Abstract
The 2009 pandemic influenza H1N1 (H1N1pdm) virus was generated by reassortment of swine influenza viruses of different lineages. This was the first influenza pandemic to emerge in over 4 decades and the first to occur after the realization that influenza pandemics arise from influenza viruses of animals. In order to understand the biological determinants of pandemic emergence, it is relevant to compare the tropism of different lineages of swine influenza viruses and reassortants derived from them with that of 2009 pandemic H1N1 (H1N1pdm) and seasonal influenza H1N1 viruses in ex vivo cultures of the human nasopharynx, bronchus, alveoli, and conjunctiva. We hypothesized that virus which can transmit efficiently between humans replicated well in the human upper airways. As previously reported, H1N1pdm and seasonal H1N1 viruses replicated efficiently in the nasopharyngeal, bronchial, and alveolar epithelium. In contrast, representative viruses from the classical swine (CS) (H1N1) lineage could not infect human respiratory epithelium; Eurasian avian-like swine (EA) (H1N1) viruses only infected alveolar epithelium and North American triple-reassortant (TRIG) viruses only infected the bronchial epithelium albeit inefficiently. Interestingly, a naturally occurring triple-reassortant swine virus, A/SW/HK/915/04 (H1N2), with a matrix gene segment of EA swine derivation (i.e., differing from H1N1pdm only in lacking a neuraminidase [NA] gene of EA derivation) readily infected and replicated in human nasopharyngeal and bronchial epithelia but not in the lung. A recombinant sw915 with the NA from H1N1pdm retained its tropism for the bronchus and acquired additional replication competence for alveolar epithelium. In contrast to H1N1pdm, none of the swine viruses tested nor seasonal H1N1 had tropism in human conjunctiva. Recombinant viruses generated by swapping the surface proteins (hemagglutinin and NA) of H1N1pdm and seasonal H1N1 virus demonstrated that these two gene segments together are key determinants of conjunctival tropism. Overall, these findings suggest that ex vivo cultures of the human respiratory tract provide a useful biological model for assessing the human health risk of swine influenza viruses.
INTRODUCTION
The influenza pandemic of 2009 was caused by a H1N1 virus of swine-origin (H1N1pdm) that emerged in Mexico and rapidly spread worldwide. Early reports from Mexico indicated significant case fatality rates associated with the pandemic. Experimental infection of mice, ferrets, and macaques suggested that the H1N1pdm virus was markedly more virulent than seasonal influenza viruses, causing lethality without prior adaptation (4). Epidemiological data illustrated the rapidity of global spread of the pandemic virus and very high infection attack rates, especially in children, but suggested that virulence of the pandemic virus was relatively modest and not very different to seasonal influenza (13). Severe complications were largely, although not exclusively, found in those with underlying respiratory, cardiac, or endocrine diseases or seen in those with pregnancy or morbid obesity (2), and increased numbers of such complications seen were attributable to the large numbers of persons infected with a novel virus. Previously, we have reported that the tissue tropism of the H1N1pdm and human seasonal H1N1 virus in primary in vitro and ex vivo cultures of the human respiratory tract was similar except that only H1N1pdm infected and replicate in human conjunctiva (1). Our studies also showed that the pandemic virus was generally comparable to seasonal influenza viruses in its intrinsic capacity for induction of host responses, including cytokine responses, and this was in marked contrast to viruses such as the avian influenza virus H5N1 (1, 5). These results demonstrate that in vitro and ex vivo cultures of the human respiratory tract provide useful information on the tropism and pathogenic potential of novel viruses.
Pandemic influenza viruses arise from animal influenza viruses. The 2009 pandemic provides the first opportunity to understand the virus genetic determinants that facilitate emergence of pandemic influenza viruses. The H1N1pdm virus was generated by reassortment of swine influenza viruses. The neuraminidase (NA) and matrix (M) gene segments of the H1N1pdm was derived from Eurasian avian-like swine (EA) viruses, while the other gene segments originated from North American triple-reassortant (TRIG) H1N1 viruses (10). The TRIG viruses themselves emerged in the 1990s and had acquired human (PB1), classical swine (CS) H1N1 (hemagglutinin [HA], NA, nucleoprotein [NP], NS, and M), and avian (polymerase basic 2 [PB2] and polymerase [PA]) virus gene segments. It is inferred from phylogenetic studies that this reassortant circulated in swine for more than 10 years before emerging as a pandemic (3, 10).
