmr1 CA, we tested this prediction with two different outgroup CA sequences using both DIALIGN and PRANK protein alignments (29, 40). These four trees all supported an origin of host SCAN from within the RSCAN family; one of the trees is shown in Fig. 2.
Host SCAN domains in the Anolis lizard.
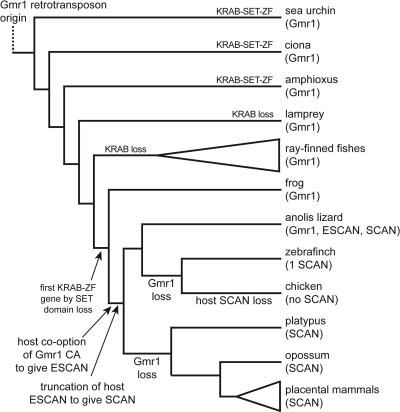
Among the genomes we analyzed, the Anolis lizard is the only one with clear host SCAN domains that span the entire length of the retrotransposon RSCAN domain. Though these sequences are closely related to Gmr1 RSCAN sequences, they clearly form a tree with the shorter host SCAN sequences from mammals, most of them are present in the genome closely upstream of tandem ZF sequences, and these putative host SCAN-ZF genes are mostly present in large gene clusters as is typical for tandem ZF genes in mammals (4, 9, 19). To distinguish these full-length host domains from the shorter SCAN domain and the retroelement RSCAN domain, we refer to them as ESCAN (extended SCAN). We interpret these evolutionary patterns to mean that the ESCAN domain predated the shortened SCAN domain and is derived from an ancestral host exaptation of an RSCAN sequence. Truncation of the ESCAN domain to give the SCAN domain presumably occurred very early, since the shorter host SCAN domain is shared by Anolis and other tetrapods (Fig. 3).
Fig. 3.
Phylogenetic model of SCAN-ZF evolution. Evolution of the ESCAN and SCAN domains and mammalian SCAN-KRAB-ZF genes from ancestral Gmr1-like elements. Gmr1-like elements are ancient, and their capsid domains were likely exapted as SCAN domains near the root of the amniote branch. Domains descended from Gmr1-like CA sequences were fused to existing KRAB-ZF genes and later trimmed to include only the C-terminal portion to form the SCAN domain. The subsequent loss of Gmr1-like elements in at least two lineages is apparent. Species designated as containing Gmr1-like elements or SCAN domains include many examples of each, except for zebra finch which has a single identifiable SCAN domain. Clear Gmr1-like retrotransposons were also found in the genomes of a Hemichordate acorn worm (Saccoglossus kowalevskii), a mollusc (Lottia gigantea), an annelid (Helobdella robusta), a sponge (Amphimedon queenslandica), the Placozoan Trichoplax adherens, and the deer tick (Ixodes scapularis), but not in hydra (Hydra magnipapillata), honeybee (Apis mellifera), mosquito (Anopheles gambiae), Daphnia pulex, Drosophila melanogaster, Caenorhabditis elegans, a Choanoflagellate (Monosiga brevicollis), all fungi, and all vascular plants (fungi and plant genomes available on NCBI nr database on 5 September 2010). The elements found had both a high-scoring RSCAN domain and the characteristic INT-RT domain order. Since the sponge and probably Trichoplax are basal metazoans, many protostomes (Arthropoda, Nematoda, etc.) have probably lost an ancestral Gmr1-like element.
Domain additions to tandem ZF genes.
On the basis of the analysis above, we conclude that the Gmr1-like RSCAN domain was exapted once by a host gene and that all subsequent host ESCAN and SCAN domains were derived from this original event. On the basis of the presence or absence on the species phylogeny, the most parsimonious phylogenetic position of this exaptation is on the branch separating amphibians and all other tetrapods (Fig. 3). Because sequence from amphibian genomes is currently limited to a single species, it remains possible that the exaptation occurred slightly earlier and that the host SCAN domain was lost in Xenopus tropicalis. Since there are genome sequences from several widely divergent ray-finned fish all lacking host SCAN domains, it is unlikely that the exaptation preceded their divergence from tetrapods. Gmr1-like retrotransposons appear to have been lost at least twice in vertebrates, once along the lineage leading to mammals and once along the lineage leading to birds.
