Abstract
Avian bornaviruses (ABV), identified in 2008, infect captive parrots and macaws worldwide. The natural reservoirs of these viruses are unknown. Reverse transcription-PCR (RT-PCR) was used to screen oropharyngeal/cloacal swab and brain samples from wild Canada geese (Branta canadensis) for ABV. Approximately 2.9% of swab samples were positive for bornavirus sequences. Fifty-two percent of brain samples from 2 urban flocks also tested positive, and brain isolates were cultured in duck embryo fibroblasts. Phylogenetic analyses placed goose isolates in an independent cluster, and more notably, important regulatory sequences present in Borna disease virus but lacking in psittacine ABVs were present in goose isolates.
TEXT
Borna disease virus (BDV) is a nonsegmented negative-strand RNA virus (order Mononegavirales, genus Bornaviridae) best known for causing neurologic disease in horses in central Europe, where the virus is likely transmitted via shrew urine (9, 15). While BDV is considered an infectious agent of mammals, there have been reports of BDV-associated infections in birds. Malkinson and colleagues identified BDV as the cause of a neurologic disease in captive ostriches (12), and Berg et al. used PCR to detect the presence of BDV sequences in mallard (Anas platyrhyncos) and jackdaw (Corvus monedula) droppings (1). The significance of these findings was unclear, however, since in neither case were the viruses cultured, leaving BDV infection of birds largely unappreciated.
Proventricular dilatation disease (PDD) was first found in large parrots, such as macaws and conures, in North America and Europe around 1977 (8, 13). PDD is primarily an encephalomyelitis that affects enteric ganglia, resulting in a loss of gut motility and enlargement of the paralyzed proventriculus (8). The cause of PDD was unresolved until 2008, when Kistler et al. (11) and Honkavuori et al. (10) detected bornaviruses during investigations into the cause of the disease and identified several distinct avian bornavirus (ABV) genotypes. Subsequent identification of ABV sequences and/or antigen in brains of birds with neurologic illness supported the initial findings (21, 22), and experimental challenge of parrots with brain homogenates (6) or ABV-infected duck embryo fibroblasts (DEF) (7, 14) caused PDD-like lesions. Thus, avian bornaviruses are now recognized as potential pathogens with widespread distribution (18).
In 1991, Daoust and colleagues reported two cases of a PDD-like disease in Canada geese (Branta canadensis) (CG) from Prince Edward Island, Canada (4), and more recently Delnatte et al. (5) used archival material from waterfowl, recorded as suffering from ill-defined neurologic disease, and detected ABV in Canada geese and swans. These findings motivated us to screen samples from apparently healthy geese. In this report, we describe the detection and recovery of a distinct bornavirus lineage from healthy Canada geese.
Samples were collected by the Wildlife Services Agency of the U.S. Department of Agriculture-Animal and Plant Health Inspection Service (USDA-APHIS) as part of an avian influenza virus (AIV) survey. Combined oropharyngeal/cloacal swab samples were placed in viral transport media, tested for AIV, and stored at −80°C. Swabs collected from 409 CG between May 2008 and November 2009 were tested for ABV. The samples (negative for AIV) were selected to broadly cover the United States and span the four major migratory bird flyways. RNA was purified from 140 μl of clarified transport medium using a viral RNA minikit (Qiagen).
Heads from 25 CG were collected in April 2011, shipped on wet ice to our laboratory and immediately frozen at −80°C. These samples were from nuisance and hunter-harvested geese collected from Newark, Union, and Somerset Counties in New Jersey. Heads were processed in a class II biological safety cabinet using sterile technique and procedures to avoid sample cross-contamination. Brain tissue (0.2 to 0.5 g) was homogenized by vortexing in RLT lysis buffer (Qiagen) and passage through a 20-gauge needle. Lysates were applied to an RNeasy minicolumn (Qiagen), washed, and eluted in 50 μl of nuclease-free water. cDNA was generated using the Applied Biosystems High Capacity cDNA reverse transcription kit (Applied Biosystems), using approximately 500 ng of RNA and random primers. PCR targeted the matrix (M) gene (1990F, 5′-GGTAATTGTTCCTGGATGGC-3′; 2322R, 5′-ACACCAATGTTCCGAAGACG-3′). PCR conditions were as follows: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and one cycle of 5 min at 72°C. PCR assays included multiple reagent controls.
