Abstract
Defining the mechanisms of chromosomal cohesion and dissolution of the cohesin complex from chromatids is important for understanding the chromosomal missegregation seen in many tumor cells. Here we report the identification of a novel cohesin-resolving protease and describe its role in chromosomal segregation. Sister chromatids are held together by cohesin, a multiprotein ring-like complex comprised of Rad21, Smc1, Smc3, and SA2 (or SA1). Cohesin is known to be removed from vertebrate chromosomes by two distinct mechanisms, namely, the prophase and anaphase pathways. First, PLK1-mediated phosphorylation of SA2 in prophase leads to release of cohesin from chromosome arms, leaving behind centromeric cohesins that continue to hold the sisters together. Then, at the onset of anaphase, activated separase cleaves the centromeric cohesin Rad21, thereby opening the cohesin ring and allowing the sister chromatids to separate. We report here that the calcium-dependent cysteine endopeptidase calpain-1 is a Rad21 peptidase and normally localizes to the interphase nuclei and chromatin. Calpain-1 cleaves Rad21 at L192, in a calcium-dependent manner. We further show that Rad21 cleavage by calpain-1 promotes separation of chromosome arms, which coincides with a calcium-induced partial loss of cohesin at several chromosomal loci. Engineered cleavage of Rad21 at the calpain-cleavable site without activation of calpain-1 can lead to a loss of sister chromatid cohesion. Collectively, our work reveals a novel function of calpain-1 and describes an additional pathway for sister chromatid separation in humans.
INTRODUCTION
DNA replication in S phase generates two identical molecules of chromosomal DNA, known as sister chromatids, which must equally segregate to the two daughter cells in the subsequent mitosis. Therefore, it is important that the sister chromatids are held together from the S phase until proper kinetochore assembly is complete. Such cohesion of the sisters is mediated by a multiprotein ring-like complex called cohesin, which is conserved from Saccharomyces cerevisiae to humans. Cohesin is comprised of four core proteins, namely, Rad21 (Scc1/Mcd1), Smc1, Smc3, and either SA2/Stag2 or SA1/Stag1 (13, 22, 25, 39; reviewed in reference 28). Cohesin undergoes a series of physical and chemical changes during the course of the cell cycle in order to orchestrate the dynamic cohesion and separation between the sister chromatids that are essential for high-fidelity transmission of the genetic material (32). The cohesin cycle includes loading cohesins onto the chromatin of the unpaired chromosome, forming cohesion between the sister chromatids in S phase, regulated freeing of chromosome arms in early mitosis, separation of sister chromatids to allow segregation to opposite poles, and recycling of cohesin molecules for subsequent usage.
Cohesin is loaded onto the chromatin in G1 phase in yeast (25) and in late telophase in vertebrates (21, 39). Loading of cohesin onto chromatin depends on two proteins, Scc2 and Scc4 (8, 46). Once it is loaded onto chromatin, cohesin undergoes acetylation on Smc3 in S phase and establishes cohesion between the sister chromatids (34, 43, 50). In early mitosis in vertebrates, cohesin is removed from chromosomal arms following phosphorylation of the cohesin component SA2 (15, 45), leaving behind a small pool of cohesins at the centromeres. Later, at metaphase-to-anaphase transition, when all the chromosomes have biorientated on the mitotic spindle, the endopeptidase separase cleaves the centromeric Rad21/Scc1 protein to finally separate the sister chromatids (45). Interestingly, yeast lacks the prophase-mediated removal of arm cohesion and all of the cohesin is removed by separase at the onset of anaphase (41, 42). The soluble pool of cohesin thus generated, which has retained acetylation on its Smc3 subunit, is then recycled in an Hos1p-dependent deacetylation reaction to complete the cohesin cycle (1, 5).
In addition to its canonical function in chromosomal cohesion, cohesin has also been implicated in DNA damage repair. In fact, the cohesin component Rad21 was first identified in a screen for sensitivity to radiation-induced DNA double-strand breaks (DSB) (2). Cohesin components Smc1 and Smc3 are phosphorylated by ATM and ATR kinases in response to DNA damage (19, 23). Following a single DSB, cohesin is rapidly recruited to all chromosomes in yeast (38, 44), whereas cohesin occupancy at preexisting sites is reinforced in human cells upon exposure to ionizing radiation (18). Cohesin is also implicated in transcriptional regulation (see reference 9 for a review). Recent investigations have revealed that cohesin cooperates with the transcriptional insulator CTCF to affect precise transcriptional regulation (30, 47), as well as collaborating with tissue-specific transcription factors (35) and the mediator complex (17) independent of CTCF. Rad21 is also implicated in apoptosis: it is cleaved in several human cell lines in response to apoptotic stimuli (7, 31). Also, the overexpression of the C-terminal apoptotic cleavage product of Rad21 promotes apoptosis in mammalian cells, indicating that Rad21 cleavage functions as both a responder to and stimulator of apoptosis (31). How cohesin participates in such diverse processes and how those functions are regulated are not clear. It is, however, clear that the failure to form or maintain cohesion results in premature separation of chromatids. Therefore, understanding the cohesin complex and the mechanisms of its dissolution from chromatids is an important step in unraveling the mechanisms of chromosomal missegregation, a key feature of tumor cells.
We reasoned that one way to better understand cohesin functions is to identify cohesin-interacting proteins. Toward that goal, we generated an affinity matrix by using in vitro-synthesized Rad21 and pulled down its interacting proteins from human cell extract. One of the interacting proteins identified from this screen was the calcium-dependent cysteine protease calpain-1. Here we present evidence that calpain-1 cleaves Rad21 and promotes resolution of sister chromatid arms, uncovering a novel pathway for effecting sister chromatid separation.
MATERIALS AND METHODS
Cell culture.
HEK293 and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), U2OS cells were grown in McCoy's medium with 15% FBS, and all other cell lines, including Molt4 and Jurkat cells, were grown in RPMI 1640 medium supplemented with 10% FBS. When needed, CaCl2 (to a final concentration of 2, 5, or 10 mM [see figure legends]), etoposide (final concentration of 5 or 20 μM), and N-[N-(N-acetyl-l-leucyl)-l-leucyl]-l-norleucine (ALLN; final concentration of 5 μM) were added to the cells for the specified periods.
Cell fractionation.
About 107 Jurkat cells were treated with 2 mM CaCl2 (final concentration) and harvested after 4, 8, and 12 h. Harvested cell pellets were washed in cold phosphate-buffered saline (PBS) and then once with cold hypotonic buffer (HB; 10 mM KCl, 10 mM Tris-Cl, pH 7.5, 1.5 mM MgCl2). Cell pellets were resuspended in HB and swollen on ice for 20 min. Cells were lysed by addition of an equal volume of HBI (HB supplemented with 0.4% Igepal, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 2× protease inhibitors [PI] [Complete Mini; Roche]) for a further incubation on ice for 30 min. The lysed cells were layered over nuclear cushion buffer (NCB; 1.2 M sucrose, 20 mM Tris-Cl, pH 7.5, 60 mM KCl, 15 mM NaCl, 1 mM β-mercaptoethanol, 1 mM PMSF, and protease inhibitors), and the nuclei were sedimented through the NCB. The upper layer was saved as the cytosolic extract. The nuclear pellet was washed in nuclear buffer (NB; same as NCB but with 0.34 M sucrose) once and then extracted with RIPA buffer supplemented with 1 mM β-mercaptoethanol, 1 mM PMSF, and protease inhibitors. The chromatin-rich nuclear debris was sedimented by centrifugation, and the supernatant (nuclear extract [NE]) was saved. Chromatin was solubilized and released from the nuclear debris by digesting the debris with 5 U micrococcal nuclease (MNase) for 15 min at room temperature (RT). Equal volumetric proportions were analyzed by immunoblotting.
Antibodies and reagents.
Anti-Rad21 polyclonal antibody (used for all immunoblot experiments; recognizes the Rad21 C terminus) and anti-Rad21 mouse monoclonal antibody (used for immunofluorescence) have been described elsewhere (31). The following other antibodies were used: anti-calpain-1 Sc-7531 (Santa Cruz) for immunoblotting and chromatin immunoprecipitation (ChIP) and ab28257 (Abcam) for immunofluorescence, antiseparase (H00009700-M01; Abnova), anti-histone H3 (ab1791; Abcam), anti-β-actin (A5316; Sigma), anti-tubulin (CP06-100UG; Oncogene), anti-histone H3 S28ph (07-145; Millipore), and anti-lamin B (ab16048; Abcam). For immunofluorescence, rhodamine-conjugated anti-mouse IgG (Invitrogen; for anti-Rad21 monoclonal antibody) and fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch Laboratories; for anti-calpain-1 polyclonal antibody) were used. Purified calpain-1 (208713) and ALLN (208719) were procured from Calbiochem. Etoposide (NDC 55390-292-01) was purchased from Bedford Laboratories, and MNase (70196Y) was purchased from USB-Affymetrix. β-Mercaptoethanol and PMSF were obtained from Sigma-Aldrich; Complete Mini protease inhibitor cocktail tablets were obtained from Roche. In ChIP experiments, normal mouse IgG (Calbiochem) was used as a control for Rad21, and normal goat IgG (Santa Cruz) was used as a control for calpain-1. Apoptotic cells were detected by staining Molt4 cells with annexin V-FITC and propidium iodide, using an apoptosis detection kit (MBL, Woburn, MA) according to the manufacturer's instructions.