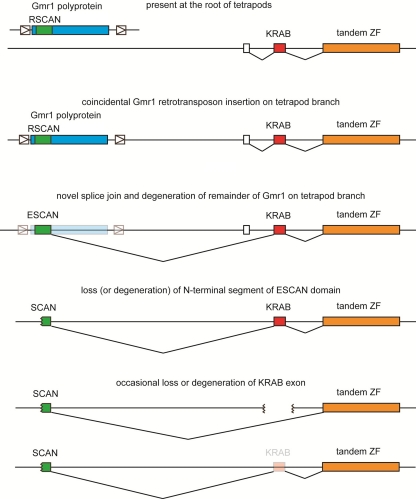
KRAB domains are also often associated with tandem ZF sequences. KRAB-ZF genes clearly arose prior to the divergence of amphibians and amniotes because the frog genome contains many KRAB-ZF genes (5, 6, 41). KRAB-ZF genes may have arisen by deletion of the SET domain from a much older KRAB-SET-ZF gene (6). A SCAN domain is often found at the N terminus of KRAB-ZF genes in mammals and in the Anolis lizard (data not shown). A simple model (Fig. 4) that explains the evolution of these associations is as follows. (i) On the amniote phylogenetic branch, a Gmr1-like Gypsy retrotransposon inserted by chance in the appropriate orientation near a KRAB-ZF gene. (ii) A novel splice donor arose that caused the RSCAN sequence in the retrotransposon to splice to the KRAB exon (or an upstream exon), producing an ESCAN-KRAB-ZF protein coding gene. This fusion might have been selectively advantageous because it conferred a novel transcription pattern or because the ESCAN domain conferred a useful function to the new fusion protein. (iii) Still prior to the separation of mammals and lizards, the remainder of the retrotransposon sequence degenerated, and in a subset of ESCAN-KRAB-ZF genes, the N-terminal segment of the ESCAN domain was lost to produce the shorter SCAN domain. For unknown reasons, only the SCAN domain is detected in sequenced mammals, but both ESCAN and SCAN domains survived on the lizard lineage. (iv) Perhaps because the SCAN (or ESCAN) domain rendered the KRAB domain superfluous in some genes, the KRAB domain was subsequently lost in some of these genes to produce the currently observed SCAN-ZF (and ESCAN-ZF) genes.
Fig. 4.
Model of SCAN-ZF derivation from Gmr1-like retrotransposons. A proposed model for the evolution of SCAN-KRAB-ZF and SCAN-ZF genes from ancestral KRAB-ZF and Gmr1-like sequences. Initial fusion of Gmr1-like CA domain to a KRAB-ZF gene by splicing is followed by gradual degradation of surrounding Gmr1-like sequence and truncation of the Gmr1-like CA to the shorter SCAN domain. The occasional subsequent loss of KRAB domain exons generates SCAN-ZF genes.
