Abstract
Human cytomegalovirus (HCMV) encodes at least 14 microRNAs (miRNAs) that act posttranscriptionally to repress gene expression. Although several HCMV miRNA targets of both cellular and viral origin have been identified, our knowledge of their function remains limited. HCMV miRNA targets, as well as phenotypes associated with HCMV miRNA mutants, have been difficult to identify since the downregulation of targets by a single miRNA is often less than 2-fold. Several factors can contribute to the strength of repression, including the mechanism of translational inhibition, the degree of complementarity between the miRNA and target mRNA, the number of binding sites for one miRNA, and cooperativity or antagonism between miRNAs. To determine the effect of multiple miRNAs on one gene, we examined the repression of a viral gene, US7. Here we demonstrate that the HCMV-encoded miRNAs miR-US5-1 and miR-US5-2 function in a highly synergistic manner to regulate US7, even at very low miRNA concentrations. Regulation of US7 involves three functional miRNA binding sites: two that are completely complementary to the 3′ untranslated region (3′UTR) and one that is imperfectly matched. Surprisingly, we observed equal contributions to inhibition from both complete and partially complementary sites, and repression was not completely abrogated until all three sites were mutated simultaneously. We also observed that the miRNA binding sites did not follow the spacing constraints for corepressive miRNAs observed in earlier reports. These results underscore the importance of evaluating the contribution of multiple miRNAs on gene regulation and shed new insight into miRNA:mRNA interactions.
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNA molecules of 19 to 24 nucleotides (nt) in length. They play an important role in a wide variety of biological processes including development, differentiation, hematopoiesis, angiogenesis, and cancer (reviewed in reference 1). Their biological significance can be attributed to the ability of each miRNA to target at least 100 genes (6, 10, 21). Although many studies have sought to define the rules that govern miRNA:mRNA interactions, a lack of complete understanding of these interactions impedes miRNA target identification (22, 34).
miRNAs direct RNA-induced silencing complexes (RISC) to target genes by binding to sites of sequence complementarity within the mRNA. Most miRNA binding sites are located within the 3′ untranslated regions (3′UTRs) of mRNAs; however, sites within 5′UTRs or coding regions have also been identified (14, 23, 25). The earliest studies indicate that most mammalian miRNAs are not completely complementary to the mRNA, yet a stretch of 6 to 8 nucleotides in the 5′ end of the miRNA that is complementary to the mRNA primarily mediates interaction (termed the seed sequence) (4, 6, 15, 21). Complete complementarity between a miRNA and an mRNA directs site-specific endonucleolytic cleavage of the mRNA, resulting in decreased abundance of the message (reviewed in references 3 and 9). Incomplete complementarity, as seen in most mammalian cells, results in translational repression due to translation inhibition and mRNA decay. In these cases, a single miRNA:mRNA interaction may have only a nominal effect (<4-fold) on gene expression (2, 31). However, stronger repression is observed when a gene contains multiple binding sites for the same miRNA (16, 30). Furthermore, multiple miRNAs can act concomitantly on the same 3′UTR to affect gene expression. Several recent reports have demonstrated cooperative and antagonistic interactions between miRNAs (11, 16, 17, 20, 24, 26, 30, 35, 37, 38). Cooperativity can lead to enhanced repression when miRNA binding sites are ideally spaced (16 to 35 nt apart) within the 3′UTR (16, 30).
The betaherpesvirus human cytomegalovirus (HCMV) is ubiquitous among the human population and, while normally benign, is a significant pathogen in immunosuppressed individuals. HCMV is also the leading infectious cause of congenital birth defects. HCMV encodes at least 14 miRNAs expressed from 11 unique RNA structures (12, 27, 36). Our lab and others have identified both viral and cellular targets of HCMV-encoded miRNAs. miR-UL112-1 regulates multiple viral genes including the major immediate early gene product IE72, a major trans-activator of viral gene transcription (13). Additional studies by authors Stern-Ginossar et al. (32) have demonstrated that miR-UL112-1 targets the major histocompatibility complex chain B, a cellular gene involved in host immune responses to viral infection. miR-UL112-1 acts in concert with cellular miRNAs to enhance repression (24). We have recently shown that miR-US25-1 targets many cellular genes associated with cell cycle control, including cyclin E2, BRCC3, EID1, MAPRE2, and CD147, through the unusual mechanism of binding to sites within the 5′UTR of mRNA transcripts (14).