Protein expression and siRNA depletion.
Wild-type (WT) Myc.Rad21 was expressed from a pCS2MT vector (pDP872) and has been described elsewhere (31). del, rep, and tev mutant constructs were generated from pDP872 by site-directed mutagenesis. Human WT Rad21 and del, rep, and tev mutants were synthesized from pCS2MT-derived constructs, either in a wheat germ extract transcription-coupled translation (TnT) system (see Fig. 1A and 2B and C) or in HEK293T cells followed by transient transfection (see Fig. 2D and E and 6B). Transient transfection, small interfering RNA (siRNA)-mediated protein depletion, and protein analyses were done as described previously (31, 51), except that RIPA buffer was used for protein extraction. Dharmacon transfection reagent 1 (DharmaFECT1) was used for all siRNA transfections. The following target sequences (5′ to 3′) were used. The three calpain-1 siRNAs were duplex A (CCACGGAACTGCTGTCAAA) (49), duplex B (AAGCTAGTGTTCGTGCACTCTGCC), and duplex C (GGGACTTGTGTACTGGTTATT). One Rad21 siRNA sequence was AGAGTTGGATAGCAAGACA (targeting the coding region [CR]), and a combination of three duplexes targeting the Rad21 3′ untranslated region (3′-UTR) were used to effectively deplete endogenous Rad21: GCAAAGTTAACACTGAAAGTTCTAG, AGCATTATAGCTAGTGTTTGATTCA, and GGATGGTATCTGAAACAACAATGGT (all custom synthesized by Integrated DNA Technologies, San Diego, CA). The separase siRNA sequence was GCTTGTGATGCCATCCTGA. Silencer negative control 1 siRNA (Applied Biosystems) was used as a control. Duplexes at a final concentration of 100 nM were used for transfection. Cells were analyzed at 48 to 72 h posttransfection, as indicated.
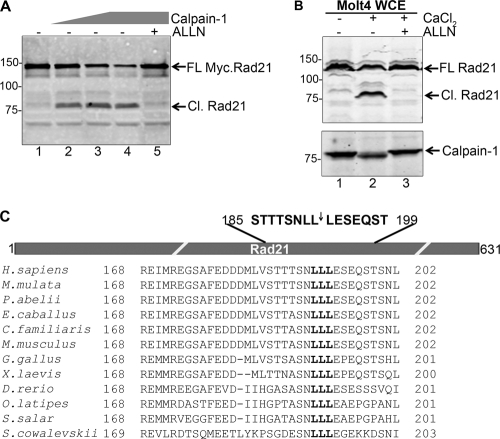
Fig. 1.
Rad21 is a substrate of calpain-1. (A) Myc.Rad21, synthesized in vitro using wheat germ extract, was incubated with 0, 0.2, 0.5, 1.25, and 1.25 mU of purified calpain-1 (lanes 1 to 5, respectively); ALLN was added to the last reaction mix at a 5 μM final concentration (lane 5). Full-length (FL) and cleaved (Cl.) Rad21 bands are indicated by arrows. (B) Molt4 cells were treated with CaCl2 (2 mM final concentration) for 5 h (lane 2) or additionally pretreated with 5 μM ALLN (lane 3). WCE was probed with the indicated antibodies. (C) (Top) Schematic of hRad21 and relative location of the calpain-1 cleavage site (not drawn to scale). The inverted arrow indicates the fissile bond. (Bottom) Phylogenetic sequence comparison of Rad21 sequences relative to the sequence of the human protein (amino acids [aa] 168 to 202). Numbers on the left of gels indicate molecular masses in kDa.
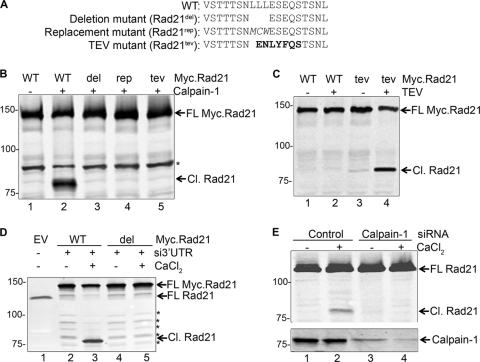
Fig. 2.
Cleavage of Rad21 by calpain-1. (A) Amino acid sequences flanking the calpain-cleavable site (relative to aa 184 to 202) in WT and mutant Rad21. Replacement residues and the TEV cleavage sequence are highlighted in italics and bold, respectively. (B) WT and mutant Myc.Rad21 proteins were synthesized in vitro and incubated with 1 mU calpain-1. A nonspecific band is marked with an asterisk. (C) WT and tev mutant Myc.Rad21 proteins were incubated with or without 0.5 U of TEV protease (TEV). (D) HEK293T cells were transfected with empty vector (EV) or a plasmid carrying WT or del mutant Myc.Rad21. Endogenous Rad21 was knocked down by siRNA duplexes targeting the hRad21 3′-UTR. At 2 days posttransfection, cells were treated with 10 mM CaCl2 for 6 h. (E) HEK293T cells were transfected with control or calpain-1-specific siRNA (duplex A) and treated with 10 mM CaCl2 for 6 h at 3 days posttransfection.
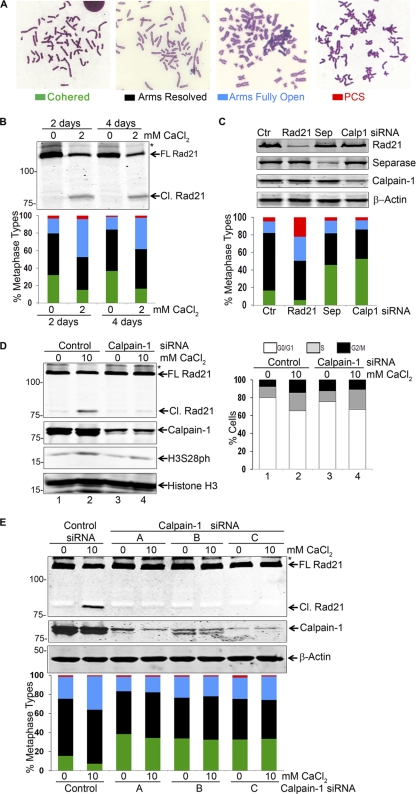
Fig. 6.
Calpain-1 affects sister chromatid separation. (A) Four major types of metaphase were taken into account. (B) Jurkat cells were treated with 2 mM CaCl2 for 2 and 4 days and then subjected to protein analysis (top) and metaphase spread analysis (bottom) along with untreated controls. Various metaphase types were scored. (C) HEK293T cells were transfected with control (Ctr) siRNA duplexes and with siRNAs targeting Rad21, separase (Sep), and calpain-1 (Calp1; duplex A). Cells were harvested at 72 h posttransfection. Protein (top) and metaphase spread (bottom) analyses are shown. (D) HeLa cells were transfected with control and calpain-1 (duplex A) siRNAs and were treated with 10 mM CaCl2 at 2 days posttransfection. Cells were analyzed for protein profiles by immunoblotting (left) and for DNA content by FACS (right). (E) HEK293T cells were transfected with either one of the three independent calpain-1 siRNAs (duplexes A, B, and C) or with control siRNA and were treated with CaCl2 at 48 h posttransfection. Protein profiles (top) and metaphase chromosome analysis (bottom) are shown. Nonspecific bands are marked with asterisks.
Calpain cleavage assay.
Calpain-1 was serially diluted in calpain buffer (CB; 50 mM Tris-HCl, 50 mM NaCl, 0.25 mM CaCl2, and 5% glycerol). One microliter of diluted calpain-1 (0.2 to 1.25 mU) was added to 2.5 μl Rad21 TnT product in a total volume of 20 μl; the volume was made up with CB. The reaction was carried out at 37°C for 30 min and was analyzed by immunoblotting as described below.
Immunoblot analysis.
Cells were harvested and washed in PBS. Whole-cell extract (WCE) was prepared by extracting cells with RIPA buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1.0% Igepal, 0.5% sodium deoxycholate, and 0.1% SDS supplemented with 1 mM β-mercaptoethanol, 1 mM PMSF, and 1× PI). Standard immunoblotting protocols were followed as described earlier (51). Total protein concentration was measured using the Bradford protein assay system (Bio-Rad). Typically, 30 μg of proteins was resolved by SDS-8% PAGE and transferred to nitrocellulose membranes. Bands were visualized in an Odyssey infrared imaging system (Li-Cor), using specific secondary antibodies (Li-Cor).
