Abstract
Polydnaviruses (PDVs) are symbionts of parasitoid wasps that function as gene delivery vehicles in the insects (hosts) that the wasps parasitize. PDVs persist in wasps as integrated proviruses but are packaged as circularized and segmented double-stranded DNAs into the virions that wasps inject into hosts. In contrast, little is known about how PDV genomic DNAs persist in host cells. Microplitis demolitor carries Microplitis demolitor bracovirus (MdBV) and parasitizes the host Pseudoplusia includens. MdBV infects primarily host hemocytes and also infects a hemocyte-derived cell line from P. includens called CiE1 cells. Here we report that all 15 genomic segments of the MdBV encapsidated genome exhibited long-term persistence in CiE1 cells. Most MdBV genes expressed in hemocytes were persistently expressed in CiE1 cells, including members of the glc gene family whose products transformed CiE1 cells into a suspension culture. PCR-based integration assays combined with cloning and sequencing of host-virus junctions confirmed that genomic segments J and C persisted in CiE1 cells by integration. These genomic DNAs also rapidly integrated into parasitized P. includens. Sequence analysis of wasp-viral junction clones showed that the integration of proviral segments in M. demolitor was associated with a wasp excision/integration motif (WIM) known from other bracoviruses. However, integration into host cells occurred in association with a previously unknown domain that we named the host integration motif (HIM). The presence of HIMs in most MdBV genomic DNAs suggests that the integration of each genomic segment into host cells occurs through a shared mechanism.
INTRODUCTION
Viruses in the family Polydnaviridae consist of two genera, Bracovirus (BV) and Ichnovirus (IV), which are symbiotically associated with parasitoid wasps (Hymenoptera) in the families Braconidae and Ichneumonidae (summarized in references 12, 31, 32, and 41). Each polydnavirus (PDV) from a given wasp species is genetically unique and exists in two forms. In the wasp, PDVs persist and are transmitted to offspring as stably integrated proviruses with replication restricted to specialized calyx cells in the ovaries of females. Replication results in the production of the encapsidated form of the virus, which accumulates in the lumen of the calyx. Wasps then inject the encapsidated form of the virus into host insects that they parasitize. For most PDV-carrying wasp species, these hosts are larval-stage Lepidoptera (moths and butterflies). Following parasitism, PDVs do not propagate themselves in the wasp's host because the encapsidated form of the genome lacks most or all of the genes required for replication. However, the encapsidated form of the genome encodes several other genes that alter the development and immune defenses of the host. These alterations in turn enable the wasp's offspring to successfully develop.
The encapsidated genomes of PDVs are segmented and consist of multiple circular double-stranded DNAs that are nonequimolar in abundance (12, 13, 27, 32, 41). In the case of Microplitis demolitor bracovirus (MdBV), the genome consists of 15 segments (named segments A to O), which are individually packaged into virions and have an aggregate size of 190 kb (5, 42). The encapsidated MdBV genome encodes 51 open reading frames (ORFs) for predicted proteins of ≥100 amino acids (7, 42). More than half of these ORFs form four multimember gene families, designated the ptp (13 members), ank (12 members), egf (3 members), and glc (2 members) genes, while the remaining ORFs are predicted single-copy genes (7, 42). Following an infection of the host Pseudoplusia (Chrysodeixis) includens, MdBV virions infect primarily hemocytes as well as other tissues, including the fat body and nervous system (5, 30). Transcripts corresponding to nearly all predicted ORFs are detected in one or more of these tissues within 2 h of infection, with expression thereafter continuing for the 7 days required for M. demolitor progeny to complete their development (7, 35, 36). Following wasp emergence, the host larva dies in 2 to 4 days.
While PDV genomes are generally thought to persist in hosts as episomes (28, 38, 41), infection of cell lines with Glyptapanteles indiensis bracovirus (GiBV), Hyposoter fugitivus ichnovirus (HfIV), and Hyposoter didymator ichnovirus (HdIV) suggests that some viral genomic segments persist for long periods by integration (14, 17, 21, 40). The sequencing of host-viral DNA junctions confirmed that one genomic segment from GiBV integrated into two cell lines from the gypsy moth, Lymantria dispar (15), while similar data showed that a genomic segment from Tranosema rostrale ichnovirus (TrIV) integrated into a cell line from the spruce budworm, Choristoneura fumiferana (11). We recently developed a cell line of hemocyte origin from P. includens, named CiE1 cells, which is highly permissive to infection by MdBV and exhibits functional alterations very similar to those of MdBV-infected hemocytes (16). Whether the MdBV genome persists in this cell line, however, is unknown. Here we report that all genomic segments of the MdBV genome persist for long periods in CiE1 cells. Studies focusing on a subset of genomic segments further indicate that persistence is due to integration and that integration also occurs in naturally parasitized P. includens larvae.
MATERIALS AND METHODS
Insects and cell lines.
M. demolitor and P. includens were reared at 27°C with a 16-h-light, 8-h-dark photoperiod as previously described (29, 33). Hosts used in the study were parasitized as third-instar larvae. A single wasp offspring emerges from the host's body on day 7 or 8 to pupate within a silken cocoon, followed by emergence into an adult 4 days later (26). CiE1 cells were cultured in Sf-900 medium (Gibco) supplemented with 5% fetal calf serum (HyClone) (16). Noninfected cells were maintained and passaged weekly as strongly adherent cells in Corning 75-cm2 tissue culture flasks. MdBV-infected cells became nonadherent but were maintained and passaged identically to noninfected cells.
MdBV collection and nomenclature.
MdBV virions were collected from the reproductive tract of adult female wasps in calyx fluid as previously described (2, 35). As is convention in the PDV literature, the amount of MdBV collected from the reproductive tract of a single adult female is defined as one wasp equivalent, which for MdBV contains on average 1 × 1010 virions (5). The encapsidated genome of MdBV was previously deposited in GenBank as individual genomic segments under accession numbers AY887894, AY875680 to AY875690, AY848690, AY842013, and DQ000240 (42). Each genomic segment is named by uppercase letters from smallest (genomic segment A, 3,433 bp) to largest (genomic segment O, 34,355 bp) (42). Nucleotide positions referred to in the study for a given segment correspond to the above-mentioned GenBank submissions, while the abundance of each genomic segment in calyx fluid was previously determined (5). Most predicted genes are named by their location on a given genomic segment (7). Thus, members of the ptp gene family consist of one predicted gene located on genomic segment D (ptp-D1), five on segment H (ptp-H1, ptp-H2, ptp-H3, ptp-H4, and ptp-H5), four on segment J (ptp-J1, ptp-J2, ptp-J3, and ptp-J4), and three on segment N (ptp-N1, ptp-N2, and ptp-N3) (25, 42). Members of the ank gene family are named similarly (ank-C1, ank-C2, ank-F4, ank-F5, ank-G3, ank-G4, ank-H4, ank-I1, ank-J4, ank-N1, ank-N4, and ank-N5), whereas all egf (egf0.4, egf1.0, and egf1.5) and glc (glc1.8 and glc3.2) genes reside on genomic segment O and are named by the size of their corresponding cDNAs (4, 36, 39, 42).
Total RNA isolation and RT-PCR assays.