HCMV-encoded miRNAs are expressed coordinately in infected human fibroblasts. We hypothesized that viral miRNAs may function together to regulate both viral and cellular gene expression. In the current study we found that HCMV tightly regulates the expression of one of its own genes, US7, by using two different miRNAs, miR-US5-1 and miR-US5-2. Regulation by miR-US5-1 and miR-US5-2 is highly synergistic, demonstrating cooperativity among the miRNAs. Both miR-US5-1 and miR-US5-2 are encoded on the strand opposing the US7 3′UTR and therefore each binds to the 3′UTR with complete complementarity. Additionally, we found a second functional miR-US5-2 binding site in the US7 3′UTR. We demonstrate that binding to the miR-US5-2 site that is completely complementary induces degradation of the mRNA, but binding to the second site does not. To our knowledge this is the first report of a gene that is regulated by two distinct miRNA-directed mechanisms. Surprisingly both miR-US5-2 sites contribute equally to regulation of the 3′UTR, despite the differences in mRNA abundance. We also observed that the distances between each set of miRNA binding sites were greater than 79 bases. This is far greater than the ideal distances seen between cooperative binding sites in previous papers (16, 30). These new findings highlight the value of using viruses to study biological processes.
MATERIALS AND METHODS
Reagents.
Double-stranded miRNA mimics were designed using published miRNA sequences (www.mirbase.org). The guide strand sequence matched the published sequence, while the passenger strand sequence contained a mismatch in the 3′ end of the strand. Sequences were as follows: for miR-US5-1, guide UGACAAGCCUGACGAGAGCGU and passenger UCGCUCUCGUCAGGCUUGUUAUG; for miR-US5-2, guide UUAUGAUAGGUGUGACGAUGUCUU and passenger GACAUCGUCACACCUAUCAUCAGA. Annealed mimics were purchased from IDT. Negative control small interfering RNA (siRNA) was purchased from Ambion.
Cloning and site-directed mutagenesis.
The 3′UTR of US7 was PCR amplified from total viral cDNA by using a US7-specific forward primer (GCGCCCTAGGATAAACTGTTAGGTTCG) and an oligo(dT) reverse primer (TCAGCACTGTCGAGCTCCTTAAGCTTTTTTTTTTTTTTTTTT) and cloned downstream of the firefly luciferase (Fluc) gene in the pMIR kanamycin (Kan) luciferase single-reporter construct (13). A PCR-based site-directed mutagenesis protocol was used to introduce mutations into potential miRNA binding sites (QuikChange site-directed mutagenesis; Stratagene). The following mutagenesis primers were used: for miR-US5-1, forward (GATGAACGCTCTCGTCAGGACTAGTATGGTCTGTAAAAGCTGCATG) and reverse (CATGCAGCTTTTACAGACCATACTAGTCCTGACGAGAGCGTTCATC); miR-US5-2 Mut 1 primer, forward (AAAGACATCGTCACACCGAATTCAGAATGCAACGCTTTCA) and reverse (TGAAAGCGTTGCATTCTGAATTCGGTGTGACGATGTCTTT); and miR-US5-2 Mut 2 primer, forward (ACTCACGTCACACGTTACCTCGAGCAACGTAGGGCGGTATTT) and reverse (AAATACCGCCCTACGTTGCTCGAGGTAACGTGTGACGTGAGT). The wild-type (WT) US7 3′UTR and the US7 3′UTRs containing the miR-US-5-2 mutations, or the miR-US5-2 mutations and the miR-US5-1 mutation, were then PCR amplified from the pMIR Kan constructs (forward [GCGGAGCTCTAAACTGTTAGGTTCGTTATAAGC] and reverse [GCGGCGGCCGCGAGACGATAAAACAGCATCAGG]) and subcloned downstream of the Renilla luciferase (Rluc) gene in the psiCHECK-2 dual reporter construct (Promega). The US7 3′UTR containing the single miR-US5-1 mutation was subcloned directly from the pMIR Kan construct into the psiCHECK-2 plasmid by using existing restriction sites.
Cells and viruses.
HEK293T and primary human fibroblasts were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin-glutamine. The AD169 strain of HCMV was grown and titered in human fibroblasts by using standard techniques. The entire 70-nt sequence coding for miR-US5-1 or miR-US5-2 was deleted from the AD169 bacterial artificial chromosome (BAC) clone by using BAC technology, as previously described (29). For the double mutant, the entire region encompassing both miRNAs was deleted. Briefly, a PCR-amplified cassette containing FLP recombination target (FRT)-flanked kanamycin was recombined into the BAC replacing the indicated sequences. miR-US5-1 deletion primers were as follows: forward, GTTGTATGATGTAACGTGTGACGTGAGTCTGATCCAACACTGTAAAACGACGGCCAG; reverse, CAAATATATGGAGTTTGTGTAATGCGTACTTCATGCCCCGATCAGGAAACAGCTATGAC. miR-US5-2 deletion primers were as follows: forward, ACAAACTCCATATATTTGTTACGATAGAATACGGAACGGAGTAAAACGACGGCCAG; reverse, GATGAATGTCATCATCACGCAAAGCAGCCGTGGGAATGGTCAGGAAACAGCTAT GAC. miR-US5-1 and miR-US5-2 deletion primers were as follows: forward, ACAAACTCCATATATTTGTTACGATAGAATACGGAACGGAGTAAAA CGACGGCCAG; reverse, CAAATATATGGAGTTTGTGTAATGCGTACTTCATGCCCCGATCAGGAAACAGCTATGAC. The kanamycin cassette was then removed by recombining the FRT sites through induction of a FLIP recombinase. The resulting BACs were isolated and electroporated into human fibroblasts to generate infectious virus.