ChIP and qPCR.
Chromatin immunoprecipitation was performed as described previously (36), with some modifications adapted from the Affymetrix chromatin immunoprecipitation assay protocol (http://media.affymetrix.com/support/downloads/manuals/chromatin_immun_ChIP.pdf). Jurkat cells were grown in RPMI 1640 medium supplemented with 10% FBS to about 106 cells per ml and then treated with 2 mM CaCl2 for 12 h. Alternatively, HEK293T cells were transfected with Myc.Rad21 (WT or del mutant), and the endogenous Rad21 was knocked down by siRNA targeting the Rad21 3′-UTR. One hundred fifty milliliters of cells was then fixed with formaldehyde (1% final concentration) for 10 min at RT and quenched with 0.125 M glycine (final concentration) for 10 min on ice. Cell pellets were washed twice in ice-cold PBS and thrice in lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% Igepal) supplemented with 1 mM PMSF. Cell pellets were resuspended in 2.5 ml pre-IP dilution buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 1.5% Igepal, 1% SDS) supplemented with 4 mM PMSF and 1× PI and then were fragmented by use of a Sonic Dismembrator 550 (Fisher Scientific) for 6 min (20 s on and 30 s off; setting 3.5). The lysate was then digested by 125 U MNase for 15 min at RT and stopped by 10 mM (final concentration) EGTA on ice. The extent of sonication and MNase digestion was optimized such that the majority of chromatin was reduced to 2 to 4 nucleosomes. The material was then centrifuged at 20,000 × g to sediment cellular debris. The chromatin supernatant was recovered, 25 μl (1%) was saved for input control, and the rest was diluted to 12.5 ml with IP dilution buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl) supplemented with 1× PI and 1 mM PMSF. The diluted chromatin was then precleared with 50 μl protein A-agarose for 30 min at 4°C (IP06 [Calbiochem] preequilibrated with IP dilution buffer). The clarified chromatin was recovered, and 4 μg of specific antibody or control nonimmune IgG was added, mixed, and rotated overnight at 4°C for immunoprecipitation. The immunoprecipitates were recovered by binding to 50 μl protein A-agarose (equilibrated with IP dilution buffer) for 3 h. Anti-Myc-agarose beads (Sigma) were used in ChIP experiments for Myc.Rad21. The precipitates (beads) were recovered by centrifugation and transferred to a 1.5-ml Eppendorf tube. The beads were washed twice each with ChIP wash 1 (IP dilution buffer with 1 mM PMSF at 4°C; all subsequent washes were performed at RT), ChIP wash 2 (20 mM Tris-HCl, pH 8, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 500 mM NaCl, 1 mM PMSF), ChIP wash 3 (10 mM Tris-HCl, pH 8, 1 mM EDTA, 0.25 M LiCl, 0.5% Igepal, 0.5% sodium deoxycholate), and TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). The bound chromatin was eluted from the beads twice with 100 μl of elution buffer (25 mM Tris-HCl, pH 7.5, 10 mM EDTA, 0.5% SDS) at 65°C for 30 min each. The input DNA saved earlier and the eluates were pooled and digested overnight with 20 μl proteinase K, and the DNA was purified using a PCR cleanup kit (Qiagen). The purified DNA was used for quantitative real-time PCR (qPCR) analysis of each locus in triplicate, using FastStart Universal SYBR green master mix (Roche) in an Eppendorf Realplex Mastercycler. Primer sequences were adopted from the work of Wendt et al. (47). Primers for nine cohesin-positive loci (cohesin binding sites [CBS]) and three cohesin-negative loci (cohesin-free sites [CFS]) were selected based on sequence specificity and PCR efficiency. Data analysis was performed as described previously (20), and % input for the ChIP fractions was calculated by linear conversion of the normalized ChIP ΔCT values. Data from at least three biological repeat experiments were analyzed.
Chromosome analysis and cell cycle.
Cells were harvested and swelled in 75 mM KCl for 15 min before being fixed with methanol-acetic acid (3:1 [vol/vol]) at 4°C. Cell pellets were washed in methanol-acetic acid at least thrice and were dropped onto slides and air dried. Chromosomes were stained with Giemsa stain as described previously (37). For each experimental set, about 100 metaphase cells were counted at a magnification of ×63. Those cells for which almost all chromosomes conferred a certain phenotype were counted. In those few instances where the chromosomes of a cell appeared to represent more than one phenotype, the cell was assigned the phenotype that was represented by >75% of the chromosomes. For cell cycle analysis by fluorescence-activated cell sorting (FACS), cells were fixed in 70% ethanol (vol/vol), treated with RNase A, washed in PBS, and stained with propidium iodide (50 μg/ml in PBS). The stained cells were analyzed by a Becton Dickinson FACScan flow cytometer.
Immunofluorescence analysis.
Immunofluorescence microscopy was performed as described previously (51). Briefly, HeLa cells were detached from plates by being treated with trypsin-EDTA and were spun onto slides. Cells were fixed with cold methanol before being permeated with 1% Triton X-100. Alternatively, cells were extracted with 1% Triton X-100 prior to fixing with methanol. The samples were blocked in 1% bovine serum albumin (BSA) and incubated with antibodies. The slides were mounted using mounting medium (Vectashield; Vector Laboratories) containing DAPI (4′,6-diamidino-2-phenylindole). Images were obtained with a microscope (E800; Nikon) equipped with Quips imaging software (Applied Imaging) and a ×100/1.4 objective lens (Nikon) at RT.
Protein identification and N-terminal sequencing.
Nuclear extract was prepared from Jurkat cells and was bound to anti-Myc-agarose (mock) or Myc-tagged human Rad21 (Myc.Rad21)–anti-Myc-agarose (Rad21 matrix) beads in a buffer containing 20 mM Tris-HCl, pH 7.5, 5% glycerol, 0.15 M NaCl, 0.5% Triton X-100, 1 mM MgCl2, 1 mM EDTA, 1 mM PMSF, and 1× PI. Unbound proteins were washed in the same buffer four times, and the bound proteins were eluted by boiling the beads in SDS sample buffer. The proteins were resolved by SDS-PAGE, and the gel slices covering the entire length were submitted to Nextgen Sciences (Ann Arbor, MI) for mass spectrometric identification of Rad21-interacting proteins. WCE was prepared from CaCl2-treated Jurkat cells, Rad21 was immunoprecipitated, transferred to a polyvinylidene difluoride (PVDF) membrane, and stained, and full-length (FL) and cleaved Rad21 bands were excised and subjected to N-terminal sequencing as described previously (31). N-terminal sequencing of the cleaved Rad21 fragment was done by the Proteomics Core Facility, Baylor College of Medicine, Houston, TX.
RESULTS
Rad21 is a calpain-1 substrate.
To obtain better insight into the roles of cohesin in various cellular processes, we sought to identify cohesin-interacting proteins. We generated Myc-tagged human Rad21 (Myc.Rad21) by in vitro transcription-coupled translation and developed a Rad21 affinity matrix by immobilizing Myc.Rad21 on anti-Myc-agarose beads. Soluble proteins extracted from the nuclei of Molt4 human acute lymphoblastic leukemia cells were bound to the Rad21 affinity matrix. The bound proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). This analysis identified 96 proteins that specifically bound to the Rad21 matrix (unpublished data). Among these, we were particularly interested in the calcium-dependent cysteine proteinase calpain-1, for which two peptides were identified (DFFLANASR and LYELIITR). We considered this finding important since Rad21 is a known substrate of proteases: it is cleaved by separase at the metaphase-to-anaphase transition (16) and is also cleaved by an unknown caspase-like protease during apoptosis (7, 31).
To verify that Rad21 is indeed a calpain-1 substrate, we procured purified calpain-1 and incubated it with Myc.Rad21 to assay Rad21 cleavage. We performed immunoblot analysis using an antibody that recognizes the C terminus of human Rad21 (hRad21). Interestingly, calpain-1 cleaved Myc.Rad21 in vitro to generate an 80-kDa C-terminal fragment in a reaction that was inhibited by the known calpain inhibitor ALLN (Fig. 1A). Since calpain-1 activation is dependent on Ca2+, we reasoned that treating cultured cells with CaCl2 would lead to an increased influx of Ca2+ into the cells and a consequent activation of calpain-1. Therefore, to assay if calpain-1 can also cleave Rad21 ex vivo, we treated Molt4 cells with 2 mM CaCl2, prepared the WCE after 6 h, and performed an immunoblot analysis. As seen before (Fig. 1A), we observed the emergence of a similar 80-kDa fragment of Rad21 upon CaCl2 treatment (Fig. 1B, lane 2). The 80-kDa fragment did not appear when Molt4 cells were treated with ALLN 30 min prior to the addition of CaCl2 (compare lane 2 to lane 3), indicating that the cleavage of Rad21 is calpain-1 specific. To verify that the calcium-induced cleavage of Rad21 is not specific to Molt4 cells only, we treated cells from several other human cell lines with CaCl2. We observed similar 80-kDa Rad21 fragments in an array of cell lines tested, including Jurkat, SupB15, KMH2, U937, Nalm6, Reh, HL60, HeLa, HEK293T, and U2OS cells, representing both solid tumors and hematological malignancies (data not shown).
