Abstract
Exquisite control of the level and activity of p53 are required in order to preserve cellular homeostasis following DNA damage. How this regulation is integrated with other key metabolic pathways in vivo is poorly understood. Here, we describe an endogenous feedback circuit for regulation of p53 through its transcriptional target gene, Redd1, a stress-induced inhibitor of TOR complex 1 (TORC1) activity. Cells and tissues of Redd1−/− mice exhibit enhanced sensitivity to ionizing radiation and chemotherapy treatment, which we demonstrate is attributable to abnormally increased p53 protein level and activity in the absence of Redd1. We find that deregulation of p53 in this setting is not due to failed DNA repair or to increased p53 stabilization but, instead, to increased p53 translation. We show that Redd1 loss leads to elevated mammalian TORC1 (mTORC1) activity, which explains the increased p53 translation and protein levels. Together, these findings suggest that REDD1-mediated suppression of mTORC1 activity exerts feedback control on p53, thereby limiting the apoptotic response and contributing to cellular survival following DNA damage. This work therefore defines a role for REDD1 in the control of p53 in vivo, with potential therapeutic implications for cancer and for the variety of genetic diseases involving TOR pathway signaling components.
INTRODUCTION
The maintenance of genome stability is a central challenge for all organisms, as evidenced by the conservation of DNA damage response and repair pathways from yeast to higher eukaryotes. In higher organisms, additional key factors orchestrate the damage response by integrating complex signaling pathways inherent in a multicellular context. The p53 transcription factor is one such central regulator of the DNA damage response and genomic integrity (33). When activated in response to DNA damage, p53 induces the transcription of a large variety of target genes, which in turn control fundamental cellular processes, including growth arrest, senescence, apoptosis, and oxidative metabolism (26). In mammals, activated p53 participates in the DNA damage response in a wide variety of tissues; among the most sensitive are the developing brain and thymus, which undergo p53-dependent growth arrest and apoptosis following DNA damage (20, 34). While the precise mechanisms which dictate the cellular consequences of p53 activation in a given context remain under investigation, higher protein levels of p53 are thought to favor the induction of apoptosis (3, 21).
As a key signal integrator, p53 is subject to multiple levels of regulation through inputs that remain under intensive investigation. Furthermore, because of its potent effects, the p53 level and activity must be kept under exquisitely tight control. The most well-described mechanisms of p53 regulation involve posttranslational modifications, including phosphorylation, acetylation, methylation, ubiquitination, and sumoylation (33). Since many of these modifications have been identified primarily through in vitro studies, in many cases, their precise contribution to p53 regulation in vivo remains to be determined (17). Among the most important regulators of p53 in vivo is the E3 ubiquitin ligase MDM-2, which binds to p53 and thereby induces its ubiquitination and subsequent proteasomal degradation. The Mdm2 gene is a direct, positively regulated transcriptional target of p53, thus defining a key negative feedback loop for p53 regulation. Several other, less direct feedback loops have been described for negative regulation of p53 stability, in most cases involving indirect effects of p53 on MDM-2 (12). More recently, p53 has been demonstrated to be regulated at the level of its protein translation (11, 31, 36). Nevertheless, the contribution of translation to homeostatic regulation of p53-mediated responses in vivo is not well defined.
The Redd1 gene (regulated in DNA damage 1, also known as RTP801, DDIT4, and Dig-2) was initially identified as a stress response gene induced following DNA damage and other cellular stress stimuli (4, 8, 29). Redd1 was found to be a direct transcriptional target of p53 (8), as it was induced following ionizing radiation in wild-type primary cells but not in cells with genetic inactivation of either p53 or the ataxia telangiectasia mutated (ATM) kinase. Furthermore, the Redd1 promoter contains a consensus p53 family binding element which is required for the induction of REDD1 by p53 (8). Work by several groups has demonstrated that Redd1 and its fly orthologues serve as essential inhibitors of TOR kinase complex 1 (TORC1) activity in both Drosophila and mammalian cells (1, 4, 25). Thus, in Redd1−/− cells, mammalian TORC1 (mTORC1) activity is not appropriately downregulated in response to cellular stress (30). These studies also showed that the effect of Redd1 on mTORC1 activity is mediated through the tuberous sclerosis tumor suppressor complex TSC1/2, which functions as a negative regulator of mTORC1 activity. The REDD1 protein functions biochemically as a molecular shuttle that disrupts the inhibitory interaction between the TSC2 and 14-3-3 proteins. Thus, the induction of REDD1 leads to dissociation of TSC2/14-3-3, which activates TSC1/2 and leads to mTORC1 inhibition (6).