The following observations are consistent with each of these steps. (i) Both Gmr1-like Gypsy retrotransposons and KRAB-ZF genes were probably abundant in the amniote ancestor, since they are both abundant in frog and lizard genomes. This situation improved the chance of an appropriately positioned retrotransposon insertion. (ii) Evidence that some sort of RSCAN fusion to a tandem ZF gene occurred is overwhelming. We suggest that the fusion resulted from a novel splice junction, because in extant mammalian genes, the entire SCAN domain resides on a single exon with the C-terminal end of the SCAN domain almost perfectly coincident with the 3′ end of the exon. (iii) Current host SCAN (and ESCAN) genes have no other associated retrotransposon sequences, so these must have been deleted, degraded, or otherwise lost at some point (though possibly this occurred before the fusion event). Many (in lizards) or all (in mammals) SCAN- or ESCAN-containing genes have only the shorter SCAN sequence, so loss of the N-terminal part of the ESCAN domain presumably occurred early. (iv) Both lizards and mammals have a substantial number of SCAN-ZF (or ESCAN-ZF) genes with no detectable KRAB domain. In principle, these could have arisen from an independent ESCAN fusion to a tandem ZF exon, but loss of the KRAB domain from SCAN-KRAB-ZF genes is supported by several observations. First, all SCAN and ESCAN domains are monophyletic, regardless of whether they appear in SCAN-ZF or SCAN-KRAB-ZF genes. Second, two human ZF genes annotated as “SCAN-only” (ZSCAN18 and ZNF498) in fact have a degenerate KRAB-encoding exon in an appropriate location and well supported by mRNA sequence data. Each has bona fide SCAN-KRAB-ZF paralogs with whom they could share a gene ancestor. We interpret these as intermediates in transition from a SCAN-KRAB-ZF gene to a SCAN-ZF gene. Third, most human SCAN-only proteins are interspersed on a sequence tree with SCAN-KRAB-ZF proteins (data not shown). The simplest explanation for this pattern is that SCAN-KRAB-ZF genes sometimes lose their KRAB domain.
Function of the SCAN domain.
We hypothesize that SCAN-ZF fusion genes were initially useful to the host because the SCAN domain targeted the ZF DNA binding domains to cytoplasmic retroelement capsids, where the ZF domains bound and sequestered newly synthesized retroelement DNA or served some other host function. In order to test this hypothesis, we matched the predicted DNA binding profiles of all putative Anolis lizard SCAN-ZF proteins to the DNA sequence of Anolis Gmr1 elements. We analyzed Anolis sequences because it is the only sequenced genome that we found to have both abundant Gmr1-like retroelements and abundant putative SCAN-ZF genes.
The lizard genome was searched for putative exons encoding multiple tandem C2H2 zinc finger domains and was additionally searched for the presence of putative SCAN (Pfam PF02023) and KRAB (Pfam PF01352) domains using RPS-BLAST software and the Pfam database of protein family sequence profiles (13). Any putative ZF exon encoding at least 4 zinc finger domains that was also within 20 kb of a putative SCAN domain in the correct orientation was considered a SCAN-ZF gene and used for further analysis. Each was given a name and number as a putative SCAN-ZF (PSZ) gene or a putative SCAN-KRAB-ZF (PSKZ) gene if a KRAB domain match was also found in the appropriate genomic position and orientation. In total, we identified and named PSZ0 through PSZ46 and PSKZ0 through PSKZ173 in this fashion (see Data S3 in the supplemental material).
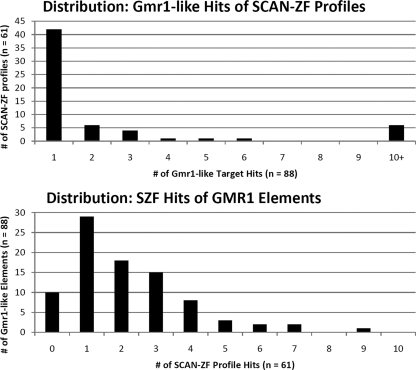
Given the set of putative SCAN-ZF genes above, we constructed a predicted DNA binding profile for each tandem ZF array in these SCAN-ZF genes as described in Materials and Methods (24, 31, 33). These SCAN-ZF genes yielded 266 tandem ZF arrays whose profiles were used to search the DNA sequences of 89 predicted Gmr1-like elements in Anolis, and the significance for binding relationships was analyzed in terms of the false discovery rate (FDR). This analysis revealed significant binding relationships between Anolis SZF and Anolis Gmr1-like elements, with 166 pairwise binding hypotheses considered significant at an FDR of 6.5% (that is, about 10.7 of the 166 hypotheses are expected to be falsely positive). Five data sets consisting of sequence-shuffled Gmr1-like elements produced no statistically significant tests (the estimated false-positive rate when considering all tests significant was 1 in these cases). There were statistically significant matches to the binding profiles of at least one putative SCAN-ZF gene in 78 out of the 89 Gmr1-like elements tested, indicating that targeted binding by SCAN-ZF genes is widespread among this family of lizard retroelements, as predicted. Figure 5 shows the distribution of how many Gmr1-like elements match the binding profile of each Anolis SCAN-ZF and vice versa. While putative binding relationships are widespread among Gmr1-like elements (a large majority have at least one predicted binding partner, and 22 out of 89 Gmr1-like elements account for half the total predicted binding relationships), putative binding matches among Anolis SCAN-ZFs are considerably more concentrated (only 56 out of 266 profiles generated one or more significant binding match, and the top 5 SCAN-ZF profiles account for half of the 166 total significant binding matches). The presence of many SCAN-ZF profiles that do not appear to match Gmr1-like elements suggests that targeting of Gmr1-like elements is not the only role for this family of Anolis lizard genes.