The 80-kDa fragment from CaCl2-treated Molt4 cells was then immunopurified using anti-Rad21 antibody, and the sequence of its N terminus was determined by Edman degradation. This analysis revealed the N-terminal sequence of the 80-kDa fragment to be LESEQSTSNL, which is identical to the sequence for residues 193 to 202 of human Rad21, mapping the cleavage site to L192 (Fig. 1C). The cleavage site conforms to the calpain-1 consensus cleavage sequence determined by MEROPS (http://merops.sanger.ac.uk/cgi-bin/pepsum?id=C02.001) (40) and is conserved in Rad21 homologs across the chordates (Fig. 1C). The above studies establish Rad21 as a novel substrate of calpain-1.
Calpain-1 is responsible for Ca2+-induced cleavage of Rad21.
To further characterize the calpain-1-mediated Rad21 cleavage, we generated three Rad21 mutants by site-directed mutagenesis. Bioinformatic analysis, as presented in MEROPS, indicated that leucine residues enjoy important statistical significance in a calpain-1-cleavable site. Since the three leucine residues are also conserved in chordates, we generated two mutants of Rad21 by either deleting these residues (del mutant; Rad21del) or replacing the three leucines with a Met-Cys-Trp sequence, which is not found at positions P2, P1, and P1′ in known calpain-1-cleavable sequences (rep mutant; Rad21rep). Furthermore, we also removed the three leucines and introduced a tobacco etch virus (TEV) protease-cleavable site, generating a tev mutant (Rad21tev) (see Fig. 2A for relevant sequences). While calpain-1 readily cleaved WT Rad21 as described before (Fig. 2B, lane 2), it failed to cleave the del, rep, and tev mutants (lanes 3 to 5). Likewise, the TEV protease efficiently cleaved the tev mutant (Rad21tev) but failed to cleave WT Rad21 (Fig. 2C, compare lanes 2 to 4). To examine if the identified cleavage site is indeed utilized ex vivo, we transfected HEK293T cells with constructs encoding WT or del mutant Myc.Rad21 and treated the cells with CaCl2. We then knocked down the endogenous Rad21 by using siRNA duplexes targeting the 3′-UTR of Rad21 in these cells to avoid the visual interference of the cleavage product from endogenous Rad21. As expected, cells transiently expressing WT Myc.Rad21 underwent Rad21 cleavage upon calcium treatment (Fig. 2D, lane 3), whereas no Rad21 cleavage was observed in cells transiently expressing Rad21del (compare lanes 3 to 5). These studies directly implicate calpain-1 in Ca2+-induced cleavage of Rad21.
To verify that calpain-1 is indeed responsible for calcium-induced cleavage of Rad21 ex vivo, we sought to deplete cells of calpain-1. We transfected HEK293T cells with control as well as calpain-1-specific siRNA duplexes. At 48 h posttransfection, the cells were treated with 10 mM CaCl2 for a further 6 h, and the WCE was analyzed for Rad21 cleavage. We were able to achieve significant knockdown of endogenous calpain-1. Calcium treatment of the cells transfected with control siRNA generated the expected cleavage product of Rad21 but failed to do so in cells depleted of calpain-1 (Fig. 2E, compare lane 2 to lane 4). These observations reveal that endogenous calpain-1 cleaves Rad21 upon stimulation by Ca2+.
Calcium-induced Rad21 cleavage is not associated with apoptosis.
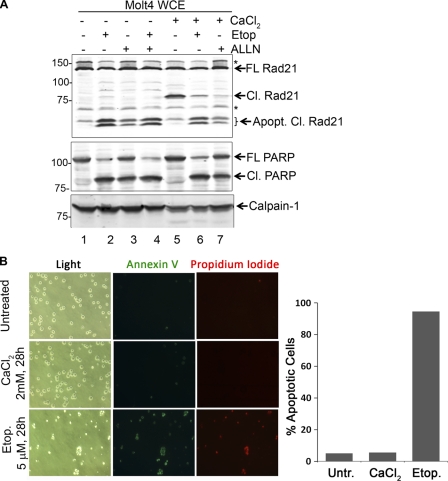
Since Rad21 is known to be cleaved during apoptosis (31) and both Ca2+ and calpains play potential roles in apoptosis (29), we first examined if calcium treatment of cells and consequent cleavage of Rad21 are associated with apoptosis. We treated Molt4 cells with etoposide to induce apoptosis and compared them to calcium-treated Molt4 cells. As a marker of apoptosis induction, we looked for cleavage of poly(ADP-ribose) polymerase (PARP). Additionally, emergence of two apoptotic cleavage products of Rad21 (∼60 and 64 kDa) also served as an internal marker of apoptosis. Upon treatment with etoposide, PARP in Molt4 cells underwent prominent proteolytic cleavage, indicating the onset of apoptosis (Fig. 3A, compare lanes 1 and 2), and this accompanied the apoptotic cleavage of Rad21 (Apopt. Cl. Rad21 bands). However, calcium treatment failed to elicit PARP cleavage, and interestingly, it led to significant cleavage of Rad21 to a C-terminal 80-kDa fragment (Cl. Rad21 band) but not to the signature apoptotic cleavage products (Fig. 3A, lane 5). We also treated Molt4 cells with CaCl2 (2 mM final concentration) for 28 h, collected samples every 4 h, and stained the cells with annexin V (for apoptosis) and propidium iodide (for necrosis and advanced cell death). As a positive control for apoptosis, we treated Molt4 cells with 5 μM etoposide and sampled the cells alongside the untreated and CaCl2-treated cells. We failed to observe any increase in apoptotic or necrotic cells upon calcium treatment for up to 28 h, while about 95% of cells turned apoptotic upon treatment with etoposide (Fig. 3B). We concluded that calcium-induced Rad21 cleavage is not associated with programmed cell death.
Fig. 3.
Calcium-induced Rad21 cleavage is not associated with apoptosis or necrosis. (A) Molt4 cells were pretreated with etoposide (Etop; 20 μM final concentration) or ALLN (5 μM final concentration) for 30 min before addition of CaCl2 (2 mM final concentration). Cells were harvested after 6 h, and WCE was prepared. Note that the calcium-induced cleavage product of Rad21 (Cl. Rad21) was different from the apoptotic cleavage products (Apopt. Cl. Rad21). Nonspecific bands are marked with asterisks. (B) Molt4 cells were treated with 2 mM CaCl2 or 5 μM etoposide for 28 h, stained with annexin V-FITC and propidium iodide, and observed using light as well as fluorescence microscopy. The graph on the right shows the number of apoptotic cells as a percentage of the total number of cells.
Calpain-1 localizes to the nucleus and to chromatin.