MdBV-infected CiE1 cells were collected by centrifugation at 200 × g, followed by the isolation of total RNA using the High Pure RNA isolation kit (Roche) according to the manufacturer's instructions. The quantification of RNA was done by using a Nanodrop spectrometer. For first-strand cDNA synthesis, 100 ng of total RNA was reverse transcribed in 20-μl reaction mixtures using random hexamers and Superscript III (Invitrogen). Reverse transcription (RT)-PCRs were run by using a Bio-Rad thermocycler and 25-μl reaction mixture volumes containing 1 μl of cDNA and 0.2 μM appropriate gene-specific primers. Primers used to amplify selected ptp family members were described previously by Pruijssers and Strand (25), primers used to amplify glc1.8/3.2 and egf1.0/1.5 were described previously by Beck and Strand (4), and primers used to amplify selected ank family members were described previously by Bitra et al. (7). Cycling conditions were as follows: an initial denaturation step at 94°C for 2 min, followed by 35 cycles at 94°C for 20 s, annealing at 50°C (ptp, glc, and egf genes) or 55°C (all ank family members) for 10 s, extension at 65°C for 30 s, and a final extension step at 72°C for 7 min. The resulting products were visualized on 1% agarose gels stained with ethidium bromide (EtBr).
Immunoblotting and immunocytochemistry.
MdBV-infected CiE1 cells were placed into lysis buffer and stored at −80°C (20). After protein concentrations were determined by use of the Micro BCA protein assay kit (Pierce), samples were resolved on 4-to-20% gradient SDS-PAGE gels (Lonza), immunoblotted onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore), and blocked (20). The membrane was probed with a murine monoclonal antibody (55F2E7) specific for Glc1.8 and Glc3.2 (1:10,000 dilution) (2, 39). The primary antibody was detected by using a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:20,000 dilution) (Jackson Laboratory), followed by visualization using a chemiluminescent substrate (ECL Advance kit; GE Healthcare) (20). MdBV-infected CiE1 cells were processed for immunofluorescence microscopy as previously outlined (3), by labeling with an anti-Glc1.8/3.2 antibody and an anti-mouse Alexafluor 564-conjugated secondary antibody. Samples were examined by using a Leica IRE2 inverted epifluorescence microscope interfaced (Compix, Cranberry, PA) with SimplePCI software and a Hamamatsu digital camera for image acquisition. Final images were assembled by using Adobe Photoshop.
DNA isolation and PCR-based detection of MdBV genomic segments.
Genomic DNA from adult male M. demolitor, MdBV-infected CiE1 cells, whole parasitized P. includens larvae (7 days postoviposition), or hemocytes from parasitized P. includens larvae (2 h to 8 days postparasitism) was isolated by using the QIAamp DNA minikit (Qiagen). For whole parasitized larvae, no M. demolitor offspring were present in the sample. MdBV genomic DNA was isolated from virions as previously described (35). Genomic DNA isolated from noninfected CiE1 cells or nonparasitized P. includens also served as controls for some experiments. The detection of each MdBV segment in infected CiE1 cells by PCR was conducted by using segment-specific primers as previously described (5). A PCR-based integration assay (1) was used to locate domains of MdBV genomic segments B, C, and J that contained the proviral excision/integration site in M. demolitor and the site of integration into CiE1 cells and P. includens larvae. Briefly, segments B and C were divided into 4 domains and segment J was divided into 5 domains by designing overlapping primer pairs that specifically amplified each region (see Table S1 in the supplemental material). PCRs were then run in 25-μl reaction mixtures containing 0.2 μM each domain-specific primer; 10 ng of MdBV, M. demolitor, CiE1, or P. includens DNA; and 1.25 units of Hotmaster Taq polymerase (5 Prime). Cycling conditions were as follows: an initial denaturation step at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 20 s, and extension at 65°C for 4 min, with a final extension step at 72°C for 7 min.
Inverse PCR.
We used inverse PCR (22) to amplify, clone, and sequence DNA junctions where a given MdBV segment had integrated and joined with flanking M. demolitor, CiE1, or P. includens chromosomal DNA. On the basis of our integration assay data (see Results), nested inverse PCR primer sets for MdBV segments C and J (see Table S2 in the supplemental material) were designed based on the domains identified as the site of integration into M. demolitor, CiE1 cells, or parasitized P. includens. We then digested 5 μg of M. demolitor, CiE1, or P. includens genomic DNA with MfeI and XbaI for the cloning of right and left virus junctions of segment B from M. demolitor, CiE1 cells and P. includens for segment C, and PciI for the left junctions for segment J. Following phenol extraction and ethanol precipitation, precipitated DNAs were resuspended in 10 mM Tris-HCl (pH 8.5), diluted to 2 ng/μl, and used for ligation reactions with T4 DNA ligase (Roche) at 10°C overnight. After ligation, the T4 DNA ligase was heat inactivated at 65°C for 10 min, and 1 μl of the reaction mixture was used as template DNA for the first of three consecutive rounds of 50-μl standard PCR amplifications employing Hotmaster Taq DNA polymerase (5 Prime) and 0.4 μM segment-specific primers (see Table S2 in the supplemental material). Cycling conditions were as follows: an initial denaturation step at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 20 s, and extension at 65°C for 3 min. After the first round of amplification employing the outer primer set, 1 μl of the PCR mixture was used to set up a second round of amplification with the same primers. One microliter from this reaction mixture was then used for a third round of amplification with the nested primer pair. The resulting PCR products from the final amplification were cloned with the StrataClone PCR cloning kit (Stratagene) and sequenced using M13 forward and reverse primers (Macrogen). Sequences were analyzed by using DNAStar (Madison, WI) and BLAST (NCBI).
qPCR and Southern blotting.
To measure the copy numbers of MdBV segments C and J per infected CiE1 cell, total genomic DNA was isolated from 1 × 106 cells as described above, followed by quantitative PCR (qPCR) analysis using segment C-specific (5′-TATGATGATTTGCCGTAAGGGTAA-3′ [forward] and 5′-AGTAGGCCATGTGGTAAGCAGTAT-3′ [reverse]) and segment J-specific (5′-CCAATTCGGAAGGGTCTCG-3′ [forward] and 5′-GGGGTAGCACTTTTGTTTGTTATCT-3′ [reverse]) primers as previously described (5). For Southern blotting, digoxigenin-labeled probes corresponding to nucleotides (nt) 4077 to 5866 on segment C and nt 5409 to 8215 on segment J were synthesized by using digoxigenin-dUTP and DIG High Prime DNA Labeling and Detection Start kit II (Roche). MdBV genomic DNA isolated from virions and CiE1 genomic DNA were digested with XbaI (segment C) or BspHI (segment J), followed by size fractionation on 0.8% agarose gels and transfer onto nylon in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Blots were then prehybridized for 30 min at 40°C in DIG Easy Hyb buffer (Roche), followed by overnight hybridization at 40°C with each probe (30 ng/ml). Blots were washed under conditions of high stringency (0.5× SSC, 0.1% SDS) at 65°C, followed by incubation with antidigoxigenin antibody (Roche) (1:10,000) and visualization using the CSP-Star ready-to-use chemiluminescent substrate (Roche).
RESULTS
MdBV persists in and functionally transforms CiE1 cells.