While the pandemic virus has not thus far undergone significant antigenic or genetic change within humans that alter its virulence or propensity to spread, there is evidence of the pandemic H1N1 virus repeatedly infecting swine, leading in some instances to genetic reassortment with other swine influenza viruses, providing an alternate milieu within which the virus may change genetically (7). Such viruses may pose novel threats to public health. From a systematic surveillance program of swine influenza viruses in southern China (8, 10), we have characterized swine influenza viruses representing the CS, EA, and TRIG virus lineages and reassortments between these lineages. In particular, A/SW/HK/915/04 (H1N2) is a naturally occurring TRIG reassortant that acquired an M gene segment from an EA lineage virus and therefore has the gene constellation of H1N1pdm with the exception of the NA gene (10). We also detected A/SW/HK/201/10 (H1N1) (11), which is a reassortant that rose within swine, which carries the NA gene of H1N1pdm, an HA of EA-lineage origin with the other gene segments being of TRIG derivation.
We investigated here the tropism of swine influenza viruses in ex vivo cultures of the human respiratory tract. We have used viruses representing the major lineages of swine influenza virus including the CS, EA, and TRIG lineages, as well as naturally occurring reassortant viruses A/SW/HK/915/04 (H1N2) and A/SW/HK/201/10 (H1N1), in comparison to H1N1pdm and seasonal influenza viruses. To further address the viral genetic determinants of tissue tropism, we generated reverse genetic viruses (14) (as listed detail in Table 1) and used them in ex vivo cultures of the human respiratory tract.
Table 1.
Influenza A viruses used in this study
| Virus | Yr | Subtype | Abbreviation | Genotype |
Corresponding figure(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |||||
| A/HongKong/54/1998 | 1998 | H1N1 | HK98/H1N1 | Human | Human | Human | Human | Human | Human | Human | Human | Fig. 3, Fig. S1 |
| A/Oklahoma/447/08 | 2008 | H1N1 | OK08/H1N1 | Human | Human | Human | Human | Human | Human | Human | Human | Fig. S1 |
| A/Swine/HongKong/4167/1999 | 1999 | H1N1 | swHK99/H1N1 | CS | CS | CS | CS | CS | CS | CS | CS | Fig. 1, Fig. 3 |
| A/Swine/HongKong/1733/2002 | 2002 | H1N1 | sw1733/H1N1 | CS | CS | CS | CS | CS | CS | CS | CS | Fig. S1 |
| A/Swine/Hong Kong/NS605/2003 | 2003 | H1N2 | sw605/H1N2 | CS | CS | CS | CS | CS | Human-N2 | CS | CS | Fig. S1 |
| A/Swine/Hong Kong/NS157/2004 | 2004 | H1N2 | sw157/H1N2 | CS | CS | CS | CS | CS | Human-N2 | CS | CS | Fig. S1 |
| A/Swine/HongKong/NS29/2009 | 2009 | H1N1 | swNS29/H1N1 | EA | EA | EA | EA | EA | EA | EA | EA | Fig. 1 |
| A/Swine/HongKong/PHK199/2009 | 2009 | H1N1 | sw199/H1N1 | EA | EA | EA | EA | EA | EA | EA | EA | Fig. S1 |
| A/Swine/HongKong/1559/2008 | 2008 | H1N1 | sw1559/H1N1 | EA | EA | EA | EA | EA | EA | EA | TRIGa | Fig. S1 |
| A/Swine/Arkansas/2976/2002 | 2002 | H1N2 | swAR02/H1N2 | TRIG | TRIG | TRIG | TRIG | TRIG | TRIG | TRIG | TRIG | Fig. 1, Fig. 3 |
| A/Swine/HongKong/PHK1110/2006 | 2006 | H1N2 | sw1110/H1N2 | TRIG | TRIG | TRIG | TRIG | TRIG | TRIG | TRIG | TRIG | Fig. S1 |
| A/HongKong/415742/2009 | 2009 | H1N1 | HK09/H1N1pdm | Avian | Human | Avian | CS | CS | EA | EA | CS | Fig. 1, Fig. 3 |
| A/OklahomaK/3052/09 | 2009 | H1N1pdm | OK09/H1N1pdm | Avian | Human | Avian | CS | CS | EA | EA | CS | Fig. S1 |