Fig. 5.
Distribution of SCAN-ZF/Gmr1-like binding relationships. (Top) Distribution of the number of predicted Gmr1-like targets (out of 88 total) for each of the 61 SCAN-ZF profiles with at least 1 predicted Gmr1-like target. Most SCAN-ZF profiles are predicted to bind only 1 Gmr1-like element, but 6 profiles are predicted to bind 10 or more Gmr1-like elements each. (Bottom) Distribution of the number of SCAN-ZF profiles predicted to bind to each of the 88 identified Anolis Gmr1-like elements. Of 88 Gmr1-like elements, 78 are predicted to be bound by at least one SCAN-ZF profile: 29 Gmr1-like elements have one predicted binding partner, and 49 Gmr1-like elements are matched by more than one predicted SCAN-ZF profile. All predicted binding relationships and associated statistics can be found in Data S6 in the supplemental material.
Eleven putative SCAN-ZF genes had predicted binding matches to three or more Gmr1-like elements, allowing us to ascertain whether each bound a single sequence element of the Gmr1-like structure consistently. Five SCAN-ZF profiles were predicted to bind three or more Gmr1-like elements and had a consistent binding pattern: PSKZ95 matches a primer binding site (PBS)-Leu (AAG/TAG) sequence (Fig. 6), PSKZ36 and PSKZ142 are predicted to bind coding sequences at similar locations between the PRO and INT domains of the pol gene, and PSKZ141 and PSKZ156 both bind a highly structured element in the 5′ untranslated region (5′ UTR) (see Fig. S3 in the supplemental material) (18). The predicted binding location of each of these five proteins is shown in Fig. 7. Six other Anolis SCAN-ZF profiles that matched three or more Gmr1-like elements had heterogeneous binding patterns, with many hits off the predicted 5′ or 3′ ends of the Gmr1-like elements themselves, which could be artifacts of the flanking sequence included in our Gmr1-like elements due to the uncertain boundaries of the elements.
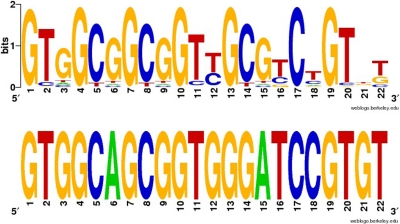
Fig. 6.
Predicted binding profile of PSKZ95. (Top) Predicted binding profile of PSKZ95 (anoCar1 scaffold_401:302793–304163). (Bottom) Consensus sequence of predicted primer binding site (PBS)-Leu (AAG/TAG) sites from Anolis carolinensis, the predicted binding target for PSKZ95. The predicted binding profile suggests that PSKZ95 specifically targets the PBS-Leu elements of many Gmr1-like elements. The logo and consensus representation were created with WebLogo (8).