Immunoprecipitations.
Infected primary human fibroblasts (multiplicity of infection [MOI] = 3) were incubated with cysteine- and methionine-free media for 60 min at the indicated times postinfection. The cells were then labeled with 125 μCi of [35S]methionine (EasyTag ExpreSS; Perkin Elmer) for 1 h prior to cell harvest. Immunoprecipitations were performed using an antibody specific for US7 (18) and glycoprotein H (gH) (5).
Luciferase assay.
Double-stranded miRNA mimics were cotransfected into HEK293T cells with either single-reporter constructs and a control construct that expressed Rluc (pRL; Promega) or the dual reporter constructs by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Rluc and Fluc expression was measured 18 h posttransfection using the dual-luciferase reporter assay (Promega). Results were normalized to the corresponding control reporter construct (Rluc in the case of single-reporter constructs and Fluc for the psiCHECK-2 system) and are reported as percent expression relative to cells transfected with a negative control miRNA.
For the isobologram analysis, HEK293T cells were seeded at 1.0 × 104 cells/well in 96-well plates. Cells were cotransfected with the 3′UTR psiCHECK-2 reporter constructs and serial dilutions of miR-US5-1 and miR-US5-2 mimics (0, 6.7, 13.3, 26.7, 53.3, 106.7, 213.3, 426.7 pM) alone or in combination using Lipofectamine 2000 (Invitrogen). Combinations of miR-US5-1/miR-US5-2 doses leading to 50% inhibition of Rluc activity (IC50) were determined from titration curves corresponding to one concentration of miR-US5-1 with increasing concentrations of miR-US5-2 and vice versa. Isobolograms were drawn by plotting each of these dose pairs on a Cartesian diagram (33).
Western blot analysis.
Cell extracts were run on an 8–12% SDS-PAGE gel, transferred to Immobilon-P transfer membranes (Milipore Corp.), and visualized with antibodies specific for luciferase (Sigma), Argonaute 2 (14), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Abcam).
RT and real-time PCR.
Cells were lysed directly in TRIzol, and total RNA was extracted following the manufacturer's recommendations (Invitrogen). For each sample, 250 ng of total RNA was treated with DNase I before being reverse transcribed using hexanucleotide primers and Multiscribe reverse transcriptase (ABI). Sybr green real-time PCR was performed in duplicate using 2 μl of each reverse transcription (RT) reaction and primer pairs specific for US7 (forward [ATCTCACACCGTCAGCTGCGTAAT], reverse [AGGCACGTTATACAAGCCACTGGT]), GAPDH (forward [TCGACAGTCAGCCGCATCTTCTTT], reverse [ACCAAATCCGTTGACTCCGACCTT]), Rluc (forward [ATTCAAGGAGAAGGGCGAGGTT], reverse [AGGCGTTGTAGTTGCGGACAAT]), and Fluc (forward [TGCAGTTCTTCATGCCAGTGCT], reverse [TAGGCTGAGAAATGCCCATGCT]). Relative quantities between samples were determined by the ΔΔCT method using GAPDH and Fluc as references for US7 and Rluc, respectively.
RESULTS
HCMV US7 is cotargeted by miR-US5-1 and miR-US5-2.
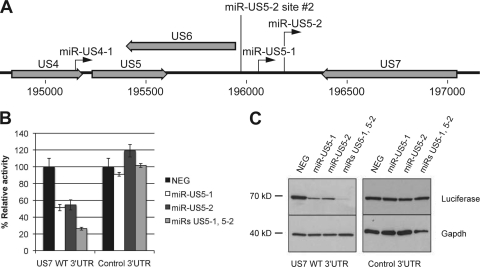
miR-US5-1 and miR-US5-2 are two HCMV-encoded miRNAs that lie within close proximity to one another in the viral genome. They are encoded on the opposing strand of an HCMV gene of unknown function called US7 (Fig. 1 A). Both miRNAs are completely complementary to the US7 3′UTR and therefore are predicted to target US7 to RISC.
Fig. 1.
US7 is cotargeted by miR-US5-1 and miR-US5-2. (A) Diagram depicting the genomic positions of miR-US5-1 and miR-US5-2. The black arrows indicate the position and direction of the miRNAs. The second miR-US5-2 binding site is labeled as miR-US5-2 site #2. Open reading frames near viral miRNAs are shown in light gray block arrows. (B) The 3′UTR of US7 or a negative control 3′UTR were cloned downstream of the Rluc gene within a dual reporter construct. The constructs were then transfected into HEK293T cells with the indicated HCMV-encoded miRNAs or a negative control miRNA (NEG). Eighteen hours posttransfection, luciferase assays were performed to determine the level of Rluc activity normalized to Fluc activity. (C) Forty-eight hours posttransfection, Western blot analyses were performed using an antibody specific for Fluc (luciferase) or GAPDH as a loading control.