| A/Swine/HongKong/PHK915/2004 | 2004 | H1N2 | sw915/H1N2 | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | TRIG | H1N1pdm | H1N1pdm | Fig. 1, Fig. 3 |
| A/Swine/Hong Kong/201/2010 | 2010 | H1N1 | sw201/H1N1 | TRIG | TRIG | TRIG | EA | TRIG | H1N1pdm | TRIG | TRIG | Fig. 1, Fig. 3 |
| rg/sw/HK/PHK915/04 × H1N1pdm-NA | NAb | H1N1 | rg915 × H1N1pdmNA | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | Fig. 2 |
| rg/HK/54/98 | NA | H1N1 | NA | Human | Human | Human | Human | Human | Human | Human | Human | Fig. 3 |
| rg/HK/415742/09 | NA | H1N1pdm | NA | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | H1N1pdm | Fig. 3 |
| rg/HK/54/98 + rg/H1N1pdm HA | NA | NA | NA | Human | Human | Human | H1N1pdm | Human | Human | Human | Human | Fig. 3 |
| rg/HK/54/98 + rg/H1N1pdm NA | NA | NA | NA | Human | Human | Human | Human | Human | H1N1pdm | Human | Human | Fig. 3 |
| rg/HK/54/98 + rg/H1N1pdm HANA | NA | NA | NA | Human | Human | Human | H1N1pdm | Human | H1N1pdm | Human | Human | Fig. 3 |
| rg/H1N1pdm + rg/HK/54/98 HANA | NA | NA | NA | H1N1pdm | H1N1pdm | H1N1pdm | Human | H1N1pdm | Human | H1N1pdm | H1N1pdm | Fig. 3 |
TRIG (PB2 and PA, avian; PB1, human, HA, NA, NP, and NS; M, CS).
NA, not applicable.
MATERIALS AND METHODS
Influenza virus preparation.
The viruses used in the present study are listed in Table 1 with their origins and abbreviations. Virus stock was propagated in Madin-Darby canine kidney (MDCK) cells. The stock was titrated as previously described (1). Recombinant viruses of HK54/H1N1 and HK415742/H1N1pdm (14) were created by reverse genetics as previously described (15), with HA and NA genes of HK415742/H1N1pdm on a sw915/H1N2 backbone and vice versa, to address the issues of tropism of the virus for the respiratory tract and conjunctiva. Replication competence and kinetics of the recombinant viruses were tested in an MDCK culture.
Ex vivo organ cultures and influenza virus infection.
Ex vivo culture of the conjunctiva, nasopharynges, bronchi, and lungs was performed as previously described (1). In brief, fresh conjunctival tissues were obtained from 20 individuals who underwent excision for pterygium during surgical management, biopsy specimens of nasopharyngeal tissues (n = 10) were obtained from patients undergoing elective nasopharyngoscopy as detailed earlier (6), and bronchi (n = 19) and lung tissues (n = 15) were obtained from patients undergoing surgical resection of lung tissue. Biopsy specimens or tissue fragments of normal nonmalignant tissue that were obtained in excess to the requirements for clinical diagnosis were used. All of the studies were approved by the Institutional Review Board of the University of Hong Kong and Hospital Authority Hong Kong West Cluster, and written informed consent was provided by each patient. The conjunctival, nasopharyngeal, and bronchial biopsy specimens were incubated at 33°C, while the bronchial and lung tissue fragments were incubated at 37°C in culture medium (F-12K nutrient mixture with l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml). Influenza viruses were used at a titer of 106 50% tissue culture infectious doses (TCID50)/ml, a titer similar to that used previously (6, 9) for infecting the ex vivo cultures. The biopsy specimens were infected for 1 h and washed with 5 ml of warm 1× phosphate-buffered saline for three times to remove unbound virus and replenished with 1 ml of culture medium at 33 or 37°C.
Immunohistochemical staining and virus titration assay.