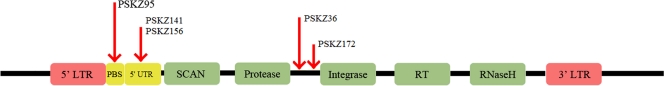
Fig. 7.
Binding targets of predicted SZF genes. A schematic of the canonical structure of Gmr1-like elements in the Anolis lizard. Boundaries of the LTR sequences and protein domains within the Gmr1 ORF are approximate and correspond to protein domain matches or LTR_FINDER results (see Materials and Methods). The predicted binding targets of SCAN-ZF genes are marked with a red arrow and represent position with respect to the canonical domain structure. One red arrow is shown per binding target: PSKZ36 and PSKZ172 are both predicted to bind between the protease and integrase domains but not at exactly the same position and not in the same set of Gmr1-like elements, while PSKZ141 and PSKZ156 bind to the same sequence feature and often in the same Gmr1-like elements.
Other SCAN genes in humans.
There are three RefSeq human genes that encode a clear SCAN domain but no tandem zinc fingers (SCAND1, SCAND3, and PGBD1). Curiously, two of these genes are comprised of an N-terminal SCAN exon and a C-terminal exon that is clearly derived from the transposase of DNA transposons (Charlie for SCAND3 and PiggyBac for PGDB1). SCAND3 has an additional internal coding exon derived from a retroviral INT domain. Both genes have well-conserved orthologs across Eutheria (placental mammals), suggesting that they have host functions (data not shown). We cannot offer a specific explanation for these genes, but the direct association of SCAN with other transposon-derived sequences is interesting.
DISCUSSION
The ultimate function of proteins bearing the SCAN domain is not well understood at present. There is abundant evidence for a common role of the SCAN domain as a mediator of protein-protein interactions, but evidence for binding partners of specific SCAN domains is limited to a small set of genes (10, 35, 36, 38, 45), and even a detailed knowledge of the homo- and heterodimers formed by SCAN-ZF proteins might not reveal their functions beyond DNA binding. Our investigation of the origins of the SCAN domain was informed by several recent results indicating a link between KRAB-ZF genes and transcriptional repression of endogenous and exogenous retroviral sequences (30, 34, 47) and linking the evolution of the KRAB-ZF gene family to LTR retrotransposons and endogenous retroviruses (42), all of which put a new and interesting perspective on the retroviral affinities of the SCAN domain (23, 36).
Building on previous evidence linking the sequence and structure of the SCAN domain to retrotransposon and retroviral sequences, we have conclusively identified the origin of the SCAN domain as an exaptation of the CA sequence of the Gmr1-like family of Gypsy retrotransposons. This finding suggests that previous results regarding the assembly of SCAN multimers may be directly applicable to at least one group of retrotransposons (23, 26). We propose that somewhere at or near the root of the amniote branch, the CA sequence of a Gmr1-like element was inserted upstream of a KRAB-ZF gene and spliced into its coding sequence to form the host ESCAN domain. This exapted sequence was then truncated from its full-length form to the shorter SCAN domain by deletion of its N terminus. The presence of both SCAN and ESCAN domains only in Anolis lizard suggests that the truncation of ESCAN to form the SCAN domain happened shortly after its fusion to a KRAB-ZF gene structure. Despite subsequent loss of Gmr1-like retrotransposons in multiple lineages, including mammals, the SCAN domain has persisted as a common feature of many ZF gene complements, though in some cases (most notably in birds), Gmr1-like elements are not detected and the SCAN domain is also rare or absent.