To determine whether miR-US5-1 and miR-US5-2 regulate the US7 transcript, cells were cotransfected with reporter constructs encoding the US7 3′UTR and double-stranded RNA mimics of miR-US5-1 and miR-US5-2. Luciferase assays revealed that each miRNA alone decreased reporter activity by approximately 50% (Fig. 1B). When miR-US5-1 and miR-US5-2 were combined, reporter activity was decreased by approximately 75%. In contrast, miR-US5-1 and miR-US5-2 had no effect on a control 3′UTR, when transfected either alone or in combination with one another. Comparable results were seen when Western blot analysis was used to analyze total luciferase protein expression (Fig. 1C).
These results show that miR-US5-1 and miR-US5-2 each target the US7 3′UTR and that targeting by two miRNAs has a more robust effect than does targeting by either miRNA alone.
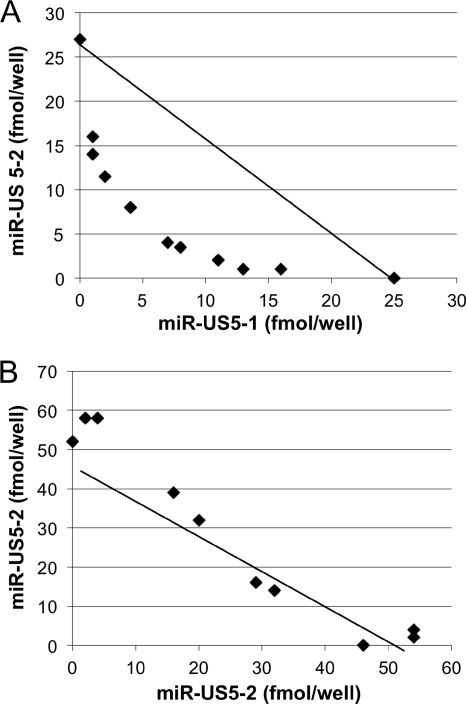
miR-US5-1 and miR-US5-2 act synergistically to regulate US7.
To determine whether miR-US5-1 and miR-US5-2 act additively or synergistically to reduce expression of US7, we used an isobologram analysis commonly utilized for drug interactions. An isobologram is defined as a graph of equally effective dose pairs (isoboles) for a single effect level. Specifically, a particular effect level is selected, such as 50% of the maximum, and doses of drug A and drug B (each alone) that give this effect are plotted as axial points in a Cartesian plot (33). The straight line connecting A and B represents the dose pairs that will produce this effect in a simply additive combination. Points located below the additivity line denote lesser quantity of each drug necessary to reach the same effect (synergistic effect). Conversely, points above the line denote antagonism between the 2 drugs.
Luciferase reporter constructs were transfected with increasing concentrations of one of the miRNA mimics while holding the second miRNA constant, and vice versa. miRNA dose pairs that caused 50% inhibition of luciferase activity (IC50) were determined from titration curves and plotted on a graph (Fig. 2 A). All data points were below the additivity line, indicating synergy between miR-US5-1 and miR-US5-2. To rule out the possibility that miRNA combinations generate isobolograms with nonspecific synergistic effect, we also performed an additivity control isobologram by combining miR-US5-2 with itself (Fig. 2B). As expected, the pairs of miR-US5-2 doses scattered along the additivity line. These data demonstrate that miR-US5-1 and miR-US5-2 have a synergistic effect on the HCMV US7 3′UTR.
Fig. 2.
miR-US5-1 and miR-US5-2 act synergistically to downregulate US7. (A) Luciferase assays were performed on cells cotransfected with the US7 WT 3′UTR luciferase reporter construct and various combinations of miR-US5-1 and miR-US5-2 as indicated in Materials and Methods. Dose pairs of miRNAs yielding 50% inhibition of luciferase activity were determined from titration curves and plotted onto a Cartesian graph. The line connecting the plots corresponding to miR-US5-1 and miR-US5-2 alone is the line of additivity. Dots located below the line of additivity indicate synergy. (B) Analysis of cells transfected with the US7 WT 3′UTR luciferase reporter construct and dilutions of miR-US5-2.
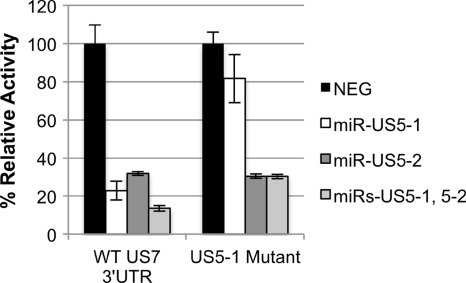
miR-US5-1 interacts with the US7 3′UTR at one site.