Immunohistochemical staining of conjunctival and respiratory tract tissue was carried out for the influenza virus nucleoprotein. The tissue sections were incubated with 0.1% Pronase (Roche) in 0.1 M Tris (pH 7.5) at 37°C for 1 min and blocked with 3% H2O2 in Tris-buffered saline for 10 min, followed by treatment with an avidin/biotin blocking kit (Vector Laboratories). After blocking with 10% normal rabbit serum for 10 min at room temperature, the sections were incubated with 1/25 (15 μg/ml) HB65 antibody for 1 h at room temperature, followed by the addition of biotinylated rabbit anti-mouse antibody (Dako Cytomation) diluted 1/100 for 30 min at room temperature. After incubation with Elite-ABC kit (Vector, PK-6100) diluted 1/50 for 30 min at room temperature, the sections were developed with a Vector NovaRed substrate kit (SK-4800). To determine productive viral replication from the infected biopsy specimens, supernatants of the infected cultures were collected at 1, 24, and/or 48 h postinfection (hpi) and stored at −80°C for virus titration using TCID50 assay in MDCK cells as described previously (1). The increasing virus titers along the time course provided evidence of productive virus replication.
Statistical analysis.
The differences of log10-transformed virus titers for different viruses at different temperatures of incubation and time points postinfection were compared using the Student t test and one-way analysis of variance, followed by the Bonferroni multiple-comparison test, respectively. Differences were considered significant at P < 0.05. Statistical analysis was carried out using GraphPad Prism 5.
RESULTS
Tissue tropism of swine and pandemic H1 viruses in human airway.
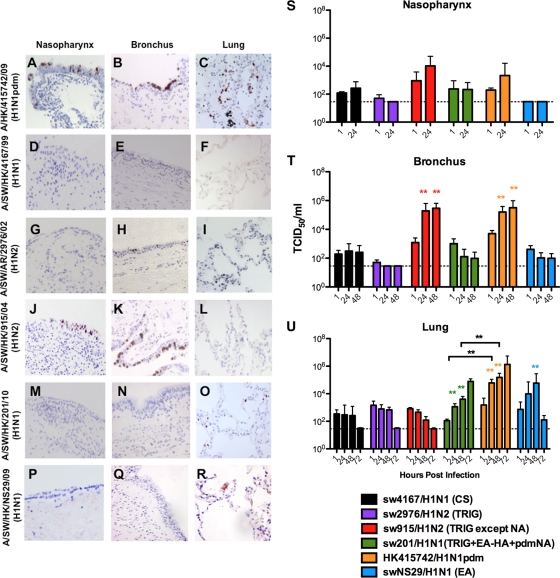
HK415742/H1N1pdm infected the human nasopharynx (Fig. 1A), bronchus (Fig. 1B), and lung (Fig. 1C) extensively and productively replicated to high virus titers (Fig. 1S to U) in accordance with our previous report (1). Classical swine sw4167/H1N1 (Fig. 1D and E) failed to infect the nasopharyngeal, bronchial, or alveolar epithelia, whereas TRIG sw2976/H1N1, the putative precursor of the 2009 pandemic virus, only showed focal infection of bronchial epithelium (Fig. 1H) with no convincing increase in virus titer (Fig. 1S to U). EA virus swNS29/H1N1 infected and replicated only in the alveolar epithelium (Fig. 1R and U) and not in the nasopharynx (Fig. 1P and S) and bronchus (Fig. 1Q and T). In contrast, we found that sw915/H1N2, a TRIG reassortant that has acquired an M gene segment of EA origin, infected the nasopharynx and bronchus, but no infection was found in the lungs (Fig. 1J to L, respectively). sw915/H1N2 and HK415742/H1N1pdm demonstrated comparable replication in the nasopharynx and bronchus (Fig. 1S and T); in contrast to HK415742/H1N1pdm, the sw915/H1N2 virus failed to replicate in the lungs (Fig. 1U). The TRIG-pandemic reassortant swine virus sw201/H1N1, which has an HA of EA derivation, NA of pandemic origin, and TRIG “internal” gene segments, failed to infect and replicate in the nasopharyngeal and bronchial epithelium but did infect and replicate in the alveolar epithelium, although less extensively than did HK09/H1N1pdm (P < 0.01) (Fig. 1M to O and S to U). We confirmed these overall patterns of virus tropism in the different swine virus lineages using additional representative of the H1N1pdm (OK3052/H1N1pdm), CS (sw1733/H1N1), TRIG (sw1110/H1N2), and EA (sw199/H1N1) virus lineages (see Fig. S1 in the supplemental material). Naturally occurring CS reassortant influenza viruses with the human N2 gene segment (sw605/H1N2 and sw157/H1N2) (12) and EA viruses with a TRIG NS gene segment (sw1599/H1N1) were also tested. As with other CS lineage viruses, sw605/H1N2 and sw157/H1N2 were not able to infect human bronchial and alveolar epithelia. As with other EA viruses, the EA reassortant virus (sw1599/H1N1) was able to infect and productively replicate in human lung but not in the bronchus (see Fig. S1 in the supplemental material).