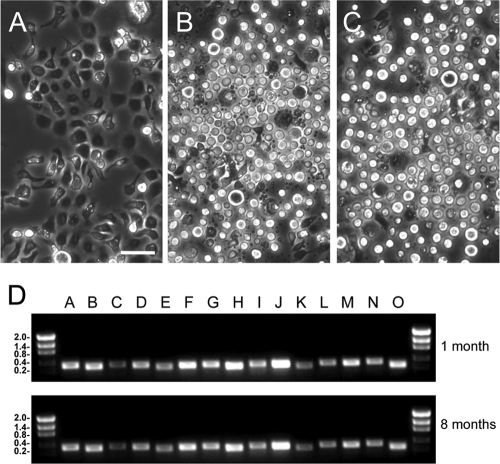
A key function of PDVs in parasitism is to prevent the host's immune system from killing the progeny (eggs and/or larvae) of parasitoids by a cellular defense response called encapsulation (summarized in references 31 and 32). MdBV disables encapsulation by preventing hemocytes called granulocytes and plasmatocytes from adhering to wasp eggs and causing some granulocytes to apoptose (33, 34, 39). The loss of adhesion is due primarily to the expression of the glc genes in infected hemocytes, which encode very similar cell surface glycoproteins (3, 4, 25), while apoptosis is associated with the expression of ptp-H2 (37). We also previously showed that MdBV infection blocks adhesion and causes some CiE1 cells to apoptose, while the RNA interference (RNAi) knockdown of glc gene expression rescues adhesion (16). At the beginning of this study, we infected CiE1 cells with MdBV at an average multiplicity of infection (MOI) of 10, which resulted in most cells becoming nonadhesive and some cells apoptosing at between 24 and 72 h, as previously reported (Fig. 1 A and B). However, apoptosis thereafter declined, with surviving cells remaining nonadhesive but also proliferating at rates comparable to those of uninfected cells. We therefore maintained these cells by passaging them weekly. Strikingly, cells remained nonadhesive after 1 month and 4 passages as well as after 8 months and 39 passages (Fig. 1C). PCR assays using DNA isolated from CiE1 cells as a template and primers specific for each MdBV genomic segment further indicated that each segment persisted over the same period (Fig. 1D).
Fig. 1.
MdBV persists in and transforms CiE1 cells. (A) Phase-contrast micrograph of uninfected CiE1 cells. Note that cells are strongly adhered to and spread on the surface of culture plates. The scale bar equals 100 μm. (B) Phase-contrast micrograph of CiE1 cells 72 h after infection by MdBV. (C) Phase-contrast micrograph of CiE1 cells 8 months after infection by MdBV. Inspection of panel B shows that most cells are rounded and that numerous small blebs are present due to apoptosis, while panel C shows that most cells remain rounded but that few blebs are present. (D) PCR products generated using DNA from infected CiE1 cells as a template and primers specific for each MdBV genomic segment (segments A to O). The top gel shows products produced from CiE1 cells 1 month postinfection, while the bottom gel shows products generated from CiE1 cells 8 months postinfection. Size markers (kb) are shown at the left.
Multiple MdBV genes are persistently expressed in CiE1 cells.
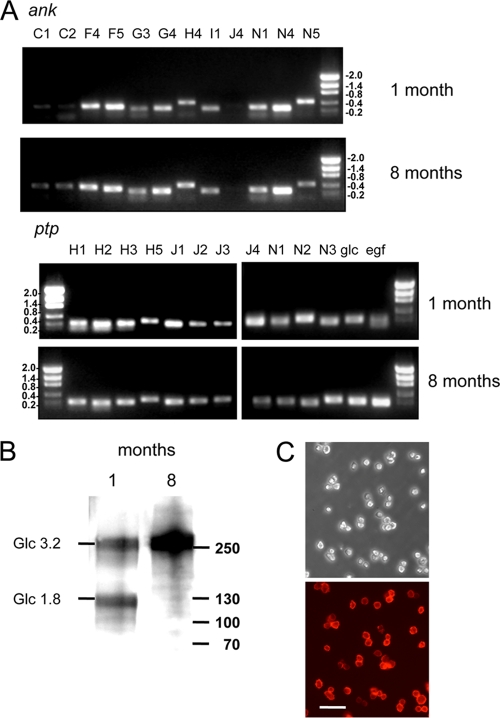
Transcriptome analysis previously showed that a majority of the MdBV ptp, ank, glc, and egf gene family members are expressed in P. includens hemocytes (7). Given that CiE1 cells were transformed from an adhesive to a nonadhesive state after infection and all MdBV genomic segments persisted, we qualitatively assessed by RT-PCR whether the persistent expression of MdBV genes also occurred. Our results showed that most viral gene family members expressed in P. includens hemocytes, including the glc genes responsible for adhesion loss, continued to be expressed in CiE1 cells after 1 and 8 months (Fig. 2 A). In contrast, no amplicons for these gene products were detected in noninfected CiE1 cells (not shown). Immunoblot analysis using an anti-Glc1.8/3.2 antibody detected the presence of both Glc1.8 and Glc3.2 in cell extracts prepared at 1 month postinfection but detected only Glc3.2 at 8 months postinfection (Fig. 2B). Immunocytochemical analysis also showed that virtually all CiE1 cells at 8 months postinfection expressed Glc3.2 on their surface (Fig. 2C).
Fig. 2.
MdBV transcripts are persistently detected in infected CiE1 cells. (A) Total RNA was isolated from CiE1 cells at 1 and 8 months postinfection, followed by RT-PCR analysis using primers specific for selected ank and ptp gene family members. Primers that amplify both glc1.8/3.2 and egf1.0/1.5 were also used. Amplicons for most genes were detected at both 1 and 8 months postinfection. Size markers (kb) are indicated to the right or left of the gels. (B) Immunoblot showing the presence of Glc1.8 and Glc3.2 in CiE1 cell extracts 1 month postinfection and the presence of only Glc3.2 in cells at 8 months postinfection. Molecular mass markers (in kDa) are indicated at the right. (C) Phase-contrast (top) and epifluorescence (bottom) micrographs of CiE1 cells at 8 months postinfection labeled with anti-Glc1.8/3.2 and visualized by using an Alexa 564 secondary antibody. The scale bar in the bottom image equals 180 μm.
MdBV genomic segments B, C, and J persist in CiE1 cells by integration.
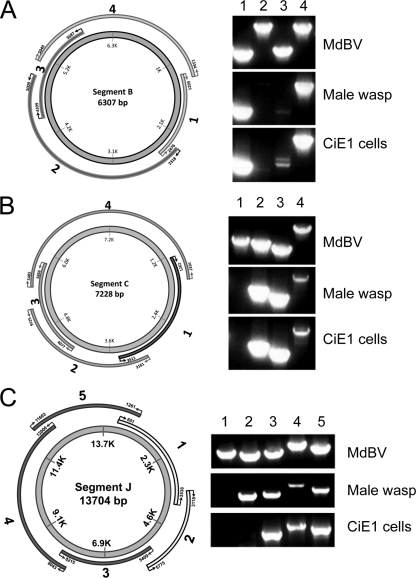
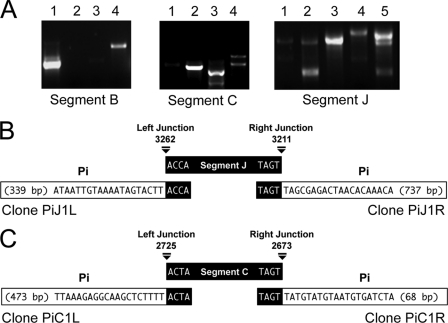
Since the encapsidated form of the MdBV genome cannot replicate, we asked whether MdBV genomic segments persisted in CiE1 cells as episomes or by integration. We selected the most (segment J) and least (segment C) abundant segments of the genome (5) plus a segment of intermediate abundance (segment B) for further study by designing overlapping primer pairs to amplify specific domains of each segment unless linearized and integrated (Fig. 3). We then compared amplification products using these primers and the following templates: (i) episomal viral DNA isolated from virions; (ii) genomic DNA from male M. demolitor wasps, which contain primarily the proviral (integrated) form of MdBV; and (iii) genomic DNA from CiE1 cells infected 21 days earlier by MdBV. As expected, we amplified each domain of segments B, C, and J using episomal viral DNA as a template (Fig. 3). In contrast, we generated no amplicons for domains 2 and 3 of segment B from male wasp DNA and MdBV-infected CiE1 DNA (Fig. 3A). For segment C, we generated no amplicon for domain 1 from male wasp and CiE1 DNAs (Fig. 3B), while for segment J, we generated no amplicon for domain 1 from male wasp DNA and no amplicons for domain 1 or 2 from CiE1 DNA (Fig. 3C). Identical results were generated by using DNA isolated from CiE1 cells at 8 months and 12 months postinfection (not shown). Taken together, these data strongly suggested that segments B, C, and J persisted in CiE1 cells by integrating within 21 days of infection and that no segments persisted as episomes. These data also suggested that integration occurred in domains on each segment that also contained the site of integration of proviral segments B, C, and J in M. demolitor.