To determine whether miR-US5-1 regulates US7 by binding to the identified site present within the 3′UTR, six bases in the miR-US5-1 seed sequence were mutated using site-directed mutagenesis. Mutation of the miR-US5-1 target site completely restored luciferase activity in the presence of miR-US5-1 (Fig. 3), suggesting that miR-US5-1 regulates US7 by binding to the one target site within the 3′UTR. As expected, mutation of the miR-US5-1 target site had no effect on miR-US5-2 regulation of the US7 3′UTR. This result demonstrates that miR-US5-1 interacts with the US7 3′UTR at one site.
Fig. 3.
miR-US5-1 interacts with the US7 3′UTR at one functional site. Site-directed mutagenesis was used to mutate the predicted miR-US5-1 target site in the US7 3′UTR luciferase reporter construct. Reporter constructs encoding the wild-type US7 3′UTR or the mutated US7 3′UTR (US5-1 Mutant) were cotransfected into cells with either a negative control miRNA (NEG) or the indicated miR(s). Dual luciferase analyses were performed to examine reporter activity 16 h posttransfection.
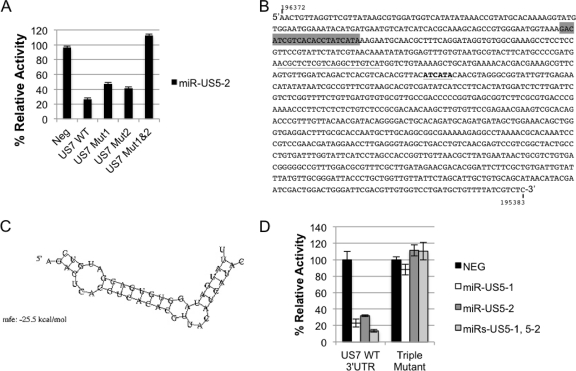
miR-US5-2 interacts with the US7 3′UTR at two sites.
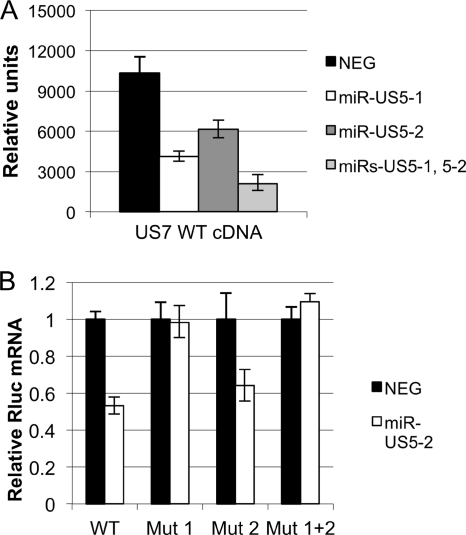
Similarly, we used site-directed mutagenesis to determine whether miR-US5-2 interacts with the US7 3′UTR by binding to the identified site. Surprisingly, we observed only partial restoration of expression when the miR-US5-2 binding site was mutated (Fig. 4 A, Mut 1). Luciferase expression was only partially restored to 50% of the levels seen after transfection of a negative control (similar results were observed by Western blotting [data not shown]).
Fig. 4.
miR-US5-2 interacts with the US7 3′UTR at two independent sites. (A) Site-directed mutagenesis was used to mutate the perfectly matched miR-US5-2 binding site (US7 Mut 1), the 6-mer miR-US5-2 binding site (US7 Mut 2), or both binding sites (US7 Mut1&2) in the US7 3′UTR luciferase reporter construct. Reporter constructs containing a negative control 3′UTR (NEG) or the WT or mutated US7 3′UTRs were cotransfected into cells with miR-US5-2. (B) The US7 3′UTR sequence. The genomic location of the sequence in AD169 is indicated (196372 to 195383). The perfectly matched miR-US5-2 binding site is highlighted in gray, the perfectly matched miR-US5-1 binding site is underlined, and the 6-mer miR-US5-2 binding site is underlined and in boldface. (C) RNAhybrid was used to predict alternate miR-US5-2:US7 3′UTR interaction sites. The predicted structure of the second miR-US5-2:US7 mRNA interaction containing the 6-mer seed sequence match is shown. The top strand is the miR-US5-2 sequence, while the bottom strand is the US7 3′UTR sequence. (D) Site-directed mutagenesis was used to mutate the single miR-US5-1 and the two miR-US5-2 binding sites simultaneously. Cells were cotransfected with either the reporter construct encoding wild-type US7 3′UTR or a 3′UTR in which all three sites were mutated (Triple Mutant) along with a negative control miRNA (NEG) or the indicated miR(s). Dual luciferase reporter assays were performed 16 h posttransfection.
We used the online RNAhybrid program to predict stable miRNA:mRNA interactions between miR-US5-2 and the US7 3′UTR (28). Six interactions with a minimum free energy (mfe) of ≤−20 kcal/mole were predicted including the completely complementary site (−42.4 kcal/mole). The second most stable interaction (−28.0 kcal/mol) used extensive G-U base pairing, which does not support stable miRNA:mRNA interactions (8). We therefore chose to mutate the bases that gave the third most stable interaction (−25.5 kcal/mol) consisting of a 6-base pair seed sequence match (termed a 7-mer seed sequence) in the US7 3′UTR, shown in Fig. 4B and C. Similar to the previous observation after mutation of the first site, we observed a partial restoration of expression (42%) upon mutation of this site (Fig. 4A, Mut 2). We therefore mutated both sites simultaneously and observed that luciferase activity was restored to 100%, demonstrating that miR-US5-2 interacts with the US7 3′UTR at two independent sites (Fig. 4A, Mut 1&2).