Fig. 1.
Expression of influenza virus nucleoprotein (reddish brown) in the upper, conducting, and lower respiratory tract (A, D, G, J, M, and P), in the nasopharynx (B, E, H, K, N, and Q), and in the bronchi and lungs (C, F, I, L, O, and R) infected with influenza virus A/HK/415742/09 (H1N1pdm), A/SW/HK/4167/99 (H1N1), A/SW/AR/2976/02 (H1N2), A/SW/HK/915/04 (H1N2), and A/SW/HK/201/10 (H1N1) at 48 h postinfection (hpi). Viral replication kinetics in ex vivo cultures of nasopharynx (S), bronchi (T), and lung biopsy specimens (U) infected with 106 TCID50 of influenza viruses/ml by virus titration at 37°C were also evaluated. The chart shows the means and standard errors of the means of the virus titer pooled from at least three independent experiments. Horizontal dotted line denotes the detection limit of the viral titration assay. Colored asterisks indicate the statistically significant increases in viral yield compared to 1 hpi, and black asterisks indicate statistically significant differences between HK415742/H1N1pdm and sw201/H1N1 in figure U. **, P < 0.005.
H1N1pdm NA confers alveolar epithelial tropism to sw915/H1N2 virus.
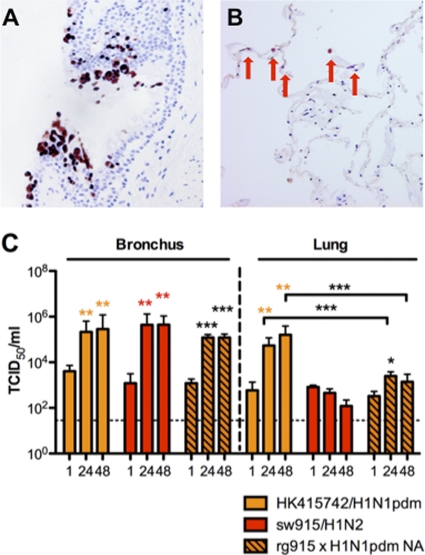
Since sw915/H1N2 virus differed from swHK09/H1N1pdm in being unable to replicate in alveolar epithelium, we used reverse genetics to generate a sw915 virus with the NA of H1N1pdm (14). The recombinant virus, sw915/H1N2, with the inclusion of NA from H1N1pdm replicated to a similar titer in MDCK cells, and it retained its tropism for the bronchus and acquired tropism for the alveolar epithelium (Fig. 2A and B). An increasing viral yield was observed in the infected lung culture at 24 hpi (P < 0.05), although its replication competence remained lower than that of the HK415742/H1N1pdm at 24 and 48 hpi (P < 0.005) (Fig. 2C). The virus replication kinetics of this recombinant virus in the bronchus was not significantly different from that of HK415742/H1N1pdm and sw915/H1N2.
Fig. 2.
Expression of influenza virus nucleoprotein (reddish brown) in the bronchus (A) and in lung biopsy specimens (B) by recombinant influenza A virus of rg sw915/H1N2 with the insertion of NA from HK415742/H1N1pdm influenza virus at 48 hpi. (C) Viral replication kinetics in ex vivo cultures of bronchi and lungs infected with 106 TCID50 of HK415742/H1N1pdm, sw915/H1N2, and rg sw915/H1N2 with the insertion of NA from HK415742/H1N1pdm influenza virus by virus titration at 37°C. Colored asterisks indicate statistically significant increases in the viral yield compared to 1 hpi, and black asterisks represent statistically significant differences between HK415742/H1N1pdm and rg sw915/H1N2 with the insertion of NA from HK415742/H1N1pdm. ***, P < 0.0005. The chart showed the means and the standard errors of the means of the virus titer pooled from three independent experiments. Asterisks indicate statistically significant increases in viral yield compare to 1 hpi: *, P < 0.05; **, P < 0.005; and ***, P < 0.0005.