Fig. 3.
MdBV genomic segments B, C, and J are integrated into adult male M. demolitor and CiE1 cells 21 days after infection with MdBV. Panels A to C show the designs and outcomes of PCR-based integration assays for segments B, C, and J, respectively. The schematics to the left show each genomic segment, with the inner circles indicating segment sizes (bp) and the outer bands indicating the locations of primers used to amplify different domains of each segment. To the right of each schematic are the PCR products generated using the domain-specific primers and DNA from MdBV, adult male M. demolitor, or 21-day-postinfection CiE1 cells. Numbers above each lane correspond to the domains shown in each schematic.
A proviral excision/integration motif identifies the site of integration of MdBV genomic DNAs into M. demolitor but not CiE1 cells.
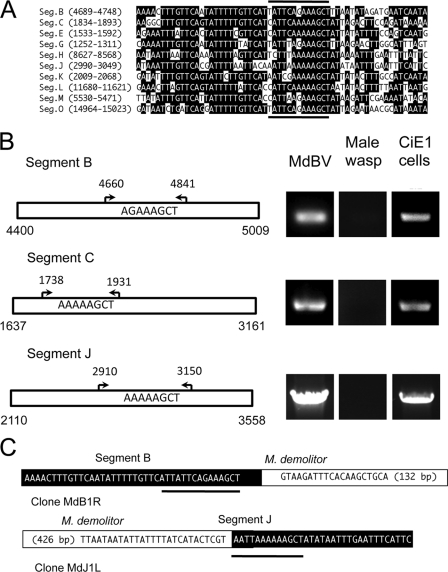
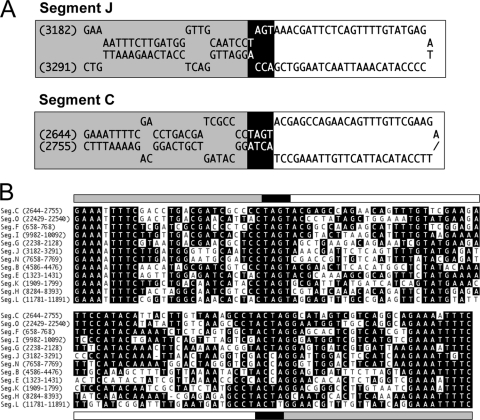
The tetramer AGCT embedded within a larger motif was previously identified as the site of excision for proviral genomic segments of GiBV and Glyptapanteles flavicoxis bracovirus (GfBV) from their associated wasps (10). Similar sequences have also been identified in some proviral genomic segments from Chelonus inanitus bracovirus (CiBV) (1). Here we refer to these domains as the predicted wasp excision/integration motif (WIM). Since all BVs evolved from a common ancestor (6, 43), we assessed whether such motifs existed within the larger domains on MdBV segments B, C, and J where integration into the genome of M. demolitor and CiE1 cells occurred. Our analysis confirmed the presence of a predicted WIM in each of these genomic segments as well as in several others (Fig. 4 A). Sequences to 36 nt upstream of the AGCT tetramer were AT rich and highly conserved among segments, whereas sequence conservation was weaker downstream of the AGCT tetramer (Fig. 4A).
Fig. 4.
A wasp excision/integration motif (WIM) identifies the site of integration of MdBV genomic segments B, C, and J in M. demolitor but not CiE1 cells. (A) Alignment of the predicted WIM on selected MdBV genomic segments. The location of the motif on each segment is indicated at the left. Identical nucleotides are indicated in black. The dark lines above and below the alignment indicate the predicted sites of integration of the corresponding proviral DNA in M. demolitor. (B) Outcome of PCR-based integration assays. Schematics to the left show larger domains on segments B, C, and J where the predicted WIM is located. Arrows and corresponding nucleotides identify the locations of flanking primers used in PCR-based integration assays. To the right of each schematic are the PCR products generated using these primers and DNA from MdBV, adult male M. demolitor, or CiE1 cells infected 21 days earlier with MdBV as a template. (C) Schematics illustrating the right segment B-M. demolitor junction sequence and left segment J-M. demolitor junction sequence cloned by inverse PCR. The MdBV sequence is highlighted in black, and the M. demolitor genomic sequence is highlighted in white. Note that the right boundary border for segment B is identified by the tetramer AGCT, while the right boundary for segment J is identified by the tetramer AATT, as shown by the black underlining in panel A. The cloned and analyzed M. demolitor sequence flanking segment B is 132 bp, while the sequence for segment J is 426 bp.
To assess whether these motifs identified the site of integration into M. demolitor and CiE1 cells, we conducted PCR-based integration assays using primers that flanked the WIMs on segments B, C, and J (Fig. 4B). In control assays, PCR products of the expected size were amplified by using episomal MdBV DNA as a template (Fig. 4B). In our treatment assays, no products were generated from male wasp DNA, whereas products were generated for each segment using CiE1 cell DNA as a template (Fig. 4B). Together, these data indicated that the predicted WIM on these MdBV segments identified the site of integration into M. demolitor but did not identify the site of integration into CiE1 cells. To confirm that integration into M. demolitor corresponded precisely with the WIM, we used nested primers; MfeI-, XbaI-, or PciI-digested M. demolitor DNA; and inverse PCR to amplify, clone, and sequence wasp-proviral junction sequences for segments B and J. Sequencing of the clone MdB1R identified the right junction for proviral segment B (Fig. 4C). The AGCT tetramer identified the boundary for proviral segment B, which was then followed by 132 bp of M. demolitor genomic sequence (Fig. 4C). Reciprocally, sequencing of the clone MdJ1L identified the left junction for proviral segment J where the tetramer AATT formed the boundary followed by 426 bp of M. demolitor genomic sequence (Fig. 4C).
Sequencing of host-viral DNA junctions confirms that segments J and C integrate into CiE1 cells and parasitized P. includens.
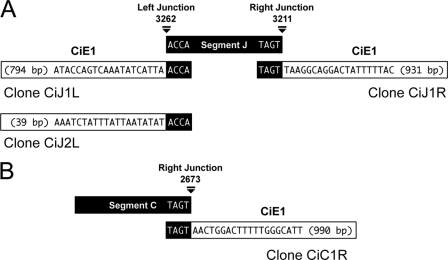
To narrow the location on segments B, C, and J where integration into CiE1 cells occurred, we used other primer pairs within the domains identified in Fig. 3 and conducted additional PCR-based integration assays. Our results indicated that segment B integrated into CiE1 cells at a region between nucleotides (nt) 4440 and 4660, segment C integrated between nt 1931 and 3161, and segment J integrated between nt 3050 and 3558 (data not shown). We then used these results to design nested primer sets for use in inverse PCRs to clone and sequence CiE1-viral DNA junction sequences. We identified two left junction clones (CiJ1L and CiJ2L) and one right junction clone (CiJ1R) that corresponded to CiE1-segment J integration sequences (Fig. 5 A). The boundaries of both left junction clones corresponded to nt 3262 on segment J and the tetramer ACCA, while the right boundary of the right junction clone corresponded to nt 3211 and the tetramer TAGT. These data also indicated that integration was associated with a ca. 50-bp deletion of segment J. BLAST analysis of the CiE1 flanking sequences for both the left and right junction clones revealed no significant homology with other sequences in current databases. We also identified one right junction clone (CiC1R) for a CiE1-segment C integration event (Fig. 5B). The boundary of this junction corresponded to nt 2673 on segment C and the tetramer TAGT, which was identical to the boundary identified for the right junction of segment J. CiE1 flanking sequences associated with segment C shared no homology with known sequences in current databases and also differed from the flanking sequences associated with the integration of segment J.