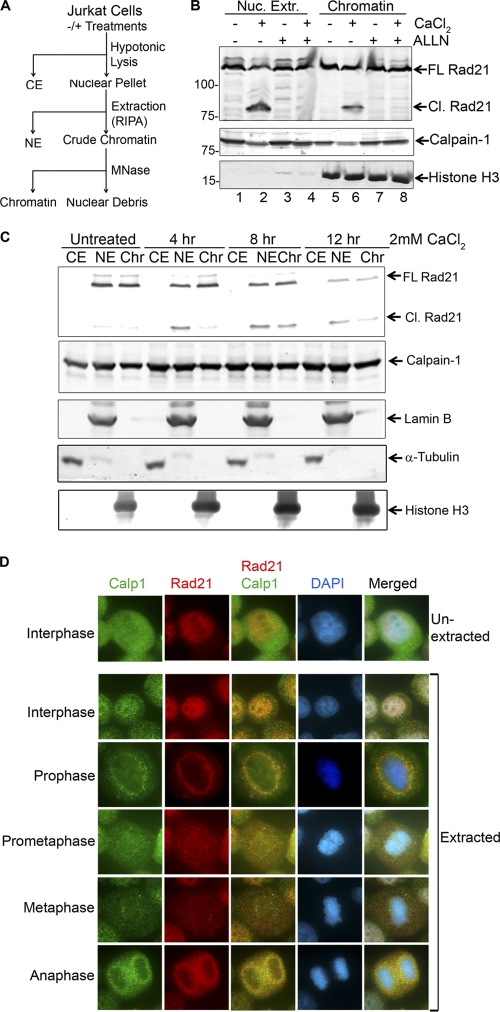
We reasoned that if calpain-1 calpain-1-mediated cleavage of Rad21 is not associated with cell death, it may have a role in the dissolution of sister chromatid cohesion. Calpain-1 is traditionally believed to be a cytosolic protease (11, 26). Intact calpain-1 or its truncation mutants reportedly localize to the nucleus only when they are overexpressed (10). However, subcellular localization of calpain-1 has not been studied systematically. Therefore, we wanted to examine if calpain-1 naturally localizes to the nucleus and if chromatin-bound Rad21 is cleaved upon calcium treatment. We treated Jurkat cells with 2 mM CaCl2 and/or ALLN and fractionated the cells into cytosolic extract (CE), nuclear extract (NE), and chromatin fractions as shown in Fig. 4A. The NE and chromatin fractions were analyzed by immunoblotting for the presence of cleaved Rad21. In addition to the nuclear extract, interestingly, we also found a substantial amount of cleaved Rad21 in the chromatin fraction upon treatment with CaCl2 (Fig. 4B, lanes 2 and 6). We also found calpain-1 present in the chromatin fraction. To better understand the calcium-induced Rad21 cleavage, we treated Jurkat cells with 2 mM CaCl2 for 4, 8, and 12 h and fractionated the cells into CE, NE, and chromatin fractions (Fig. 4C). At any given time in an asynchronous population of human cells, nearly half of all Rad21 is present in a soluble pool in the nucleus, with the rest remaining chromatin bound. Cleavage of the soluble Rad21 was observed within 4 h of treatment with CaCl2, whereas cleaved Rad21 appeared on chromatin by 8 h. It is possible that when it is activated, calpain-1 cleaves soluble nuclear Rad21 first, before acting upon the chromatin-bound Rad21 (Fig. 4C). As an additional investigation of calpain-1 localization in the cell, we conducted immunofluorescence staining of HeLa cells. Consistent with its reputation of being a cytoplasmic protein (11), we observed the majority of calpain-1 in the cytoplasm (Fig. 4D, top panel). To verify if calpain-1 also localizes to chromatin and/or the nucleus, we extracted HeLa cells with Triton X-100 before fixing them with ice-cold methanol prior to immunostaining (Fig. 4D, bottom panel). Interestingly, we observed calpain-1 localized to chromatin in interphase cells and to perichromatic territory in later stages of the cell cycle (e.g., prophase, metaphase, and anaphase). Importantly, we observed colocalization of calpain-1 with Rad21 but did not observe any alteration in its localization in response to calcium treatment (data not shown). From these studies, we inferred that calpain-1 also localizes to chromatin and cleaves chromatin-bound Rad21 when activated by an increase in nuclear Ca2+ concentration ([Ca2+]).
Fig. 4.
Calpain-1 localizes to chromatin and cleaves chromatin-bound Rad21. (A) Scheme of cell fractionation. Cells were pretreated with ALLN (final concentration, 5 μM) for 30 min before the addition of CaCl2 (2 mM) for 10 h. (B) Nuclear extract (Nuc. Extr.) and chromatin fractions were resolved and probed with the indicated antibodies. (C) Fractionation of Rad21 cleavage upon calcium treatment. Jurkat cells, treated in the presence or absence of 2 mM CaCl2 for the indicated times (hours), were fractionated into cytosolic extract (CE), nuclear extract (NE), and chromatin (Chr) fractions. Proteins of equal volumetric proportions were analyzed by immunoblotting. Full-length (FL) and cleaved (Cl.) Rad21 proteins are indicated. α-Tubulin, lamin B, and histone H3 serve as markers for cell fractionation. (D) Immunofluorescence staining of HeLa cells by use of the indicated antibodies before (top) and after (bottom) extraction with Triton X-100.
Cleavage by calpain-1 causes loss of Rad21 from chromatin.
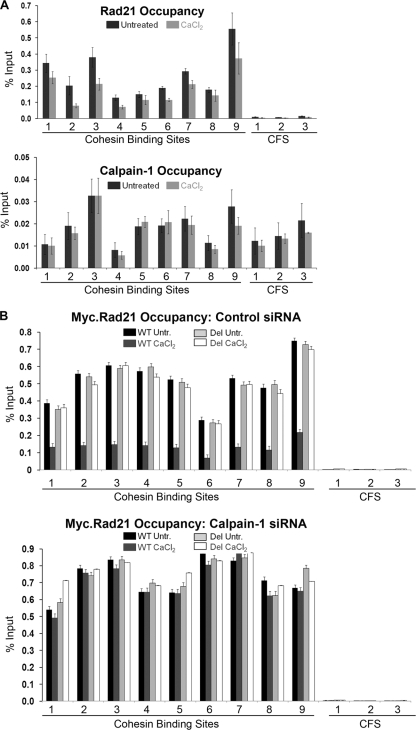
Since calpain-1 localizes to and cleaves Rad21 on chromatin, we reasoned that calpain-1-mediated Rad21 cleavage may affect cohesin occupancy on the chromatin. To address this possibility, we performed ChIP assays with Rad21 from Jurkat cells and looked at Rad21 occupancy at several cohesin-bound sites (CBS) by qPCR. We selected 9 CBS and 3 cohesin-free sites (CFS), across chromosomes 5, 6, 7, 9, 11, 15, 21, and 22, from a previous report of genomic cohesin occupancy in human cells (47) (Table 1). As reported before (47), we detected high levels of Rad21 at each of the nine CBS but very little Rad21 at the three CFS (Fig. 5A, top panel). Interestingly, the level of Rad21 occupancy at all CBS examined decreased following a 12-h treatment with 2 mM CaCl2 (P < 0.05 for each CBS, based on paired Student's t test analysis of three independent experiments). This reduction of Rad21 occupancy was coincident with the appearance of cleaved Rad21 in chromatin fractions, as shown in Fig. 4. We also performed ChIP with calpain-1 and detected low levels of calpain-1 at both the CBS and CFS (Fig. 5A, bottom panel). We did not detect any significant CaCl2-dependent change in calpain-1 occupancy. It is possible that a low level of calpain-1 is always present on the chromatin and that it cleaves Rad21 when activated by an increase in nuclear [Ca2+].
Table 1.
Primers used in this studya
| Locus | Primer sequence (5′ → 3′) | Coordinates (chromosome: nucleotide positions) |
|---|---|---|
| CBS1 | AGTACACAGAGCGTAGCCTCC | Chr5: 131,890,348–131,890,535 |
| CCTCTAGTGGCTGATGCAGG | ||
| CBS2 | CTCACTTCAGGTGGCAGAGG | Chr5: 141,908,579–141,908,800 |
| ATGGTCCAGTGTGGCATTC | ||
| CBS3 | CAGCTCTGTGTCCTGTCTTATCC | Chr6: 132,642,584–132,642,808 |
| CAGCTATAATTGATGAAGAGGCG | ||
| CBS4 | CCCATCTTGCTGACCTCAC | Chr11: 1,980,758–1,980,922 |
| AGACCTGGGACGTTTCTGTG | ||
| CBS5 | GGCAGTTCCAGCACATCC | Chr15: 41,983,359–41,983,571 |
| GAATGGCGACACCGAAC | ||
| CBS6 | GTGATCGGTCCAGTGCATAG | Chr16: 55,716–55,937 |
| CTGGCATGTCATGGTAGAGC | ||
| CBS7 | TGAAGCCACCACCTGCTTAG | Chr21: 34,218,281–34,218,465 |
| TCTTGCATGTGAGCGAGAAC | ||
| CBS8 | GGAGACACCATTCTGGTACGAC | Chr22: 30,696,211–30,696,409 |
| GAGCGAGCAGCTGAGGAC | ||
| CBS9 | ACATGTGGCCAGCTCAGG | Chr22: 31,522,929–31,523,153 |
| GGCGCTATAAGCCAGAGAAC | ||
| CFS1 | CCACTGCTCAGCCTTAGAGGAA | Chr7: 27,159,330–27,159,485 |
| GTTTGTCTGGTTTTTGCGTGTG | ||
| CFS2 | TGCCATGCGTTGAAAATATCC | Chr7: 27,213,317–27,213,491 |
| TGCTTTCTGAAGTTGCCAAGC | ||
| CFS3 | GCTCTGCTGACTGGCACC | Chr9: 128,979,088–128,979,307 |
| CTCACCTTGAAGGAGCAATAAG |
Sequences and coordinates are as described by Wendt et al. (47).
Fig. 5.
Calpain-1 activation affects Rad21 occupancy. (A) Chromatin occupancy of Rad21 and calpain-1 in Jurkat cells. Rad21 and calpain-1 occupancy is shown for nine cohesin binding sites and three cohesin-free sites (CFS). (B) Chromatin occupancy of Myc-tagged WT and del mutant Rad21 in HEK293T cells transfected with control (top) or calpain-1 (duplex A) (bottom) siRNA. Average threshold cycle (CT) values for each primer pair for three independent biological replicates (each run in triplicate) were analyzed. Error bars represent standard errors of the means.