Tissue tropism of swine influenza viruses for conjunctival epithelium.
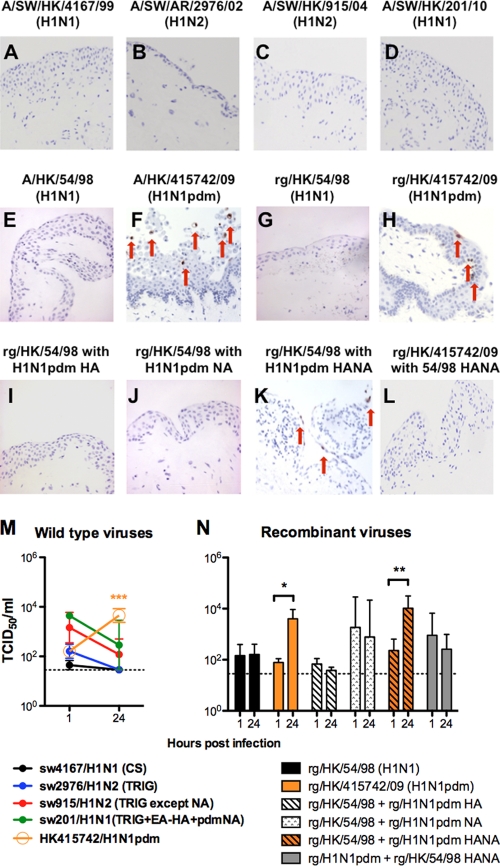
We investigated whether swine viruses also possessed conjunctival tropism. Immunohistochemistry for viral nucleoprotein (Fig. 3A to D) revealed that the classical swine virus sw4167/H1N1 (Fig. 3A), the triple-reassortant swine viruses sw2976/H1N2 (Fig. 3B) and sw915/H1N2 (which shares a common origin for seven gene segments with H1N1pdm swine virus) (Fig. 3C), and the swine H1N1pdm reassortment virus sw201/H1N1 (Fig. 3D) were not able to infect and replicate in human conjunctiva ex vivo culture (Fig. 3M). As previously reported (1), the HK415742/H1N1pdm (Fig. 3F) but not the human HK54/H1N1 virus (Fig. 3E) infected and productively replicated in the human conjunctiva (P < 0.0001) (Fig. 3M).
Fig. 3.
Ex vivo organ cultures of conjunctiva infected with wild-type influenza A/SW/HK/4167/99 (H1N1) (A), A/SW/AR/2976/02 (H1N2) (B), A/SW/HK/915/04 (H1N2) (C), A/SW/HK/201/10 (H1N1) (D), A/HK/54/98 (H1N1) (E), and A/HK/415742/09 (H1N1pdm) (F) viruses and the recombinant influenza viruses rg/HK/54/98 (H1N1) (G), rg/HK/415742/09 (H1N1pdm) (F), rg/HK/54/98 with the HA of A/HK/415742/09 (H1N1pdm) (I), rg/HK/54/98 with the NA of A/HK/415742/09 (H1N1pdm) (J), rg/HK/54/98 with the HANA of A/HK/415742/09 (H1N1pdm) (K), and rg/HK/415742/09 (H1N1pdm) with the HANA of A/HK/54/98 (H1N1) (L) at 24 hpi and stained for influenza virus A nucleoprotein in reddish brown, as indicated by arrows. (M and N) Wild-type (M) and recombinant (N) influenza virus yield at 24 hpi after infection with 106 TCID50 of the virus/ml at 33°C. The chart shows the means and the standard errors of the virus titers pooled from three independent experiments. Horizontal dotted line denotes detection limit of the viral titration. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Both HA and NA genes of HK415742/H1N1pdm are essential for conjunctival tropism.