Fig. 5.
Inverse PCR clones confirm that MdBV segments J and C integrate into the genome of CiE1 cells. (A) Schematic illustrating the two left (CiJ1L and CiJ2L) and one right (CiJ1R) segment J-CiE1 junction sequences cloned by inverse PCR. Each junction clone is aligned with segment J linearized at nt 3262 (left) and nt 3211 (right). The MdBV sequence in each junction clone is highlighted in black, and the CiE1 genomic sequence is highlighted in white. Note that the segment J boundary for both left clones corresponds to nt 3262 and the tetramer ACCA, while the boundary for the right junction clone corresponds to nt 3211 and the tetramer TAGT. The analyzed CiE1 sequences for the two left junction clones are 794 and 39 bp long, respectively, while the CiE1 sequence for the right junction clone is 931 bp. (B) Schematic illustrating the one right (CiC1R) segment C-CiE1 junction sequence cloned by inverse PCR. The schematic is organized as described above (A). The CiE1 sequence for the clone is 990 bp.
We assessed whether MdBV genomic segments integrated into naturally parasitized P. includens by first isolating genomic DNA from host hemocytes at day 7 postparasitism and conducting PCR-based integration assays with segments B, C, and J as described above for CiE1 cells. Our results suggested that segments B and C had integrated due to the absence of amplicons for domains 2 and 3 with segment B and an absence of domain 1 for segment C (Fig. 6 A). In contrast, the detection of amplicons for each domain of segment J suggested that episomal DNA remained present (Fig. 6A). However, the reduced band intensity for domains 1 and 2 also suggested that some copies of segment J had potentially integrated (Fig. 6A). Additional assays conducted with segment B and DNA isolated from host hemocytes at earlier time points postparasitism suggested that integration had begun by 36 h, as evidenced by the reduced band intensities for domains 2 and 3 (not shown). By 120 h (5 days) postinfection, however, all copies of segment B appeared to be integrated, as evidenced by a failure to amplify domains 2 and 3, as shown in Fig. 6A. We then isolated genomic DNA from whole host larvae at day 7 postparasitism, followed by the cloning and sequencing of inverse PCR products under reaction conditions that were identical to those used for CiE1 cells. Note that no wasp offspring were present in these samples. One left junction sequence (clones PiJ1L and PiC1L) and one right junction sequence (clones PiJ1R and PiC1R) were identified for both segments J and C (Fig. 6B and C). As with CiE1 cells, the left and right boundaries for segment J corresponded to nt 3262 and 3211, while the left and right boundaries for segment C corresponded to nt 2725 and 2673. Host flanking sequence data also shared no homology with known sequences in databases.
Fig. 6.
PCR-based integration assays and inverse PCR clones confirm that MdBV segments integrate into the genome of parasitized P. includens. (A) PCR products amplified using domain-specific primers for segments B, C, and J and hemocyte genomic DNA collected from parasitized P. includens larvae (day 7 postparasitism). Domains correspond to the domains shown in Fig. 3. (B) Schematic illustrating the one left (PiJ1L) and one right (PiJ1R) segment J-P. includens junction sequences cloned by inverse PCR. The schematic is organized as described in the legend of Fig. 5. The P. includens (Pi) sequence for the left junction clone is 339 bp long, while the P. includens sequence for the right junction clone is 737 bp. (C) Schematic illustrating the one left (PiC1L) and one right (PiC1R) segment C-P. includens junction sequences cloned by inverse PCR. The P. includens sequence for the left junction clone is 473 bp long, while the P. includens sequence for the right junction clone is 68 bp.
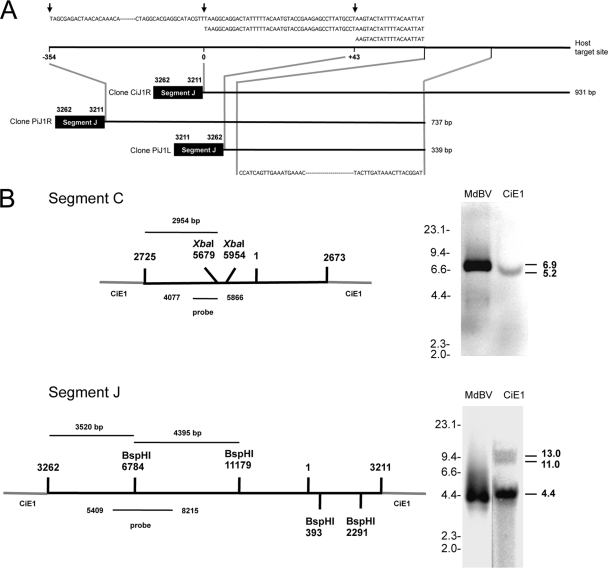
However, by comparing the cloned junctions for segment J to one another, we determined that a portion of clone CiJ1R from CiE1 cells was identical to clones PiJ1R and PiJ1L from parasitized P. includens. We aligned these clones with one another in Fig. 7 A, which shows that three copies of segment J integrated within a 1,285-bp domain in the genome of CiE1 cells and parasitized P. includens that we call a putative host target site. By arbitrarily designating position 0 the site of insertion for segment J in the right junction clone from CiE1 cells (CiJ1R), Fig. 7A shows that segment J for the right junction clone (PiJ1R) from P. includens was integrated 354 bp upstream, while segment J for the left junction clone (PiJ1L) from P. includens was integrated 43 bp downstream. Figure 7A also shows that segment J was integrated in one orientation (flip) for the right junction clones from CiE1 cells and P. includens but was integrated in the opposite orientation (flop) for the left junction clone from P. includens. We assembled the sequence of the presumptive host target site from the flanking sequence data of the three clones. The region overall exhibited a high A+T content (64%) and showed that each copy of segment J was inserted at sites identified by the sequence TA or TTA but shared no obvious homology with the boundary sequences for segment J (Fig. 7A). Examination of the host sequences for the two left junction clones (CiJ1L and CiJ2L) (Fig. 5) for segment J from CiE1 cells indicated that they shared no sequence homology with one another or the host target site shown in Fig. 7A. The host sequences from the two junction clones for segment C (PiC1L and PiC1R) (Fig. 5) from P. includens also shared no homology with one another or the putative host target site for segment J.
Fig. 7.