Bornavirus sequences were detected in 12 of 409 (2.9%) swab samples. Positive samples were identified in flocks from 5 of the 12 U.S. states represented (Table 1). Bornavirus was also detected in 11 of 25 (52%) brain samples. Seven positive CG were from a nuisance flock, while 4 were hunter harvested. Viral sequences were detected in the forebrain and cerebellum or forebrain only from 7 and 4 CG, respectively. The difference between the detection rate for brain tissue and that for swab material was expected, since we have demonstrated that fecal shedding is intermittent for experimentally and naturally infected cockatiels, African gray parrots, and mallards (14, 19; I. Tizard and G. Jianhua, unpublished data). Birds may shed detectable (by PCR) amounts of ABV on a single occasion and then test negative for months. However, since positive swab samples from only 5 of 12 states were identified, and the brain samples originated with Canada geese harvested in New Jersey, it is possible that the high level of ABV detected in brain samples is particular to the two urban flocks tested.
Table 1.
Sampling sites yielding confirmed CG bornavirus sequences
| Sexa | Sample | Location | No. of ABV-positive samples |
|---|---|---|---|
| ND | O/Cb | Queens, NY | 2 |
| F | O/C | Cheshire, NH | 1 |
| M | O/C | Island, WA | 1 |
| F | O/C | King, WA | 1 |
| F | O/C | Multnomah, OR | 2 |
| M | O/C | Multnomah, OR | 3 |
| F | O/C | Tillamook, OR | 1 |
| F | O/C | Ramsey, MN | 1 |
| ND | Brain | Newark Airport, NJ | 7 |
| ND | Brain | Union and Somerset Counties, NJ | 4 |
ND, not determined; F, female; M, male.
O/C, combined oropharyngeal/cloacal swab.
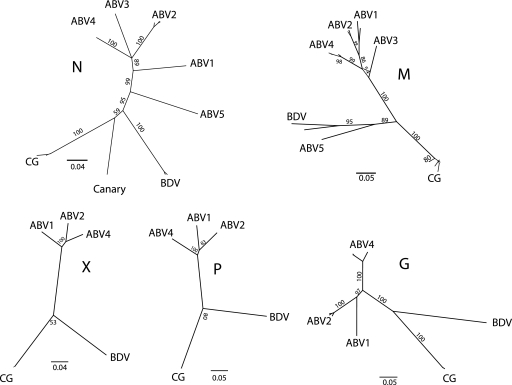
A 200-nucleotide (nt) region of the M gene was aligned for 24 CG samples. As determined using pairwise comparisons, the nucleotide identities between the samples ranged from 95.8% to 100%. In contrast, CG-derived sequences shared only 68% (most divergent pairing) to 73% (most conserved pairing) nucleotide identity with parrot ABVs. Comparison of GC-derived bornaviruses with BDV revealed nucleotide identities ranging from 67% to 69%. A phylogeny generated using M sequences placed the CG-derived samples in a cluster distinct from other ABVs (Fig. 1).
Fig. 1.
Phylogeny based on partial M sequences of bornaviruses from birds and mammals. A consensus tree was generated using a neighbor-joining algorithm, with no outgroup assigned. To generate the consensus tree, 1,000 bootstrap replicates were generated. The bootstrap values are shown at the major nodes. Previously identified psittacine ABV groups 1 to 5 are in green. The CG cluster is red; mammalian bornaviruses are in blue. Isolates are identified by accession number: FJ620690 (ABV2, 6609), EU781967 (ABV2, bil), HM998710 (ABV2NM-CT15), GU24959 (ABV1, NM-M25), FJ002329 (ABV1, VTH1561/06), FJ002328 (ABV3, KD), FJ002331 (ABV4, VTH1688/07), HQ123584 (strain CG_2002), NM-20 (JN014949), NM-06 (JN014948), FJ002334 (ABV5, ABRC-98-512), NC_001607 (BDV), and AJ311524 (BDV, strain No/98). Canada goose viruses are as follows: JN251048 (CG-NY86-2008), JN251049 (CG-NH1502008), JN251050 (CG-OR409-2009), JN251051 (CG-NY89-2008), JN251052 (CG-OR377-2009), JN251053 (CG-OR380-2009), JN251054 (CG-OR332-2009), JN251055 (CG-OR-378-2009), JN251056 (CG-EWR64-2011), JN251057 (CG-EWR62-2011), JN251058 (CG-EWR72-2011), JN251059 (CG-NJ81-2011), JN251060 (CG-NJ82-2011), JN251061 (CG-MN-2008), JN251062 (CGWA400-2009), JN251063 (CG-WA363-2008), JN251064 (CG-OR381-2009), JN251065(CG-EWR71-2011), JN251066 (CG-EWR66-2011), JN251067 (CG-EWR65-2011), JN251068 (CG-EWR76-2011), JN251069 (CG-EWR68-2011), JN251070 (CGNJ83-2011), JN251071 (CGNJ84-2011), JN251072 (CG-EWR64-2011), JN251073 (CG-EWR-64-2011), JN251074 (CG-NJ82-2011), and JN251075 (CG-NJ82-2011).