We generated a set of recombinant HK54/H1N1 viruses wherein HA alone, NA alone, or both HA and NA HK54/H1N1 were exchanged with that from HK415742/H1N1pdm. The recombinant HK54/H1N1 (Fig. 3G) and HK415742/H1N1pdm (Fig. 3H) retained the tropism manifested by their wild-type counterpart. We observed that the combination of HA and NA genes from HK415742/H1N1pdm was essential for the recombinant HK54/H1N1 virus to acquire conjunctival tropism (Fig. 3K and N). The recombinant viruses with either HA alone (Fig. 3I) or NA alone (Fig. 3J) from H1N1pdm did not have an efficient tropism in human conjunctiva. Conversely, a recombinant HK415742/H1N1pdm virus with HA and NA derived from seasonal HK54/H1N1 failed to replicate in the conjunctiva (Fig. 3L).
DISCUSSION
We used representative swine influenza viruses of different lineages isolated in our 13-year systematic surveillance of swine influenza viruses in southern China to investigate the hypothesis that viral tropism for the human upper respiratory tract and bronchial epithelium correlates with transmissibility in humans and aerosol transmission in ferrets. Although H1N1pdm and seasonal influenza viruses replicated efficiently in the human upper airways and bronchus, swine influenza viruses generally failed to do so. A reassortant sw201/H1N1 virus with TRIG internal genes, HA of EA derivation and NA derived from H1N1pdm (11), was also unable to replicate in the upper airways or bronchus. Interestingly, a naturally occurring reassortant swine virus, sw915/H1N2 (10), which has a triple reassortant virus genes with a matrix gene segment of EA swine derivation, thus differing from H1N1pdm only in lacking a NA gene of EA derivation, did replicate in the nasopharynx and bronchus. These findings suggest that the H1N1pdm-like sw915/H1N2 virus had acquired tropism for the human upper respiratory tract and bronchus and might have potential for transmission in humans. Interestingly, these same swine viruses were investigated in a ferret experimental infection model for aerosol transmission competence and sw915/H1N2 was the only virus to demonstrate (though weak) transmission competence, a striking correlation (14).
Unlike the H1N1pdm virus, the sw915/H1N2 virus failed to replicate in the alveolar epithelium. A recombinant sw915/H1N2 virus with the NA gene segment of EA derivation acquired tropism for the alveolar epithelium, thereby confirming the contribution of M and NA gene segments of EA origin in mimicking the unique tropism of the H1N1pdm virus. In contrast, sw201/H1N1 which was a TRIG virus with an HA of EA derivation and an NA of H1N1pdm derivation, thus differing from H1N1pdm in its derivation of the M and HA gene segments, replicated poorly in the nasopharynx and bronchus and was unlikely to spread efficiently in humans. However, given its tropism for the lower respiratory tract, sw201/H1N1 had its potential for severe lung pathology if the virus gained access to the lower respiratory tract. Swine viruses of pure EA origin or EA reassortants with the NS gene segment of TRIG origin both infected and replicated in the alveolar epithelium but not in the upper respiratory tract or bronchus. Recent surveillance data from southern China demonstrate the increasing dominance of the EA reassortants with a TRIG NS gene (12). The lack of tropism for the nasopharynx and bronchus may indicate that these viruses are not a pandemic threat. However, these EA viruses, like the highly pathogenic avian influenza virus H5N1, have tropism for the human alveolar epithelium and thus may have the potential to cause severe disease in the rare event that these viruses gain access to the alveolar spaces through zoonotic transmission. As with H5N1 viruses, the EA-like swine viruses have HA and NA of avian derivation, and this may explain their tropism for the alveolar epithelium. Avian influenza viruses are believed to have preferential binding to α2-3 sialic acid receptors found in the alveolar epithelium, and this may explain the alveolar tropism of EA-lineage viruses.
In order to investigate the viral genetic determinants that contribute to viral tropism of H1N1pdm, a recombinant sw915/H1N2 virus with NA of H1N1pdm derivation was rescued by plasmid-based reverse genetics (14). This recombinant virus had expanded tropism for the lower lung, suggesting that the NA gene of the H1Npdm has an important contribution to the tissue tropism of the H1N1pdm virus. The NA gene of H1N1pdm origin is required for sw915/H1N2 to recapitulate the full tissue tropism exhibited by the pandemic virus, suggesting that the balance between the HA and the NA is important in this regard. However, the replication competence of this recombinant virus in human lung was lower than that of the wild-type H1N1pdm.