We presume that the involvement of KRAB through KAP1 and SETDB1 in retroelement repression is ancestral on the basis of the role of KAP1 and SETDB in endogenous retroviral repression in mice and the widespread conservation of these genes. Additionally, while the KRAB domain is best known for transcriptional repression of genomic DNA target sequences, recent work has demonstrated that KRAB-mediated repression can affect transcription of episomal retroviral genes, and additionally, the KRAB domain has been shown to inhibit the genomic integration of HIV-1 (1, 3). These results suggest that in some cases it may be selectively advantageous for an organism to target KRAB domains to retroviral DNA sequences even before integration. We therefore speculate that the fusion of a retrotransposon capsid sequence to KRAB-ZF genes initially targeted these gene products to Gmr1-like capsid structures (and possibly to other retroviral or retrotransposon particles) by heteromultimerization with other CA proteins, which might allow for interaction between KRAB-ZF proteins and newly synthesized retrotransposon DNA before genomic integration and be selected for on that basis.
This hypothesis implies that at least some SCAN-ZF proteins should bind LTR retrotransposon, and particularly Gmr1-like, sequences. We analyzed the binding patterns of Anolis SCAN-ZF proteins, since it is the only sequenced species with both SCAN-ZF proteins and Gmr1-like elements, and demonstrated statistically significant matches between SCAN-ZF transcription factors and Gmr1-like element targets. Five Anolis SCAN-ZF genes are clearly and consistently predicted to bind various sites within Gmr1-like retroelements, including the primer binding site, the 5′ UTR, and two sites in the coding sequence of the pol gene. These results are in accordance with the known targeting of mouse Zfp809 to the murine leukemia virus (MLV) PBS (47) but also suggest other sites that may be effective for transcriptional regulation of retroelements. KRAB-mediated transcriptional silencing is known to spread many kilobases from the initial site of DNA binding, so precise targeting of specific functional elements may not be required for the ultimate function of some KRAB-ZF and SCAN-KRAB-ZF genes (16).
Our results provide more evidence that tandem ZF proteins specifically target endogenous retroelements and suggest that such a role may have been important in the recruitment and original function of the SCAN domain. If this is so, then the large ZF gene family might represent a very substantial system dedicated to host control of retroelements. However, there are many Anolis SCAN-ZF proteins that are not predicted to bind Gmr1-like retrotransposons, and extant mammalian SCAN-ZF proteins cannot possibly interact with Gmr1-like retrotransposons since they were lost early on the mammalian lineage. One possibility is that the SCAN domain can interact with capsids from other classes of retrotransposons and retroviruses, but further experiments will be necessary to fully elucidate the role of the SCAN domain in tandem ZF genes.
Supplementary Material
ACKNOWLEDGMENT
No extramural funding was provided for this work.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Allouch A., et al. 2011. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9: 484–495 [DOI] [PubMed] [Google Scholar]
- 2. Anisimova M., Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate and powerful alternative. Syst. Biol. 55: 539–552 [DOI] [PubMed] [Google Scholar]
- 3. Barde I., et al. 2009. Regulation of episomal gene expression by KRAB/KAP1-mediated histone modifications. J. Virol. 83: 5574–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bellefroid E. J., et al. 1993. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 12: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bellefroid E. J., et al. 1991. The evolutionarily conserved Krüppel-associated box domain defined a subfamily of eukaryotic multifingered proteins. Proc. Natl. Acad. Sci. U. S. A. 88: 3608–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birtle Z., Ponting C. P. 2006. Meisetz and the birth of the KRAB motif. Bioinformatics 22: 2841–2845 [DOI] [PubMed] [Google Scholar]
- 7. Carlson K. A., et al. 2004. Molecular characterization of a putative antiretroviral transcriptional factor, OTK18. J. Immunol. 172: 381–391 [DOI] [PubMed] [Google Scholar]
- 8. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dehal P., et al. 2001. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science 293: 104–111 [DOI] [PubMed] [Google Scholar]
- 10. Edelstein L. C., Collins T. 2005. The SCAN domain family of zinc finger transcription factors. Gene 359: 1–17 [DOI] [PubMed] [Google Scholar]