Integration of MdBV genomic DNAs is nonrandom. (A) Schematic showing that segment J-host junction clones CiJ1R (931 bp), PiJ1R (737 bp), and PiJ1L (339 bp) integrate into the same target site in CiE1 cells and P. includens. The integration of segment J from clone CiJ1R is shown at position 0 in the host target site. Segment J from clone PiJ1R is inserted at position −354, while segment J from clone PiJ1L is inserted at position +43. Above each clone is the sequence of the deduced host target site, with arrows indicating where each copy of segment J integrated. Below each clone is the region in each clone that was fully identical. (B) Southern blot analysis of MdBV and CiE1 genomic DNAs probed with a segment C-specific (above) or segment J-specific (below) probe. The schematics at the left show segments C and J integrated into CiE1 genomic DNA, as determined by the sequencing of junction clones (Fig. 5 and 6). XbaI (segment C) and BspHI (segment J) sites are indicated, as is the site within each segment that corresponds to the probes that we synthesized. At the right are Southern blots of XbaI-digested MdBV and CiE1 genomic DNAs hybridized with the segment C probe (above) or BspHI-digested MdBV and CiE1 genomic DNAs hybridized with the segment J probe (below). Size markers (kb) are indicated to the left of each blot, while the estimated sizes (kb) of the fragments recognized by each probe are indicated at the right.
To further characterize integration into CiE1 cells, we conducted qPCR assays to estimate the average copy numbers per cell for segments C and J and Southern blotting experiments to determine whether integration occurred in one or more locations in the genome. Our qPCR results indicated that 1.03 ± 0.13 copies of segment C were present on average per CiE1 cell at 8 months postinfection, while 21.03 ± 0.1 copies of segment J were present. If these segments randomly integrated into the genome of CiE1 cells, we would expect to observe a large number of bands or a smear on Southern blots. We digested MdBV and CiE1 genomic DNAs with XbaI, for which there are two predicted cut sites in segment C (Fig. 7B). In turn, the segment C probe that we generated should hybridize to a 6.9-kb fragment of segment C if not integrated but should recognize a 2.95-kb region of segment C plus an unknown length of flanking CiE1 genomic DNA if integrated as shown in Fig. 5 and 6. Our results indicated that this probe recognized the predicted 6.9-kb fragment of nonintegrated segment C when hybridized to MdBV genomic DNA and a single 5.2-kb fragment when hybridized to CiE1 genomic DNA (Fig. 7B). We then digested CiE1 and MdBV genomic DNAs with BspHI, for which there are four predicted cut sites in segment J (Fig. 7B). The segment J probe that we generated should hybridize to two 4.4-kb fragments of MdBV segment J if not integrated but should recognize a 4.4-kb fragment and a 3.5-kb region of segment J plus an unknown length of flanking CiE1 genomic DNA. Our results showed that this probe recognized the predicted 4.4 kb-fragments of nonintegrated segment J when hybridized to MdBV genomic DNA (Fig. 7B). When hybridized to CiE1 genomic DNA, the probe also recognized the predicted 4.4-kb fragment plus two other bands of approximately 13.0 and 11.0 kb (Fig. 7B). Overall, these results clearly indicated that segments J and C integrate into both CiE1 cells and parasitized P. includens larvae and that integration is associated with near-identical boundary sequences on each of these viral segments. In addition, these results also suggested that segments C and J do not integrate randomly into the P. includens genome.
MdBV segments share a host integration motif.
Given the near-identical boundary sequences associated with the integration of segments J and C into host cells, we asked whether similarities also existed in terms of where on each segment integration occurred. Indeed, the site of integration on each segment corresponded to a 110- or 111-nt domain consisting of two imperfect inverted repeats, which formed predicted stem-loop structures using Mfold 3.2 (45), to optimize base pairing (Fig. 8 A). A comparison of these stem-loop structures to our junction sequence data further showed that the integration boundary for each segment and every clone that we sequenced corresponded with the deletion of the 50-nt (segment J) or 51-nt (segment C) loop. This deletion in turn resulted in the tetramer ACCA or ACTA forming the left border with the host genome and TAGT forming the right border. An examination of other MdBV genomic segments revealed the presence of similar domains comprised of conserved inverted repeats and a more variable loop region (Fig. 8B). We therefore named this domain the MdBV host integration motif (HIM). Consistent with our PCR-based integration assay data (Fig. 3), the HIM and WIM were located in close proximity (109 to 168 nt) to one another on segments B and J as well as most other MdBV genomic DNAs (Table 1). However, these motifs were 770 and 836 nt apart in segments C and G, respectively, while segment O contained two predicted WIMs that were separated from the predicted HIM by more than 7 kb (Table 1).
Fig. 8.
MdBV genomic DNAs contain similar host integration motifs (HIMs). (A) Predicted stem-loop structures for the segment J and C HIMs generated by Mfold. Gray highlights nucleotides that form the base of the stem, black highlights the tetramers that identify the boundary site of integration of each segment into host cells, and white highlights the predicted loop domain that is deleted with integration into the host genome. (B) Sequence alignment of the HIMs from selected MdBV genomic segments. The position of the motif on each segment is indicated at the left. Identical nucleotides are indicated in black. The gray, black, and white lines above and below the alignment correspond to the stem and loop regions shown in panel A.
Table 1.
Location of the predicted host integration motif compared to the predicted wasp excision/integration motif on selected MdBV genomic segmentsa
| Segment (size [nt]) | Positions of host integration motif | Positions of wasp excision/integration motif | Proximity (nt) |
|---|---|---|---|
| B (6307) | 4586–4476 | 4699–4728 | 112 |
| C (7228) | 2644–2755 | 1844–1873 | 770 |
| E (8443) | 1323–1431 | 1543–157 | 111 |
| G (10,790) | 2238–2128 | 1262–1291 | 836 |
| H (11,238) | 8284–8393 | 8562–8591 | 168 |
| J (13,704) | 3182–3291 | 3055–3026 | 126 |
| K (15,058) | 1909–1799 | 2019–2048 | 109 |
| L (15,096) | 11781–11891 | 11670–11641 | 110 |
| M (15,218) | 5697–5798 | 5520–5491 | 176 |
| O (34,334) | 22429–22540 | 14974–15003 | 7,426 |
| 30126–30155 | 7,586 |
The positions of the host and wasp motifs on each segment correspond to sequences submitted to GenBank (see Materials and Methods for accession numbers).
DISCUSSION
PDVs are specialized symbionts of wasps that function as replication-defective gene delivery vehicles in the insects (hosts) that the wasps parasitize. Our understanding of how PDVs alter the physiology of hosts for the benefit of wasp offspring has increased greatly in recent years (summarized in references 23, 32, and 41), yet little is known about the fate of PDV genomes in infected host cells. This reflects in large measure that parasitism results in the relatively rapid death of permissive hosts, which are usually consumed by the wasp's offspring 7 to 14 days after the adult oviposits (23). Thus, based on limited experimental data (28, 38), the assumption has generally been that PDV genomes persist in hosts as episomes, which is adequate for the transient expression of the viral gene products required for successful parasitism (41). However, the long-term detection of some PDV genomic DNAs in continuous cell lines (11, 14, 15, 17, 21, 40), together with data showing that genomic DNAs from GiBV and TrIV integrate into continuous cell lines of host origin (11, 15), raises the possibility that PDV DNAs may not persist as episomes in hosts. However, it is unclear from these cell culture studies whether all or only some PDV genomic segments are capable of integrating, how quickly integration occurs, and whether the expression of viral gene products responsible for specific functional alterations continues after integration. It is also unknown whether PDVs actually integrate into hosts after parasitism.