To better understand the role of calcium-induced Rad21 cleavage in cohesin occupancy, we transiently expressed Myc-tagged WT and del mutant Rad21 in 293T cells, knocked down the endogenous Rad21 by use of an siRNA targeting its 3′-UTR (as shown in Fig. 2D), and treated the cells with CaCl2 for 12 h. We performed Rad21 ChIP by using anti-Myc-agarose beads. The ChIP-qPCR analysis revealed a 3- to 4-fold reduction in WT Myc.Rad21 occupancy upon CaCl2 treatment, whereas no such reduction was observed for the del mutant (Fig. 5B, top panel). When the same experiment was conducted with calpain-1-depleted cells, no calcium-induced loss of Myc.Rad21 was observed (Fig. 5B, bottom panel). These observations reveal that calpain-1 cleaves Rad21 and causes a loss of cohesin from chromatin.
Calcium treatment of cells results in increased sister chromatid separation.
After verifying both the presence of calpain-1 on chromatin and a decreased level of Rad21 at several genomic loci upon CaCl2 treatment, we sought to address the possibility that calcium-induced cleavage of Rad21 might impair sister chromatid cohesion. We cultured Jurkat cells in the presence of 2 mM CaCl2 for 2 and 4 days and prepared metaphase spreads. While analyzing the cells, we classified the metaphase spreads into four categories according to the following major phenotypes of chromosomes: (i) cohered, where the two sister chromatids are perfectly together with no discernible separation; (ii) arms resolved, where the sisters are separated by a thin hairline separation along the arms but are held together at the centromere (railroad phenotype); (iii) arms fully open, where sister arms are fully separate but held at centromeres (X phenotype); and (iv) precocious sister chromatid separation (PCS), where the majority of sisters are not only completely separated but scattered randomly, with no evident pairing. Representatives of these chromosome types are depicted in Fig. 6A. We observed that the number of cohered sister chromatids decreased nearly 2-fold upon calcium treatment, with a concomitant 2-fold increase in sister chromatids with fully open arms (Fig. 6B), indicating a loss of arm cohesion between the sisters.
To further validate the above observations, we sought to deplete calpain-1 in cells by using siRNA and to conduct chromosome spread analysis. In this analysis, we also included the depletion of Rad21 and separase as positive controls effecting sister chromatid cohesion. As shown in Fig. 6C, we achieved considerable depletion of the target proteins. Depletion of separase led to a considerable increase in the number of cohered sister chromatids. This observation is consistent with a report that in addition to centromeric cohesin cleavage, separase is also required for complete removal of cohesin from chromosome arms (27). Conversely, and as anticipated, depletion of Rad21 led to a decrease in cohered chromosomes and a significant increase in the number of sister chromatids with resolved arms as well as in the number of PCS events. Interestingly, the metaphase spread analysis revealed a nearly 3-fold increase in the number of cohered chromosomes upon calpain-1 depletion, with a concomitant decrease in the number of sister chromatids with resolved arms (Fig. 6C, bottom panel). This observation complements the results obtained by treating Jurkat cells with CaCl2 (Fig. 6B), suggesting that calpain-1 promotes dissolution of arm cohesion.
To verify if calcium treatment and/or calpain-1 depletion somehow arrested the cells at a certain stage so as to accentuate the chromosomal phenotype observed, we depleted calpain-1 from HeLa cells, treated the cells with CaCl2 for 12 h, and analyzed the cell cycle profile by FACS (Fig. 6D). We observed a slight increase (7%) in the number of G2/M cells upon calcium treatment, but no such effect was observed when calpain-1 was depleted. Interestingly, the level of phosphorylated histone H3 (at Ser 28 [H3S28ph]), a known mitosis marker, appeared to increase slightly upon calcium treatment, corroborating the FACS analysis. To rule out any off-target effect of calpain-1 depletion by the calpain-1 siRNA (duplex A), we treated HEK293T cells with two additional independent calpain-1 siRNAs (duplex B and duplex C) (Fig. 6E) and with CaCl2 and then analyzed the protein and metaphase profiles. As shown in Fig. 6E, the effects of calpain-1 depletion by all three siRNA duplexes were similar, indicating the specificity of the calpain-1 phenotype.
Role of calpain-1-induced cleavage of Rad21 in mitosis.
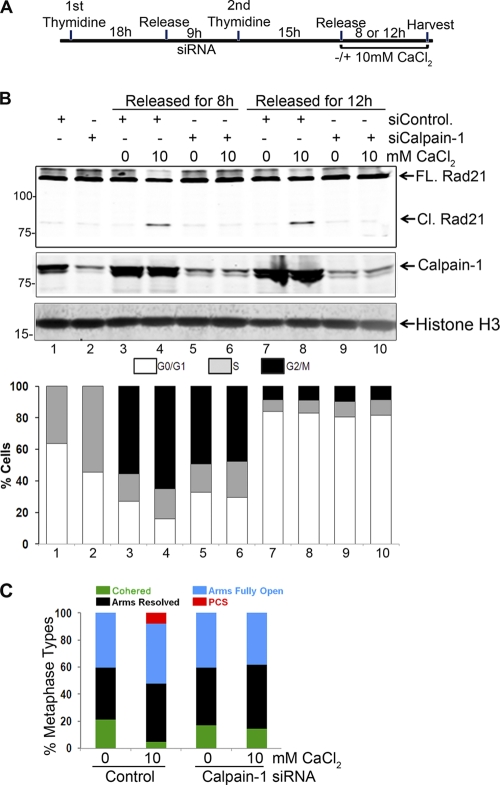
To extend our investigation of calpain-1-mediated Rad21 cleavage and its role in the dissolution of sister chromatid cohesion during mitosis, we synchronized HeLa cells by double thymidine blocking and transfected the cells with control or calpain-1 siRNA duplexes after the first thymidine block (see schematic in Fig. 7A). We released the cells from the second thymidine block for 8 h or 12 h in the presence or absence of additional 10 mM CaCl2 and then examined the chromosome features and protein and cell cycle profiles. FACS analysis revealed that over 55% of cells were in the G2/M stage 8 h after release from the double thymidine block (Fig. 7B, bottom panel). Interestingly, with CaCl2 treatment, about 10% more (for a total of 65%) cells were found in G2/M (4n DNA content) at 8 h postrelease (Fig. 7B, bottom panel, compare bars 3 and 4). Upon calpain-1 depletion, however, no such effect of additional CaCl2 was observed (compare bars 3 and 4 to bars 5 and 6), indicating that calpain-1 was required for calcium-induced enrichment of cells at metaphase. This enrichment was unlikely to be a case of metaphase arrest, since all cells exited mitosis by 12 h (bars 7 to 10), with the majority of cells having a 2n DNA content (G0/G1). Also, no effect of calcium treatment and/or calpain-1 depletion was observed on mitotic exit.
Fig. 7.
(A) Schematic of synchronization and release of HeLa cells following double thymidine block. (B) HeLa cells were synchronized by double thymidine arrest, transfected with calpain-1 siRNA duplex A after the first thymidine block, and finally released for 8 h with or without an additional 10 mM CaCl2. Protein profiles (for whole-cell lysates) (top) and percentages of cells as analyzed by FACS (bottom) are shown. Histone H3 is shown as a loading control. Lanes 1 and 2 represent cells arrested at the G1/S transition. (C) Metaphase spread analysis of cells harvested 8 h after release from the double thymidine block. Cells from a single 10-cm plate were split equally and processed for protein, FACS, and metaphase spread analyses.
As shown in Fig. 7C, release of cells from the G1/S block in the presence of an additional 10 mM CaCl2 caused a significant decrease in the number of cohered chromosomes due to an enhanced loss of arm cohesion. We did not observe a similar effect of CaCl2 when the cells were depleted of calpain-1. It is possible that the kinetics of resolution of chromosomal arms is linked to progression of cells to metaphase.
Engineered cleavage of Rad21 at the calpain-1-cleavable site can resolve sister chromatid cohesion.