To evaluate the contribution of all miR-US5-1 and miR-US5-2 sites together, all three target sites were mutated simultaneously and tested. We found that mutation of all three sites resulted in the complete restoration of expression of the reporter compared to the negative control in the presence of both miRNAs (Fig. 4D). This demonstrates that miR-US5-1 and miR-US5-2 coregulate US7 through three functional sites within the 3′UTR. miR-US5-1 regulates US7 through one functional site, while miR-US5-2 regulates US7 through two functional sites.
miR-US5-2 downregulates US7 using two distinct mechanisms.
Surprisingly, we noted that each miR-US5-2 target site within the US7 3′UTR contributed equally to repression mediated by miR-US5-2 (∼50% by each) despite the fact that one site is fully complementary to the miRNA while the other has only a 7-mer seed sequence match. This effect was seen either when miR-US5-2 was tested by itself or upon coregulation with miR-US5-1. In mammalian cells, miRNAs with complete complementarity to the target mRNA behave like an siRNA and induce rapid mRNA degradation through endonucleolytic cleavage. Incomplete base pairing between miRNA:mRNA hybrids, however, results in translational repression through a variety of mechanisms that do not necessarily involve RNA degradation or decay (reviewed in reference 9). To determine the mechanism(s) of miR-US5-1 and miR-US5-2 inhibition, we measured US7 mRNA levels after cotransfection with miR-US5-1 or miR-US5-2. As shown in Fig. 5 A, US7 mRNA was reduced in the presence of miR-US5-1 and miR-US5-2, suggesting that both miRNAs repress US7 at the RNA level. In a separate experiment, we used the Rluc construct containing the US7 3′UTR to determine the contribution of each miR-US5-2 binding site to the stability of the mRNA. We observed a decrease in Rluc transcript abundance when a construct containing the wild-type 3′UTR was used (Fig. 5B). However, when the site with complete complementarity to miR-US5-2 was mutated, the Rluc transcript was stabilized. Mutation of both sites also resulted in stabilization of the transcript. In contrast, mutation of the imperfectly matched site failed to stabilize the transcript. This demonstrates that interaction between miR-US5-2 and its fully complementary target site induces transcript degradation, similar to what is seen with miR-US5-1. In contrast, interaction through the imperfectly matched site has no effect on transcript stability.
Fig. 5.
miR-US5-2 downregulates US7 using two distinct mechanisms. (A) Plasmid containing the US7 open reading frame and predicted 3′UTR or empty plasmid was transfected into cells with either negative control miRNA or the indicated miR(s). Twenty-four hours after transfection, RNA was isolated and RT-PCR analysis was performed using primer probe sets specific for US7 and GAPDH (for ΔΔCT normalization). Relative amounts of US7 mRNA are shown. (B) Reporter constructs containing the wild-type US7 3′UTR or 3′UTRs with the perfectly matched miR-US5-2 site (Mut 1), the imperfectly matched 6-mer miR-US5-2 site (Mut 2), or both (Mut 1 + 2) were mutated were transfected into cells. Twenty-four hours after transfection, RNA was isolated and RT-PCR analysis was performed using primer probe sets specific to Rluc and Fluc. Rluc expression was normalized to Fluc by ΔΔCT, and relative Rluc mRNA is shown.
Taken together, these results suggest that miR-US5-2 modulates expression of US7 by two distinct but equally efficient mechanisms.
miR-US5-1 and miR-US5-2 downregulate US7 expression at later times in infection.