Our results indicate that all 15 genomic segments of the MdBV encapsidated genome exhibit long-term persistence in CiE1 cells. Our results also reveal that many MdBV genes continue to be expressed, including members of the glc gene family whose products block adhesion and cause persistently infected CiE1 cells to grow as a suspension culture. Lastly, our sequencing of multiple host-viral DNA junction clones demonstrates that segments C and J integrate into parasitized P. includens, while our PCR-based assays suggest that integration begins within 2 days of parasitism. Previous studies suggested that the integration of proviral DNAs into wasps and the integration of episomal DNAs into host cell lines could be regulated through a shared integration site (15, 40). In characterizing integration into CiE1 cells, we determined that MdBV genomic segments contain a motif that has been identified in other BV isolates as the site of proviral DNA excision during replication in wasps (1, 10). Our results show that this WIM identifies the location on segments B, C, and J where integration into M. demolitor occurs but does not identify the site of integration into CiE1 cells or P. includens. Instead, our sequence data for segments C and J indicate that integration into hosts occurs at a second site that we call an HIM. The presence of an HIM in other MdBV segments further suggests that most, if not all, MdBV genomic DNAs also integrate into hosts, which is further supported by the long-term persistence of all MdBV genomic segments in CiE1 cells.
The single host-virus junction site for TrIV segment F cloned from spruce budworm-derived CF-124T cells provided no insights into how integration occurs (11). Somewhat similar to our findings, though, the integration of segment F is associated with a 33-nt deletion from the episomal DNA. The single junction site identified for integrated segment 25 from GiBV (formerly segment F [10]) in gypsy moth IPLB-LdEp cells showed that the viral boundary was defined by the sequence CATGGTAC (15). This sequence is palindromic, as are the sequences on each end of the HIMs identified in MdBV segments (Fig. 8). However, our own examination of GiBV segment 25 in the region associated with integration into IPLB-LdEp cells failed to identify a motif with homology to an MdBV HIM or that differed in sequence but formed a predicted stem-loop structure. We also did not identify a homologous HIM in other genomic DNAs from BVs of wasps in other genera that have been submitted to GenBank. However, genomic sequence data that we recently generated for BVs from other species of Microplitis do identify the presence of HIMs that are similar to those identified in MdBV (M. H. Beck, G. R. Burke, and M. R. Strand, unpublished data). Taken together, the similarity in WIMs among BVs from wasps in diverse subfamilies suggests an ancient origin and long-term conservation of proviral excision/integration motifs that may date back to the evolution of BVs from a nudivirus ancestor (6, 43). In contrast, our finding of MdBV-like HIMs on genomic segments of BVs from wasps in the genus Microplitis but not other genera suggests that either the common ancestor of Microplitis BVs acquired this domain or BVs in other taxa have partially or fully lost it.
Dissimilarities in the junction sequences that we cloned indicate that the integration of segments J and C may not be fully specific at the nucleotide level. On the other hand, our finding that three junction clones for segment J integrate into a similar location in the P. includens genome together with our qPCR and Southern blotting data also suggest that integration is not random and does exhibit a level of site specificity for a particular host target domain. The detection of multiple copies of segment J per CiE1 cell genome but only two novel hybridization signals on Southern blots suggests the possibility that the host target sequence that we identified exists in multiple copies and/or that segment J also integrates into another target site. In contrast, the detection of only one copy of segment C per host cell genome together with a single novel fragment on Southern blots suggests that this viral DNA may integrate into a unique host target site. It is also possible that the higher copy number of segment J per host genome than that of segment C could reflect a greater molar abundance of segment J than that of segment C at the time of infection.
Our results suggest that MdBV segments integrate into the host genome through a similar mechanism. This mechanism could involve MdBV gene products expressed in P. includens (7). The absence of any predicted recombinase or endonuclease in the encapsidated genome, however, argues that integration also involves unknown host factors interacting with the HIM on different MdBV genomic segments and potentially specific host target sequences. Currently, the only known animal DNA virus to exhibit site-directed integration is adeno-associated virus (AAV), where nonhomologous recombination into different locations of the AAVS1 domain in human chromosome 19 involves interactions between virus-encoded proteins, palindrome-forming terminal inverted repeats in the viral genome, and host cell machinery (9, 18, 19, 24, 44). The integration of another group of parvoviruses (densoviruses) that infect insects may also involve unknown host cell factors interacting with viral sequences (8).
The inverted repeats of the HIMs on different MdBV genomic segments are clearly similar, but the loop domains differ (Fig. 8B). We speculate that these differences may play a role in where different MdBV segments integrate into the host genome. In contrast, our comparative data for host target sequences are insufficient to discern whether any common features exist. The junction site identified for GiBV segment 25 in IPLB-LdEp cells contained a small region with homology to a Pao-like retrotransposon and an open reading frame with homology to a polymerase, which Gundersen-Rindal and Lynn (15) speculated could play a role in integration. In contrast, we identified no significant homology in the host junction sequences from P. includens with any transposable element or genes in current databases. However, the host target site shown in Fig. 7A for segment J is strongly A+T rich, which is sometimes associated with nonhomologous recombination events in mammalian cells (18). Key future needs thus include determining what features of the HIM are required for integration, whether HIMs on different segments differ in their targeting preferences, and how this motif interacts with the recombination machinery of the host. In situ analysis will also be important in generating insights into the spatial distribution of different MdBV genomic DNA insertions into host chromosomes.
Since MdBV genomic segments integrate into P. includens before wasp offspring complete their development, we conclude that the integration of this PDV is a legitimate feature of parasitism and is not solely an event that occurs in continuous cell lines. However, at present, we can only speculate as to its functional significance. One previously proposed idea is that the integration of viral DNAs could alter or disrupt the expression of host genes that otherwise would adversely affect the survival of wasp offspring (15). A second idea is that integration ensures that viral DNAs persist and that associated viral gene products are produced in the latter stages of parasitism, which could also be important for the successful development of the wasp. While the progeny of most PDV-carrying wasps develop relatively rapidly, many species also diapause in their host, which extends the period required for progeny development in a host to a period of months. The integration of the PDV genome under such circumstances could thus be vital for the long-term persistence and expression of gene products required for successful parasitism. A third possibility is that the association between some PDVs and wasps may not be fully beneficial, with some PDV DNAs possessing selfish elements that facilitate horizontal movement in the absence of replication. Since PDV-carrying wasps sometimes parasitize nonpermissive or semipermissive hosts that can survive parasitism (23, 32), the ability of PDV genomic DNAs to rapidly integrate creates the potential for dissemination and persistence independent of the wasp.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by grants from the U.S. Department of Agriculture and the National Science Foundation to M.R.S.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Annaheim M., Lanzrein B. 2007. Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers, and abundance of proviral and excised segments. J. Gen. Virol. 8: 450–457 [DOI] [PubMed] [Google Scholar]