Upon finding that CG M sequences formed an independent cluster, partial N, X, P, and G sequences were amplified from two brain samples with primers: 634F (5′-CCTCATGAGGCTATTGATTGG-3′) and 991R (5′-AGTAGAATGCCGCAGAAGC-3′) for N, 634F and 2304R (5′-ACACCAATGTTCCGAAGACG-3′) for N-X-P-M, and 2068F (5′-AAGGAACCGCTCCAACTC-3′) and 3238R (5′-GCCARAYAACMCCDAYYCCATT-3′) for M-G. Additional sequencing primers were LC__CGseq1 (5′ GAAGACGTGAGGTGACTAGAGG 3′) and LC__CGseq2 (5′ AGAGACAATCCAGGCTATTC 3′).
Sequences were generated with an ABI PRISM 3100 genetic analyzer (Applied Biosystems) and edited using the Sequencher 4.1 and Geneious Pro 5.1.7 software programs. Pairwise sequence comparison of virus from these samples showed 97.7% nucleotide identity for all regions, and these isolates also shared 97% nucleotide identity with N sequences from an earlier CG virus (CG_2002; GenBank Accession no. HQ123584). Phylogenies were generated using CG brain isolates and a selection of other bornaviruses (Fig. 2). The N, X, P, and G phylogenies showed the CG bornaviruses forming a group distinct from psittacine ABVs. Viruses from psittacine ABV groups 3 and 5 were not included in some trees because sequences were not available. The N phylogeny included a previously reported canary sequence (22) that was more closely related to the CG bornaviruses than the psittacine ABVs.
Fig. 2.
Phylogenies generated from partial N, X, P, M, and G sequences of avian and mammalian bornaviruses. Accession numbers of sequences used to develop these phylogenies are as follows: FJ620690 (ABV2, 6609), EU781967 (ABV2, bil), HM998710 (ABV2 NM-CT15), GU24959 (ABV1 NM-M25), FJ002329 (ABV1 VTH1561/06), FJ002328 (ABV3, KD), FJ002331 (ABV4, VTH1688/07), JN035149 (ABV4, NM-01), FJ002334 (ABV5, ABRC-98-512), NC_001607 (BDV), HQ123584 (ABV CG_2002), and GQ161095 (canary). Sequences used to generate the M tree are provided in Fig. 1.
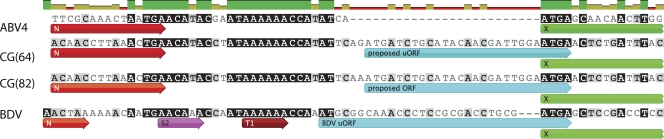
An alignment of the N/X intergenic region of two CG bornaviruses with ABV group 4 and BDV unexpectedly revealed that the CG bornaviruses were configured much like BDV (Fig. 3). The N/X intergenic region of BDV is 56 nt long and contains, just upstream of the X gene start codon, a 9-amino-acid-encoding open reading frame (uORF) that affects X expression (20). The N/X intergenic region of the two CG bornaviruses was 54 nt long, with an 8-amino-acid-encoding uORF whose termination codon overlaps the X start codon, as does the BDV uORF. As reported previously, the N-X intergenic region of psittacine ABVs is 27 nt long and lacks the BDV regulatory uORF (11, 16).
Fig. 3.
Comparison of bornavirus N-X intergenic regions. The end of the N gene (red) and the start of the X gene (green) are shown. The BDV uORF and proposed CG bornavirus uORFs are indicated in blue. The S2 transcription initiation site and the T1 termination site for BDV are also shown (2, 3, 17).
To recover CG isolates, sections of brain were homogenized in minimum essential medium with Earle's balanced salt solution (MEM-Earle's) and filtered. Filtrates were added to DEF in MEM-Earle's containing 10% fetal bovine serum and 50 μg/ml penicillin-streptomycin. Viral sequences were detected by PCR after 2 passages (data not shown), and viral antigen was detected by indirect fluorescent-antibody assay (IFA) after 3 cell passages, using as primary antibody serum from a macaw with confirmed PDD (data not shown).
PDD was first found in large captive parrots and macaws in the late 1970s (10–12), suggesting that ABV may have moved into this population from an unknown natural reservoir. Sporadic reports of PDD-like illness and/or BDV infection of wild birds (1, 4) suggested that waterfowl might be that reservoir. However, those reports did not suggest that bornavirus infection is common in these species or that healthy birds might serve as carriers, as we have demonstrated. In addition, we have shown that CG isolates are more closely related to mammalian bornaviruses than to other avian isolates, sharing important regulatory sequences with BDV.
Acknowledgments
We thank Darrel Styles of the USDA/APHIS/Veterinary Services for his contributions to this project, as well as the many wildlife disease biologists from the USDA/APHIS/WS National Wildlife Disease Program for collecting the field samples. The USDA/APHIS/WS Wild Bird Tissue Archive was invaluable to the success and cost-effectiveness of this study. We thank L. Ramirez for expert technical assistance.