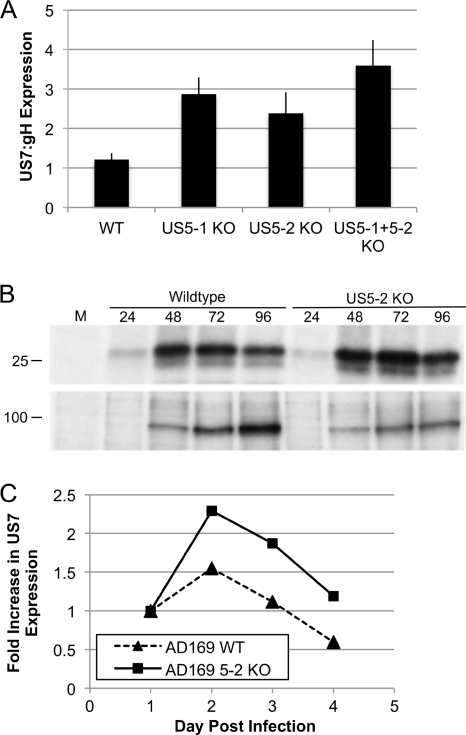
To determine whether miR-US5-1 and miR-US5-2 regulate endogenous US7 expression after infection of cells with HCMV, we constructed viral mutants lacking the 70 nucleotides coding for the miR-US5-1 or miR-US5-2 pre-miRNAs. In addition, we constructed a virus deleted for 193 nucleotides encoding both miR-US5-1 and miR-US5-2 (unpublished data). All 3 viruses demonstrated a small but reproducible growth defect in primary fibroblasts, which was mainly attributable to a defect in the release of virus. The same growth defect was observed after independent construction of 3 viruses lacking miR-US5-2, suggesting that the growth defect was not due to a second site mutation (data not shown). We infected cells with each of the mutant viruses or wild-type virus, metabolically labeled the cells, and performed immunoprecipitations using antibodies specific for US7. We normalized US7 expression to the expression of glycoprotein H (gH) as a control. As shown in Fig. 6, we observed an increase in the expression of endogenous US7 in the absence of the miRNAs as compared to wild-type virus. Densitometry revealed a 2- to 3-fold increase in US7 expression in the absence of miR-US5-1 or miR-US5-2 and an almost 4-fold increase in the absence of both. A second experiment using the miR-US5-2-deleted virus showed that the US7 overabundance was not observed until after 48 h of infection, a time at which miRNAs begin to be expressed (12).
Fig. 6.
miR-US5-1 and miR-US5-2 downregulate US7 during infection. (A) Primary human fibroblasts were infected with either wild-type, miR-US5-1 (US5-1 KO), miR-US5-2 (US5-2 KO), or miR-US5-1 plus US5-2 (US5-1+5-2 KO) mutant viruses. Ninety-six hours postinfection, the cells were metabolically labeled with [35S]methionine-cysteine for 60 min prior to the performance of immunoprecipitations using antibodies specific for US7 or glycoprotein H (gH). The intensity of the bands for each protein at each time point was quantitated using ImageQuant. US7 expression was normalized to gH expression. (B and C) Primary human fibroblasts were infected with either AD169 wild-type virus or miR-US5-2 (US5-2 KO) mutant viruses. At the indicated hours postinfection, cells were metabolically labeled with [35S]methionine-cysteine for 60 min prior to the performance of immunoprecipitations using antibodies specific for US7 or glycoprotein H (gH). (B) Representative gel. M, mock infection. Numbers at top represent hours postinfection. (C) The intensity of the bands for each protein at each time point was quantitated using ImageQuant. US7 expression was normalized to gH, and the fold increase in US7 expression relative to gH is shown over time.
DISCUSSION
In this study we identify a new target of HCMV-encoded miRNAs, the viral US7 gene. We show that the regulation of US7 is complex, involving two miRNAs, miR-US5-1 and miR-US5-2. miR-US5-1 regulates US7 by binding to a completely complementary site within the US7 3′UTR, resulting in strong repression of a reporter construct containing this 3′UTR. miR-US5-2 binds to both a completely complementary site and an additional site composed of 6 nucleotides within the seed sequence. Surprisingly, site-directed mutagenesis of each site demonstrated that both were comparable in directing the repression of a reporter construct. This is especially noteworthy given that binding of miR-US5-2 to the completely complementary site resulted in a decrease in transcript abundance, while binding of miR-US5-2 to the imperfectly matched site did not. This suggests that translational repression due to incomplete miRNA:mRNA pairing can exert an effect equal to that of repression resulting from complete pairing. To our knowledge, this is the first demonstration of a gene that is regulated through both mechanisms.
miRNAs have a robust effect on gene expression, yet analysis of the effect of a single miRNA on its target often shows only modest changes in expression. Global bioinformatic analysis of miRNAs and predicted target genes have suggested that tandem binding sites for one miRNA and binding sites for multiple miRNAs may affect the strength of repression (16, 20, 35). A few experimental studies have supported this (11, 17, 20, 24, 37, 38). However, detailed analyses into the rules governing coregulation by miRNAs are lacking. We used HCMV-encoded miRNAs, miR-US5-1 and miR-US5-2, to examine the coregulation of a viral gene, US7.