- 2. Beck M., Strand M. R. 2003. RNA interference silences Microplitis demolitor bracovirus genes and implicates glc1.8 in disruption of adhesion in infected host cells. Virology 314: 521–535 [DOI] [PubMed] [Google Scholar]
- 3. Beck M. H., Strand M. R. 2005. Glc1.8 from Microplitis demolitor bracovirus induces a loss of adhesion and phagocytosis in insect high five and S2 cells. J. Virol. 79: 1861–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beck M. H., Strand M. R. 2007. A novel polydnavirus protein inhibits the insect prophenoloxidase activation pathway. Proc. Natl. Acad. Sci. U. S. A. 104: 19267–19272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck M. H., Inman R. B., Strand M. R. 2007. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 359: 179–189 [DOI] [PubMed] [Google Scholar]
- 6. Bezier A., et al. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323: 926–930 [DOI] [PubMed] [Google Scholar]
- 7. Bitra K., Zhang S., Strand M. R. 2011. Transcriptomic profiling of Microplitis demolitor bracovirus reveals host, tissue, and stage-specific patterns of activity. J. Gen. Virol. 92: 2060–2071 [DOI] [PubMed] [Google Scholar]
- 8. Bossin H., et al. 2003. Junonia coenia densovirus-based vectors for stable transgene expression in Sf9 cells: influence of the densovirus sequences on genomic integration. J. Virol. 77: 11060–11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daya S., Cortez N., Berns K. I. 2009. Adeno-associated virus site-specific integration is mediated by proteins of the nonhomologous end-joining pathway. J. Virol. 83: 11655–11664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desjardins C. A., et al. 2008. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doucet D., et al. 2007. In vitro integration of an ichnovirus genome segment into the genomic DNA of lepidopteran cells. J. Gen. Virol. 88: 105–113 [DOI] [PubMed] [Google Scholar]
- 12. Dupuy C., Huguet E., Drezen J.-M. 2006. Unfolding the evolutionary history of polydnaviruses. Virus Res. 117: 81–89 [DOI] [PubMed] [Google Scholar]
- 13. Fleming J. G. W. 1992. Polydnaviruses: mutualists and pathogens. Annu. Rev. Entomol. 37: 401–425 [DOI] [PubMed] [Google Scholar]
- 14. Gundersen-Rindal D., Dougherty E. M. 2000. Evidence for integration of Glyptapanteles indiensis polydnavirus DNA into the chromosome of Lymantria dispar in vitro. Virus Res. 66: 27–37 [DOI] [PubMed] [Google Scholar]
- 15. Gundersen-Rindal D. E., Lynn D. E. 2003. Polydnavirus integration in lepidopteran cells in vitro. J. Insect Physiol. 49: 453–462 [DOI] [PubMed] [Google Scholar]
- 16. Johnson J. A., et al. 2010. The UGA-CiE1 cell line from Chrysodeixis includens exhibits characteristics of granulocytes and is permissive to infection by two viruses. Insect Biochem. Mol. Biol. 40: 394–404 [DOI] [PubMed] [Google Scholar]
- 17. Kim M. K., Sisson G., Stoltz D. 1996. Ichnovirus infection of an established gypsy moth cell line. J. Gen. Virol. 77: 2321–2328 [DOI] [PubMed] [Google Scholar]
- 18. Kotin R. M., Linden R. M., Berns K. I. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11: 5071–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linden M. R., Ward P., Giraud C. C., Winocour E., Berns K. I. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 93: 11288–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu Z., Beck M. H., Jiang H., Wang Y., Strand M. R. 2008. The viral protein Egf1.0 is a dual activity inhibitor of prophenoloxidase activating proteinases 1 and 3 from Manduca sexta. J. Biol. Chem. 283: 21325–21333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKelvey T. A., et al. 1996. Transformation of gypsy moth cell lines by infection with Glyptapanteles indiensis polydnavirus. Biochem. Biophys. Res. Commun. 224: 764–770 [DOI] [PubMed] [Google Scholar]
- 22. Ochman H., Ayala F. J., Hartl D. L. 1993. Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 218: 309–321 [DOI] [PubMed] [Google Scholar]
- 23. Pennacchio F., Strand M. R. 2006. Evolution of developmental strategies in parasitic Hymenoptera. Annu. Rev. Entomol. 51: 233–258 [DOI] [PubMed] [Google Scholar]
- 24. Philpott N. J., et al. 2002. Efficient integration of recombinant adeno-associated virus DNA vectors requires a p5-rep sequence in cis. J. Virol. 76: 5411–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pruijssers A. J., Strand M. R. 2007. PTP-H2 and PTP-H3 from Microplitis demolitor bracovirus localize to focal adhesions and are antiphagocytic in insect immune cells. J. Virol. 81: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pruijssers A. J., et al. 2009. Infection by a symbiotic polydnavirus induces wasting and inhibits metamorphosis of the moth Pseudoplusia includens. J. Exp. Biol. 212: 2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stoltz D. B. 1993. The PDV life cycle, p. 167–187 In Thompson S. N., Federici B. A., Beckage N. E. (ed.), Parasites and pathogens of insects, vol. 1 Parasites. Academic Press, San Diego, CA [Google Scholar]
- 28. Stoltz D. B., Guzo D., Cook D. 1986. Studies of polydnavirus transmission. Virology 155: 120–131 [DOI] [PubMed] [Google Scholar]
- 29. Strand M. R. 1990. Characterization of larval development in Pseudoplusia includens (Lepidoptera, Noctuidae). Ann. Entomol. Soc. Am. 83: 538–544 [Google Scholar]
- 30. Strand M. R. 1994. Microplitis demolitor polydnavirus infects and expresses in specific morphotypes of Pseudoplusia includens haemocytes. J. Gen. Virol. 75: 3007–3020 [DOI] [PubMed] [Google Scholar]
- 31. Strand M. R. 2009. The interactions between polydnavirus-carrying parasitoids and their lepidopteran hosts, p. 321–336 In Goldsmith M. R., Marec F. (ed.), Molecular biology and genetics of the Lepidoptera. CRC Press, Boca Raton, FL [Google Scholar]
- 32. Strand M. R. 2010. Polydnaviruses, p. 171–197 In Asgari S., Johnson K. N. (ed.), Insect virology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 33. Strand M. R., Noda T. 1991. Alterations in the haemocytes of Pseudoplusia includens after parasitism by Microplitis demolitor. J. Insect Physiol. 37: 839–850 [Google Scholar]
- 34. Strand M. R., Pech L. L. 1995. Microplitis demolitor polydnavirus induces apoptosis of a specific haemocyte morphotype in Pseudoplusia includens. J. Gen. Virol. 76: 283–291 [DOI] [PubMed] [Google Scholar]
- 35. Strand M. R., McKenzie D. I., Grassl V., Dover B. A., Aiken J. M. 1992. Persistence and expression of Microplitis demolitor PDV in Pseudoplusia includens. J. Gen. Virol. 73: 1627–1635 [DOI] [PubMed] [Google Scholar]
- 36. Strand M. R., Witherell R. A., Trudeau D. 1997. Two Microplitis demolitor PDV mRNAs expressed in hemocytes of Pseudoplusia includens contain a common cysteine-rich domain. J. Virol. 71: 2146–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suderman R. J., Pruijssers A. J., Strand M. R. 2008. Protein tyrosine phosphatase-H2 from a polydnavirus induces apoptosis of insect cells. J. Gen. Virol. 89: 1411–1420 [DOI] [PubMed] [Google Scholar]
- 38. Theilmann D. A., Summers M. D. 1986. Molecular analysis of Campoletis sonorensis virus DNA in the lepidopteran host Heliothis virescens. J. Gen. Virol. 67: 1961–1969 [DOI] [PubMed] [Google Scholar]
- 39. Trudeau D., Witherell R. A., Strand M. R. 2000. Characterization of two novel Microplitis demolitor PDV mRNAs expressed in Pseudoplusia includens haemocytes. J. Gen. Virol. 81: 3049–3058 [DOI] [PubMed] [Google Scholar]
- 40. Volkoff A.-N., et al. 2001. Persistent expression of a newly characterized Hyposoter didymator polydnavirus gene in long-term infected lepidopteran cell lines. J. Gen. Virol. 82: 963–969 [DOI] [PubMed] [Google Scholar]
- 41. Webb B. A., Strand M. R. 2005. The biology and genomics of polydnaviruses, p. 323–360 In Gilbert L. I., Iatrou K., Gill S. S. (ed.), Comprehensive molecular insect science, vol. 6 Elsevier, San Diego, CA [Google Scholar]
- 42. Webb B. A., et al. 2006. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347: 160–174 [DOI] [PubMed] [Google Scholar]
- 43. Whitfield J. B. 2002. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc. Natl. Acad. Sci. U. S. A. 99: 7508–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang C. C., et al. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vitro and in vivo. J. Virol. 71: 9231–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.