The induction of REDD1 has pleiotropic effects on cell survival, either inhibiting or promoting apoptosis in different experimental contexts (8, 27, 29). We therefore developed a genetic model to determine the contribution of endogenous REDD1 to the DNA damage response in vivo. We find that Redd1−/− tissues and cells exhibit increased sensitivity to ionizing radiation and chemotherapy treatment, both in vitro and in vivo. Remarkably, we show that this increased sensitivity in the absence of REDD1 is attributable to an increase in p53 protein levels, an effect which is due to abnormally elevated mTORC1-dependent translation of p53 itself. Thus, endogenous REDD1 limits p53 translation and thereby contributes to cellular survival following p53 activation. These findings define a new and physiologically important mechanism for negative feedback regulation of p53 through its transcriptional target gene Redd1.
MATERIALS AND METHODS
Tissue culture.
The Redd1−/− allele was generated as previously described (30). Primary mouse embryo fibroblasts (MEFs) were prepared from embryonic day 13.5 wild-type and Redd1−/− littermate embryos. Cells at passages 1 to 4 were used for experiments. MEFs were retrovirally transduced with the E1A oncogene or along with dominant negative p53 and selected with puromycin (1 μg/ml). U2OS human osteosarcoma cells expressing tetracycline-regulated wild-type REDD1 or the REDD1-RPAA mutant were described previously (6). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. Primary thymocytes were isolated from 5- to 6-wk-old littermate mice and cultured short term in RPMI 1640.
Immunoblot and QRT-PCR analysis.
Total protein extracts were prepared using radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitor (Roche) and phosphatase inhibitor cocktails (Sigma). Blots of SDS-PAGE gels were incubated with antibodies recognizing the following proteins: mouse p53 (CM5; Vector Laboratories), human p53 (DO1; Santa Cruz Biotechnologies), cleaved caspase 3 (Cell Signaling), poly(ADP-ribose) polymerase 1 (PARP1; Cell Signaling), β-actin (Santa Cruz Biotechnologies), β-tubulin (Santa Cruz Biotechnologies), γ-H2AX (Upstate Biotechnologies), histone H3 (Cell Signaling), phospho-S6 S235/S236 (Cell Signaling), total S6 (Cell Signaling), phospho-p70 S6 kinase T389 (Cell Signaling), phospho-4E-BP1 T70 (Cell Signaling), phospho-AKT S473 (Cell Signaling), total AKT (Cell Signaling), and hemagglutinin (HA) epitope (Covance). Total RNA extraction, cDNA synthesis, and quantitative reverse transcription-PCR (QRT-PCR) were performed as described earlier (13). The expression of each gene was normalized to that of β-actin.
S35 pulse-chase labeling.
Primary MEFs were starved for 1 h in methionine/cysteine-free DMEM, pulse-labeled for 30 min with 0.25 mCi 35S per ml (EXPRE35S 35S-protein labeling mix; PerkinElmer) in methionine/cysteine-free medium, washed with phosphate-buffered saline (PBS), and then chased as indicated with medium containing 2 mM methionine and 2 mM cysteine. Cells were lysed in RIPA buffer, and then equal amounts of total lysates were immunoprecipitated with anti-p53 antibody and subjected to SDS-PAGE. Following gel drying, labeled proteins were visualized by exposure to X-ray film (Biomax XAR; Kodak).
Polysome profiling.