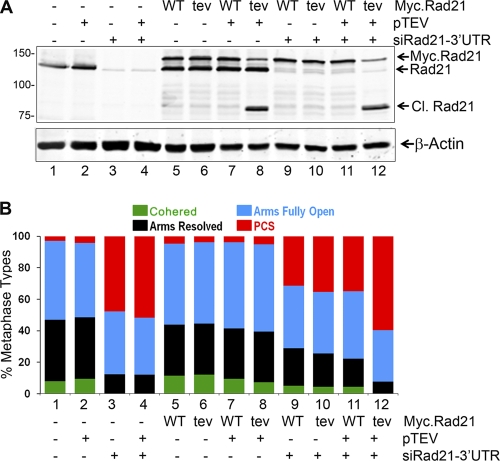
The only proteolytic cleavage of Rad21 known thus far to dissolve sister chromatid cohesion is caused by separase, which cleaves human Rad21 at R172 and R450 (16). Our results described above reveal the cleavage of Rad21 at L192 by calpain-1 as a novel proteolytic event to promote sister chromatid separation. To ascertain the role of this cleavage in separation of sister chromatids, we sought to engineer cleavage of Rad21 without calpain-1 activation. We transiently expressed WT Myc.Rad21 and the Myc.Rad21tev mutant protein in HEK293T cells and verified that the ectopically expressed WT and mutant Myc.Rad21 proteins associated with other cohesin components and arguably formed active cohesin complexes (data not shown). We then selectively knocked down the endogenous Rad21 by using siRad21-3′-UTR and obtained a population of cells where almost all of the Rad21 present was the ectopically expressed Myc.Rad21 protein (Fig. 8A, lanes 9 and 10, for WT and tev mutant proteins, respectively). Transient expression of the TEV protease in cells expressing the tev mutant, but not in those expressing WT Myc.Rad21, resulted in substantial cleavage of Myc.Rad21tev, generating the predominant 80-kDa C-terminal fragment (Fig. 8A, compare lanes 11 and 12 to lanes 7 and 8, where endogenous Rad21 is unaffected). When the metaphase chromosomes from the transfected cells were analyzed, we observed a slight increase in cohered chromosomes upon transient overexpression of both Myc.Rad21 and Myc.Rad21tev, and the concomitant expression of TEV protease largely had no effect (Fig. 8B). Depletion of endogenous Rad21 by siRad21-3′-UTR had a dramatic effect in cells, with the entire sister pairs falling apart (Fig. 8B, compare bars 3 and 4 to bars 1 and 2), and this phenotype was partially rescued by expressing ectopic Rad21 (bars 9 and 10). However, expressing the TEV protease in cells where the sole source of Rad21 was the ectopic tev mutant led to complete separation of the sisters, resembling the chromosome features of cells depleted of Rad21 altogether (compare bar 12 to bars 3 and 4). It is important that such complete separation of sisters did not occur when WT Myc.Rad21 was expressed instead, along with TEV protease and depletion of endogenous Rad21 (compare bars 11 and 12). This observation lends unambiguous support to the previous results showing that cleavage of Rad21 at L192 by calpain-1 promotes sister chromatid separation. These observations also suggest that cleavage of Rad21 at sites other than those cleaved by separase can lead to dissolution of sister chromatid cohesion.
Fig. 8.
HEK293T cells were first transfected with a plasmid for WT Myc.Rad21 (WT) or Myc.Rad21tev (tev) along with pCS2MT-TEV (pTEV), as indicated. Twenty-four hours later, cells were transfected with siRNA duplexes targeting the 3′-UTR of Rad21. Cells were grown for a further 48 h before harvesting. Protein profile (A) and metaphase spread (B) analyses are shown.
DISCUSSION
Calpain-1 is a novel cohesin proteinase.
In an ongoing effort to understand diverse functions of cohesin, we sought to identify protein interactors of Rad21 by using a Rad21 affinity matrix, which resulted in the identification of the calcium-dependent cysteine proteinase calpain-1. This prompted us to speculate that Rad21 is a substrate of calpain-1. Our results demonstrate that calpain-1 proteolytically cleaves Rad21 at residue L192 in vitro, and also ex vivo in several human cell lines, to generate a precise C-terminal fragment of about 80 kDa. Rad21 is a known substrate of proteases: it is cleaved at the metaphase-to-anaphase transition by separase, at R172 and R450 (16), and also is cleaved by an unknown caspase-like proteinase during apoptosis, at D279 (7, 31). The current work has uncovered a novel cleavage site, L192, on Rad21 and establishes calpain-1 as a new cohesin-proteinase. We found that calpain-1-mediated cleavage at L192 was not associated with cell death (Fig. 3), which prompted us to think that calpain-1-mediated cleavage of Rad21 may be involved in the regulation of sister chromatid cohesion. This possibility assumes significance when the Rad21 sequence flanking the calpain-1 cleavage site is considered. The cleavage site lies within three consecutive leucine residues housed in a short motif (SNLLLE) that is conserved from hemichordates (represented by Saccoglossus kowalevskii) to vertebrates but not found in Rad21 homologs from invertebrates, indicating that the chordates acquired this motif in Rad21 early in evolution. Arguably, calpain-1-mediated cleavage of Rad21 would resolve sister chromatid cohesion, and to do so, calpain-1 has to localize to the nucleus or chromatin and its activity needs to be regulated. Three independent approaches—biochemical fractionation, immunolocalization, and ChIP—demonstrated that calpain-1 localizes to the nucleus. Immunolocalization and ChIP experiments also revealed that calpain-1 colocalizes with chromatin-bound Rad21, supporting a role in chromosomal cohesion. While the levels of calpain-1 on chromatin remained unaffected, Rad21 occupancy decreased at all tested Rad21-positive loci upon calcium treatment. These observations suggest that calpain-1 normally localizes to the chromatin and cleaves chromatin-bound Rad21 when activated by an increased nuclear [Ca2+].
Calpain-1 regulates sister chromatid cohesion.
When we analyzed the chromosome spread experiments, we observed a loss of arm cohesion between sister chromatids in multiple human cell lines treated with CaCl2, and this coincided with significant cleavage of Rad21. When calpain-1 was knocked down, we observed the reverse: an increase in cohered chromosomes and a concomitant decrease in chromosomes with uncohered arms were seen. Release of HeLa cells from G1/S arrest in the presence of additional CaCl2 (and associated cleavage of Rad21) showed a marked reduction in the number of cohered chromatids, whereas no significant effect of additional CaCl2 was noticed when calpain-1 was depleted from the cells. Interestingly, releasing cells from G1/S block in the presence of additional CaCl2 led to an about 10% increase in the G2/M cell population compared to that for untreated controls. However, no such effect was observed when calpain-1 was depleted, indicating a role of calcium-activated calpain-1 in mitosis. Furthermore, we also showed that engineered cleavage of Rad21, which mimics calpain-1 cleavage, can result in a dramatic loss of sister chromatid cohesion. This study argues against a possible role of calcium treatment—other than Rad21 cleavage—in mediating the dissolution of sister chromatid cohesion. Collectively, the observations presented here prompted us to suggest that calpain-1 has a previously unsuspected physiological function in mediating sister chromatid resolution.
So what causes calpain-1 activation in the nucleus? Our experiments rely upon supplementing the cell culture medium with additional CaCl2. The culture medium contains a battery of growth factors, some of which are regulators of calcium transport. Thus, an increase in extracellular [Ca2+] is expected to increase the intracellular [Ca2+] (4). Increased cytoplasmic Ca2+ is likely to diffuse into the nucleus, resulting in increased nuclear [Ca2+] (3, 6). This can consequently lead to activation of nuclear calpain-1. However, the normal physiological circumstances under which the cell experiences an increased nuclear [Ca2+], consequent calpain-1 activation, and Rad21 cleavage await elucidation. Importantly, nuclear Ca2+ is reportedly essential for cell cycle passage through prophase (33), the stage when cohesion along chromosomal arms is resolved. It is notable that a transient increase in intracellular [Ca2+] in anaphase has been implicated in sister chromatid separation in sea urchin embryos (12), suggesting that calcium can play crucial roles at distinct stages during the cell cycle. It is also interesting that calpain-2, the other ubiquitous calpain in mammalian cells, was recently reported to be required for sister chromatid cohesion in mouse cells (24). While both calpain-1 and calpain-2 share the same regulatory subunit, CAPNS1, it is possible that they undertake very different sets of actions. Collectively, the above observations clearly argue for a role of nuclear calcium signaling in the regulation of sister chromatid cohesion and separation.
We previously proposed a two-ring handcuff model for cohesin (51). If both Rad21 molecules in the handcuff are cleaved by calpain-1, the entire cohesin complex will unravel, come off the chromatin, and result in sister chromatid separation (Fig. 9). Our results demonstrate that calcium-induced cleavage of Rad21 is modest, but it leads to significant separation of sister arms. Our model (Fig. 9) also predicts an equal loss of sister chromatin cohesion even when only one Rad21 molecule in the handcuff is cleaved. This will also lead to a partial loss of cohesin from chromatin: while it will come off one sister, it may still hang onto the other. A protein-DNA catenation by the cohesin complex has also been proposed (14), but how the catenating activity of the cohesin complex is achieved is an open area of research. It is possible that calpain-1-mediated cleavage of Rad21 may play a role in resolving such catenation. We suggest that calpain-1-mediated cleavage of Rad21 aids in the prophase pathway of sister chromatid separation by both of these mechanisms (Fig. 9). Additionally, it has been proposed that a calcium spike and the resulting Ca2+-CaM complex may play a role in chromosomal segregation during mitosis (48). Therefore, a role of calpain-mediated cleavage of cohesin in affecting the concatenase activity of the cohesin complex could be considered.
Fig. 9.