- 11. Eickbush T. H., Jamburuthugoda V. K. 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134: 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emerson R. O., Thomas J. H. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 5: e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedman J. R., et al. 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10: 2067–2078 [DOI] [PubMed] [Google Scholar]
- 15. Goodwin T. J., Poulter R. T. 2002. A group of deuterostome Ty3/gypsy-like retrotransposons with Ty1/copia-like pol-domain orders. Mol. Genet. Genomics 267: 481–491 [DOI] [PubMed] [Google Scholar]
- 16. Groner A. C., et al. 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatic spreading. PLoS Genet. 6: e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guindon S., Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- 18. Hamada M., et al. 2009. Predictions of RNA secondary structure using generalized centroid estimators. Bioinformatics 25: 465–473 [DOI] [PubMed] [Google Scholar]
- 19. Hamilton A. T., et al. 2006. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 16: 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horiba M., et al. 2007. OTK18, a zinc-finger protein, regulates human immunodeficiency virus type 1 long terminal repeat through two distinct regulatory regions. J. Gen. Virol. 88: 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huson D. H., et al. 2007. Dendroscope–an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Itokawa Y., et al. 2009. KAP1-independent transcriptional repression of SCAN-KRAB-containing zinc finger proteins. Biochem. Biophys. Res. Commun. 388: 689–694 [DOI] [PubMed] [Google Scholar]
- 23. Ivanov D., et al. 2005. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol. Cell 17: 137–143 [DOI] [PubMed] [Google Scholar]
- 24. Joachims T. 1999. Making large-scale SVM learning practical. In Schölkopf B., Burges C., Smola A. (ed.), Advances in kernel methods: support vector learning. MIT Press, Cambridge, MA. [Google Scholar]
- 25. Jurka J., et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110: 462–467 [DOI] [PubMed] [Google Scholar]
- 26. Kingston R. L., Vogt V. M. 2005. Domain swapping and retroviral assembly. Mol. Cell 17: 166–176 [DOI] [PubMed] [Google Scholar]
- 27. Lander E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- 28. Reference deleted.
- 29. Loytynoja A., Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. U. S. A. 102: 10557–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsui T., et al. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- 31. Myers S., et al. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pengue G., et al. 1994. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 22: 2908–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Persikov A. V., Osada R., Singh M. 2009. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics 25: 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rowe H. M., et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- 35. Sander T. L., Haas A. L., Peterson M. J., Morris J. F. 2000. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein association. J. Biol. Chem. 275: 12857–12867 [DOI] [PubMed] [Google Scholar]
- 36. Sander T. L., et al. 2003. The SCAN domain defines a large family of zinc finger transcription factors. Gene 310: 29–38 [DOI] [PubMed] [Google Scholar]
- 37. Schultz D. C., et al. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schumacher C., et al. 2000. The SCAN domain mediates selective oligomerization. J. Biol. Chem. 275: 17173–17179 [DOI] [PubMed] [Google Scholar]
- 39. Storey J. D., Taylor J. E., Siegmund D. 2004. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: a unified approach. J. Royal Stat. Soc. B 66: 187–205 [Google Scholar]
- 40. Subramanian A. R., Kaufmann M., Morgenstern B. 2008. DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol. Biol. 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas J. H., Emerson R. O. 2009. Evolution of C2H2-zinc finger genes revisited. BMC Evol. Biol. 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas J. H., Schneider S. E.Coevolution of retroelements and tandem zinc finger genes. Genome Res. [22 July 2011.]. doi: 10.1101/gr.121749.111. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 43. Touzet H., Varre J. S. 2007. Efficient and accurate P-value computation for Position Weight Matrices. Algorithms Mol. Biol. 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Venter J. C., et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- 45. Williams A. J., Blacklow S. C., Collins T. 1999. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell. Biol. 19: 8526–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williams A. J., Khachigian L. M., Shows T., Collins T. 1995. Isolation and characterization of a novel zinc-finger protein with transcriptional repressor activity. J. Biol. Chem. 270: 22143–22152 [DOI] [PubMed] [Google Scholar]
- 47. Wolf D., Goff S. P. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu Z., Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35: W265–W268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.