With the exception of H1N1pdm virus, none of the other investigated swine viruses was able to infect human conjunctiva, suggesting that H1N1pdm had a unique tropism for this epithelial site. We had previously demonstrated that the conjunctival tropism of H1N1pdm was dependent on the cell surface sialic acids (1). In the present study, we used plasmid-based virus reverse genetics to demonstrate that both HA and NA of the H1N1pdm virus act synergistically and are critical to confer conjunctival tropism to HK54/H1N1; either of these gene segments alone was unable to do so. This further emphasizes the role of the HA and NA balance required for manifesting distinctive viral tropism. The receptors that allow pandemic H1N1pdm to infect the conjunctiva remain to be defined.
The cocirculation of multiple swine influenza virus lineages and their ongoing reassortment in swine, taken together with the establishment of H1N1pdm in swine, emphasize the importance of systematic surveillance of influenza viruses in swine and the need to evaluate their pandemic risk. Our findings suggest that naturally occurring swine influenza viruses and their natural reassortants have differing tropism for the human respiratory tract. Of a range of swine influenza viruses representing different genetic lineages tested in the present study, only the naturally occurring reassortant sw915/H1N2, which shares seven genes of common origin with H1N1pdm, replicated well in the human nasopharynx and bronchus. It is possible that tropism for these tissues is a prerequisite for influenza virus to acquire efficient transmissibility in humans. While the sw915/H1N2 virus was not the direct ancestor of the H1N1pdm that emerged in Mexico, it is possible that similar intermediate viruses may have been generated through reassortment between EA and TRIG viruses in the Americas during the genesis of the 2009 pandemic H1N1 virus. Our findings suggest that ex vivo cultures of the human respiratory tract may therefore provide a useful biological model for risk assessment of the zoonotic and pandemic potential of such swine influenza viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kevin Fung for help with the immunohistochemistry staining. We thank Richard Webby for providing the influenza A/SW/AR/2976/02 (H1N2) virus.
We acknowledge financial support by a commissioned study (to J.S.M.P.) and RFCID grants (reference numbers 10090202 and 10091132 to J.M.N. and R.W.Y.C., respectively) by the Research Fund for Control of Infectious Disease, Food and Health Bureau, Hong Kong SAR Government, and the General Research Fund (HKU 7612/08 M to M.C.W.C.), Research Grants Council, Hong Kong SAR Government; the National Institutes of Health (NIAID contract HHSN266200700005C); and AoE Funding (AoE/M-12/06) from the Area of Excellence Scheme of the University Grants Committee, Hong Kong SAR Government. Additional funding was provided by a grant from the European Commission (FP7-GA258084).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 31 August 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Chan M. C., et al. 2010. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am. J. Pathol. 176: 1828–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dawood F. S., et al. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360: 2605–2615 [DOI] [PubMed] [Google Scholar]
- 3. Garten R. J., et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325: 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ilyushina N. A., et al. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84: 8607–8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S. M., et al. 2010. Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I-like alveolar epithelial cells in vitro. Respir. Res. 11: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicholls J. M., et al. 2007. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 13: 147–149 [DOI] [PubMed] [Google Scholar]
- 7. Pasma T., Joseph T. 2009. Pandemic (H1N1) infection in swine herds, Manitoba, Canada. Emerg. Infect. Dis. 16: 706–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peiris J. S., et al. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75: 9679–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shinya K., et al. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440: 435–436 [DOI] [PubMed] [Google Scholar]
- 10. Smith G. J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459: 1122–1125 [DOI] [PubMed] [Google Scholar]
- 11. Vijaykrishna D., et al. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328: 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vijaykrishna D., et al. 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473: 519–522 [DOI] [PubMed] [Google Scholar]
- 13. Wu J. T., et al. 2010. The infection attack rate and severity of 2009 pandemic H1N1 influenza in Hong Kong. Clin. Infect. Dis. 51: 1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yen H., et al. 2011. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc. Natl. Acad. Sci. U. S. A. 108: 14264–14269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yen H. L., et al. 2009. Changes in H5N1 influenza virus hemagglutinin receptor binding domain affect systemic spread. Proc. Natl. Acad. Sci. U. S. A. 106: 286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.