This project was supported by the Richard M. Schubot Endowment at Texas A&M University.
Footnotes
Published ahead of print on 7 September 2011.
REFERENCES
- 1. Berg M., Johansson M., Montell H., Berg A. L. 2001. Wild birds as a possible natural reservoir of Borna disease virus. Epidemiol. Infect. 127: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Briese T., et al. 1994. Genomic Organization of Borna-Disease Virus. Proc. Natl. Acad. Sci. U. S. A. 91: 4362–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cubitt B., Oldstone C., de la Torre J. C. 1994. Sequence and genome organization of Borna disease virus. J. Virol. 68: 1382–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daoust P. Y., Julian R. J., Yason C. V., Artsob H. 1991. Proventricular impaction associated with nonsuppurative encephalomyelitis and ganglioneuritis in 2 Canada geese. J. Wildl. Dis. 27: 513–517 [DOI] [PubMed] [Google Scholar]
- 5. Delnatte P., et al. 2011. New genotype of avian bornavirus in wild geese and trumpeter swans in Canada. Vet. Rec. 169: 108. [DOI] [PubMed] [Google Scholar]
- 6. Gancz A. Y., et al. 2009. Experimental induction of proventricular dilatation disease in cockatiels (Nymphicus hollandicus) inoculated with brain homogenates containing avian bornavirus 4. Virol. J. 6: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gray P., et al. 2010. Use of avian bornavirus isolates to induce proventricular dilatation disease in conures. Emerg. Infect. Dis. 16: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gregory C., et al. 1994. A review of proventricular dilatation syndrome. J. Assoc. Avian Vet. 8: 69–75 [Google Scholar]
- 9. Hilbe M., et al. 2006. Shrews as reservoir hosts of Borna disease virus. Emerg. Infect. Dis. 12: 675–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Honkavuori K. S., et al. 2008. Novel borna virus in psittacine birds with proventricular dilatation disease. Emerg. Infect. Dis. 14: 1883–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kistler A. L., et al. 2008. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol. J. 5: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malkinson M., Weisman Y., Ashash E., Bode L., Ludwig H. 1993. Borna disease in ostriches. Vet. Rec. 133: 304. [DOI] [PubMed] [Google Scholar]
- 13. Mannl A., Gerlach H., Leipold R. 1987. Neuropathic gastric dilatation in psittaciformes. Avian Dis. 31: 214–221 [PubMed] [Google Scholar]
- 14. Payne S., et al. 2011. Unusual and severe lesions of proventricular dilatation disease in cockatiels (Nymphicus hollandicus) acting as healthy carriers of avian bornavirus (ABV) and subsequently infected with a virulent strain of ABV. Avian Pathol. 40: 15–22 [DOI] [PubMed] [Google Scholar]
- 15. Puorger M. E., et al. 2010. Distribution of Borna disease virus antigen and RNA in tissues of naturally infected bicolored white-toothed shrews, Crocidura leucodon, supporting their role as reservoir host species. Vet. Pathol. 47: 236–244 [DOI] [PubMed] [Google Scholar]
- 16. Rinder M., et al. 2009. Broad tissue and cell tropism of avian bornavirus in parrots with proventricular dilatation disease. J. Virol. 83: 5401–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schneemann A., Schneider P. A., Kim S., Lipkin W. I. 1994. Identification of signal sequences that control transcription of Borna-disease virus, a nonsegmented, negative-strand RNA virus. J. Virol. 68: 6514–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Staeheli P., Rinder M., Kaspers B. 2010. Avian bornavirus associated with fatal disease in psittacine birds. J. Virol. 84: 6269–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Villanueva I., et al. 2010. The diagnosis of proventricular dilatation disease: use of a Western blot assay to detect antibodies against avian Borna virus. Vet. Microbiol. 143: 196–201 [DOI] [PubMed] [Google Scholar]
- 20. Watanabe Y., Ohtaki N., Hayashi Y., Ikuta K., Tomonaga K. 2009. Autogenous translational regulation of the Borna disease virus negative-control factor X from polycistronic mRNA using host RNA helicases. Plos Pathog. 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weissenbock H., et al. 2009. Avian bornaviruses in psittacine birds from Europe and Australia with proventricular dilatation disease. Emerg. Infect. Dis. 15: 1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weissenbock H., Sekulin K., Bakonyi T., Hogler S., Nowotny N. 2009. Novel avian bornavirus in a nonpsittacine species (Canary; Serinus canaria) with enteric ganglioneuritis and encephalitis. J. Virol. 83: 11367–11371 [DOI] [PMC free article] [PubMed] [Google Scholar]