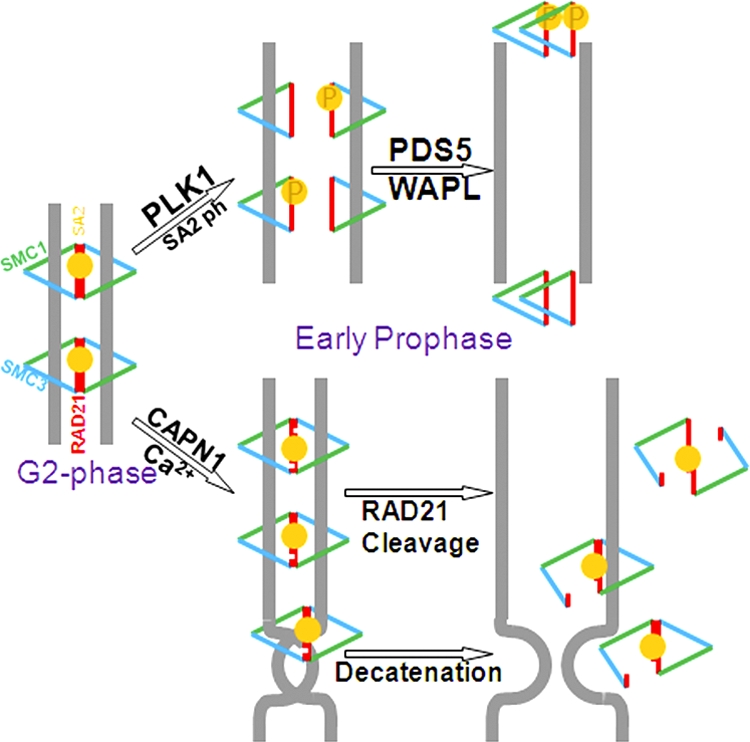
Model for calpain-1-mediated Rad21 cleavage and sister chromatid separation. Cleavage of either one or both Rad21 molecules (represented by white incisions) in the cohesin handcuff by calpain-1 (CAPN1) can equally lead to a loss of sister chromatid cohesion and can also relieve cohesin-mediated catenation of sisters. Cohesin cleavage by calpain-1 may function as an additional mechanism to separate the sisters, aiding the Plk1-mediated prophase pathway.
In summary, we have identified calpain-1 as a novel cohesin-proteinase. We also uncovered an additional pathway for regulation of sister chromatid cohesion in mammalian cells. Elucidation of the precise regulation of calpain-1 activity during mitosis and the resultant effects on sister chromatid cohesion and separation in light of cell cycle progression awaits further investigation.
ACKNOWLEDGMENTS
We thank Pulivarthi H. Rao (Baylor College of Medicine, Houston, TX) for assistance in cytogenetic analysis of the metaphase spreads.
This study was supported by grants awarded to D. Pati from the National Cancer Institute (1RO1 CA109478).
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Beckouet F., et al. 2010. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell 39:689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birkenbihl R. P., Subramani S. 1992. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20:6605–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bootman M. D., Fearnley C., Smyrnias I., MacDonald F., Roderick H. L. 2009. An update on nuclear calcium signalling. J. Cell Sci. 122:2337–2350 [DOI] [PubMed] [Google Scholar]
- 4. Bootman M. D., Young K. W., Young J. M., Moreton R. B., Berridge M. J. 1996. Extracellular calcium concentration controls the frequency of intracellular calcium spiking independently of inositol 1,4,5-trisphosphate production in HeLa cells. Biochem. J. 314:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borges V., et al. 2010. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol. Cell 39:677–688 [DOI] [PubMed] [Google Scholar]
- 6. Chamero P., Villalobos C., Alonso M. T., Garcia-Sancho J. 2002. Dampening of cytosolic Ca2+ oscillations on propagation to nucleus. J. Biol. Chem. 277:50226–50229 [DOI] [PubMed] [Google Scholar]
- 7. Chen F., et al. 2002. Caspase proteolysis of the cohesin component RAD21 promotes apoptosis. J. Biol. Chem. 277:16775–16781 [DOI] [PubMed] [Google Scholar]
- 8. Ciosk R., et al. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243–254 [DOI] [PubMed] [Google Scholar]
- 9. Dorsett D. 2007. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma 116:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gil-Parrado S., et al. 2003. Subcellular localization and in vivo subunit interactions of ubiquitous mu-calpain. J. Biol. Chem. 278:16336–16346 [DOI] [PubMed] [Google Scholar]
- 11. Goll D. E., Thompson V. F., Li H., Wei W., Cong J. 2003. The calpain system. Physiol. Rev. 83:731–801 [DOI] [PubMed] [Google Scholar]
- 12. Groigno L., Whitaker M. 1998. An anaphase calcium signal controls chromosome disjunction in early sea urchin embryos. Cell 92:193–204 [DOI] [PubMed] [Google Scholar]
- 13. Guacci V., Koshland D., Strunnikov A. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haering C. H., Farcas A. M., Arumugam P., Metson J., Nasmyth K. 2008. The cohesin ring concatenates sister DNA molecules. Nature 454:297–301 [DOI] [PubMed] [Google Scholar]
- 15. Hauf S., et al. 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hauf S., Waizenegger I. C., Peters J. M. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293:1320–1323 [DOI] [PubMed] [Google Scholar]
- 17. Kagey M. H., et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim B. J. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J. Biol. Chem. 285:22784–22792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S. T., Xu B., Kastan M. B. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 21. Losada A., Hirano M., Hirano T. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Losada A., Yokochi T., Kobayashi R., Hirano T. 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo H., et al. 2008. Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J. Biol. Chem. 283:19176–19183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magnaghi-Jaulin L., et al. 2010. Calpain 2 is required for sister chromatid cohesion. Chromosoma 119:267–274 [DOI] [PubMed] [Google Scholar]
- 25. Michaelis C., Ciosk R., Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35–45 [DOI] [PubMed] [Google Scholar]
- 26. Molinari M., Carafoli E. 1997. Calpain: a cytosolic proteinase active at the membranes. J. Membr. Biol. 156:1–8 [DOI] [PubMed] [Google Scholar]
- 27. Nakajima M., et al. 2007. The complete removal of cohesin from chromosome arms depends on separase. J. Cell Sci. 120:4188–4196 [DOI] [PubMed] [Google Scholar]
- 28. Nasmyth K., Haering C. H. 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43:525–558 [DOI] [PubMed] [Google Scholar]
- 29. Orrenius S., Zhivotovsky B., Nicotera P. 2003. Regulation of cell death: the calcium-apoptosis link. Nat. Rev. Mol. Cell. Biol. 4:552–565 [DOI] [PubMed] [Google Scholar]
- 30. Parelho V., et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422–433 [DOI] [PubMed] [Google Scholar]
- 31. Pati D., Zhang N., Plon S. E. 2002. Linking sister chromatid cohesion and apoptosis: role of Rad21. Mol. Cell. Biol. 22:8267–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peters J. M., Tedeschi A., Schmitz J. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev. 22:3089–3114 [DOI] [PubMed] [Google Scholar]
- 33. Rodrigues M. A., et al. 2007. Nucleoplasmic calcium is required for cell proliferation. J. Biol. Chem. 282:17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rolef Ben-Shahar T., et al. 2008. EcoI-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321:563–566 [DOI] [PubMed] [Google Scholar]
- 35. Schmidt D., et al. 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20:578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spencer V. A., Sun J. M., Li L., Davie J. R. 2003. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 31:67–75 [DOI] [PubMed] [Google Scholar]
- 37. Stein C. K. 1998. Modified giemsa-11 staining protocol for chromosomes of human and hybrid cells. Somat. Cell Mol. Genet. 24:191–195 [DOI] [PubMed] [Google Scholar]
- 38. Strom L., et al. 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317:242–245 [DOI] [PubMed] [Google Scholar]
- 39. Sumara I., Vorlaufer E., Gieffers C., Peters B. H., Peters J. M. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151:749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tompa P., et al. 2004. On the sequential determinants of calpain cleavage. J. Biol. Chem. 279:20775–20785 [DOI] [PubMed] [Google Scholar]
- 41. Uhlmann F., Lottspeich F., Nasmyth K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37–42 [DOI] [PubMed] [Google Scholar]
- 42. Uhlmann F., Wernic D., Poupart M. A., Koonin E. V., Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103:375–386 [DOI] [PubMed] [Google Scholar]
- 43. Unal E., et al. 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321:566–569 [DOI] [PubMed] [Google Scholar]
- 44. Unal E., Heidinger-Pauli J. M., Koshland D. 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through EcoI (Ctf7). Science 317:245–248 [DOI] [PubMed] [Google Scholar]
- 45. Waizenegger I. C., Hauf S., Meinke A., Peters J. M. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103:399–410 [DOI] [PubMed] [Google Scholar]
- 46. Watrin E., et al. 2006. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 16:863–874 [DOI] [PubMed] [Google Scholar]
- 47. Wendt K. S., et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801 [DOI] [PubMed] [Google Scholar]
- 48. Whitfield J. F. 1995. Calcium in cell cycles and cancer, 2nd ed. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 49. Xia H. G., et al. 2010. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy 6:61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang J., et al. 2008. Acetylation of Smc3 by EcoI is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31:143–151 [DOI] [PubMed] [Google Scholar]
- 51. Zhang N., et al. 2008. A handcuff model for the cohesin complex. J. Cell Biol. 183:1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]