We used an isobologram analysis to show that miR-US5-1 and miR-US5-2 have a highly synergistic effect on the US7 3′UTR. This effect is seen even at very low concentrations of miRNAs. The effect is functional in the context of a viral infection, as viruses lacking miR-US5-1 or miR-US5-2 express higher levels of US7, while a virus lacking both expresses even greater amounts of US7. Previous reports have identified spacing constraints between miRNA binding sites that affect cooperativity between miRNAs (15, 16, 30). Sites spaced between 16 and 35 nucleotides apart (measured from the beginning of each binding site) resulted in cooperativity between the miRNAs, while shorter or longer distances inhibited cooperativity. In the US7 3′UTR, there is a perfectly matched miR-US5-2 site followed by a perfectly matched miR-US5-1 site followed by a second imperfectly matched miR-US5-2 site. There are 128 bases between the first two sites and 79 bases between the second two sites. None of these spacings predict cooperativity, suggesting that longer spacings between miRNAs can also result in synergistic effects. One important difference with previous studies on miRNA cooperativity is that the US7 3′UTR contains a mix of perfectly and imperfectly matched target sites that regulate US7 expression through at least two distinct mechanisms. It is possible that synergy between miRNAs targeting imperfectly matched sites occurs only when they are closely spaced due to the more efficient recruitment of shared cofactors, similar to what has been described for response elements and transcription factors (7, 19). In contrast, when different mechanisms are at play like in the US7 situation, the synergy between miRNAs might result from a different set of interactions that do not require closely spaced sites. It will be interesting to determine whether miR-US5-1 acts cooperatively with miR-US5-2 through either or both of the miR-US5-2 binding sites.
Although the primary RNA that encodes miR-US5-1 and miR-US5-2 has not yet been identified, it is likely they are derived from the same transcript due to their close proximity, thus ensuring coexpression and tight regulation of US7. Bioinformatics has suggested that mammalian genes targeted by multiple miRNAs are involved in cellular processes that cause disease when regulation is perturbed (e.g., cell cycle progression genes) (16). The US7 protein is an endoplasmic reticulum (ER)-retained protein of unknown function. A positional homolog for HCMV US7 exists in the rhesus CMV (RhCMV) genome (Rh186). We have determined that Rh186 is also strongly regulated by RhCMV-encoded miRNAs (unpublished data), highlighting the importance of the regulation of these genes. Studies are under way to determine the role of US7 during viral infection.
ACKNOWLEDGMENTS
We thank David Johnson for providing antibodies to US7.
This work was supported by NIH grant A121640 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 7 September 2011.
REFERENCES
- 1. Ambros V. 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- 2. Baek D., et al. 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- 4. Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bogner E., et al. 1992. Recognition of compartmentalized intracellular analogs of glycoprotein H of human cytomegalovirus. Arch. Virol. 126: 67–80 [DOI] [PubMed] [Google Scholar]
- 6. Brennecke J., Stark A., Russell R. B., Cohen S. M. 2005. Principles of microRNA-target recognition. PLoS Biol. 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doench J. G., Petersen C. P., Sharp P. A. 2003. siRNAs can function as miRNAs. Genes Dev. 17: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doench J. G., Sharp P. A. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18: 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fabian M. R., Sonenberg N., Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79: 351–379 [DOI] [PubMed] [Google Scholar]
- 10. Farh K. K., et al. 2005. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- 11. Forrest A. R., et al. 2010. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 24: 460–466 [DOI] [PubMed] [Google Scholar]
- 12. Grey F., et al. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79: 12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grey F., Meyers H., White E. A., Spector D. H., Nelson J. 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3: e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grey F., et al. 2010. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog. 6: e1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grimson A., et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hon L. S., Zhang Z. 2007. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 8: R166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hua Z., et al. 2006. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huber M. T., Tomazin R., Wisner T., Boname J., Johnson D. C. 2002. Human cytomegalovirus US7, US8, US9, and US10 are cytoplasmic glycoproteins, not found at cell surfaces, and US9 does not mediate cell-to-cell spread. J. Virol. 76: 5748–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kadonaga J. T. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257 [DOI] [PubMed] [Google Scholar]
- 20. Krek A., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500 [DOI] [PubMed] [Google Scholar]
- 21. Lewis B. P., Burge C. B., Bartel D. P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- 22. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- 23. Lin H. R., Ganem D. 2011. Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc. Natl. Acad. Sci. U. S. A. 108: 5148–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nachmani D., Lankry D., Wolf D. G., Mandelboim O. 2010. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 11: 806–813 [DOI] [PubMed] [Google Scholar]
- 25. Orom U. A., Nielsen F. C., Lund A. H. 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- 26. Peter M. E. 2010. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 29: 2161–2164 [DOI] [PubMed] [Google Scholar]
- 27. Pfeffer S., et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- 28. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA. 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rue C. A., et al. 2004. A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tropism. J. Virol. 78: 12529–12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saetrom P., et al. 2007. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 35: 2333–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selbach M., et al. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- 32. Stern-Ginossar N., et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tallarida R. J. 2001. Drug synergism: its detection and applications. J. Pharmacol. Exp. Ther. 298: 865–872 [PubMed] [Google Scholar]
- 34. Thomas M., Lieberman J., Lal A. 2010. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- 35. Tsang J. S., Ebert M. S., van Oudenaarden A. 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell 38: 140–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tuddenham L., Pfeffer S. 21 April 2011. Roles and regulation of microRNAs in cytomegalovirus infection. Biochim. Biophys. Acta. doi:10.1016/j.bbagrm.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 37. Wu S., et al. 2010. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 29: 2302–2308 [DOI] [PubMed] [Google Scholar]
- 38. Ziegelbauer J. M., Sullivan C. S., Ganem D. 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 41: 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]