Wild-type and Redd1−/− primary MEFs were preincubated with 100 μg/ml cycloheximide for 10 min at 37°C and then washed with PBS containing cycloheximide. Cytoplasmic extracts were prepared by Dounce homogenization in polysome lysis buffer (10 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 10 mM NaCl, 0.12% sodium deoxycholate, 1.2% Triton X-100, 100 μg/ml heparin, and 100 μg/ml cycloheximide). Extracts were layered over 15-to-45% sucrose gradients and centrifuged for 2.5 h at 37,000 rpm at 4°C in a Beckman SW41Ti rotor. Gradients were fractionated using an ISCO gradient fractionation system connected to a UV detector for continuous measurement of the absorbance at 254 nm. The polysome region of the gradient was divided into light, intermediate, and heavy fractions based on sedimentation, representing the bottom 25%, middle 50%, and top 25% of volume collected, respectively. Total RNA was isolated from each fraction, and cDNA synthesis was performed on pooled RNA fractions. QRT-PCR was performed to assess the distribution of p53 mRNA or β-actin mRNA normalized to 18S mRNA.
Damage response, cell viability, and apoptosis.
Quantitative γ-H2AX analysis was performed using a phospho-H2AX primary antibody (Upstate Biotechnology) and fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) followed by analysis using a FACSCalibur (BD Biosciences) and CellQuest software. Cell viability was quantified following treatment in the absence or presence of doxorubicin, using trypan blue dye exclusion, the standard MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] method, or crystal violet staining. Apoptotic cell death was measured by annexin V-propidium iodide (PI) staining (BD Biosciences) followed by flow cytometry analysis as described above, as well as by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay using an ApopTag peroxidase in situ apoptosis detection kit (Millipore).
Mouse irradiation and caspase 3 immunohistochemistry.
Newborn mice were subjected to whole-body irradiation from a 137Cs irradiator (Mark I model, JL Shepherd & Associates). Brain and thymus tissues were harvested, and total lysates were prepared for Western blot analysis. For immunohistochemistry, tissues were formalin fixed and paraffin embedded. Tissue sections were stained with hematoxylin and eosin (H&E) and anti-cleaved caspase 3 antibody (51A; Cell Signaling).
Statistical analysis.
Statistical analyses were performed using Student's t test, and data are presented as the mean ± standard deviation (SD). A P value of <0.05 was considered statistically significant.
RESULTS
Increased DNA damage sensitivity of Redd1−/− cells in vitro and in vivo.
In order to test the contribution of the p53 target gene REDD1 to the DNA damage response, we first established a quantitative, cell-based model using primary wild-type and Redd1−/− MEFs immortalized by transduction of a retrovirus expressing the adenoviral E1A gene. E1A-expressing MEFs are a well-established DNA damage sensitivity model (19). Consistently, Redd1−/− cells exhibited substantially increased sensitivity to DNA damage. In a quantitative dose-response analysis, the 50% lethal dose (50% inhibitory concentration [IC50]) of the DNA-damaging agent doxorubicin was 4-fold lower in E1A-Redd1−/− cells than in matched wild-type cells (Fig. 1A). This effect was attributable to increased apoptosis following DNA damage in Redd1−/− cells, evidenced by an increase in annexin V-positive cells (Fig. 1B) and by elevated activated (cleaved) caspase 3 (Fig. 1C). Similarly, following UV radiation, Redd1−/− E1A-expressing MEFs demonstrated an increase in both cell death (Fig. 1D) and DNA fragmentation as assessed by TUNEL assay (Fig. 1E), a hallmark of apoptotic cell death. Thus, Redd1−/− cells demonstrate a physiologically significant increase in chemosensitivity and DNA damage-induced apoptosis.
Fig. 1.
Increased chemosensitivity and apoptosis in Redd1−/− tissues and cells following DNA damage. (A) Dose-response curve showing 4-fold lower IC50 for doxorubicin in Redd1−/− cells than in wild-type cells. Viable cell numbers were determined by MTT assay. (B) Increased annexin V-PI staining of Redd1−/− cells following doxorubicin treatment (Dox, 0.2 μM), assessed by FACS analysis. Error bars show the SD for triplicate plates in a representative experiment. (C) Immunoblots showing increased activated (cleaved) caspase 3 (denoted by *) in Redd1−/− cells treated as described for panel B. β-Tubulin (TUB.) serves as a loading control. (D) Dose-response curve showing increased UV sensitivity of Redd1−/− cells compared to that of wild-type cells. (E) Representative image from TUNEL assay showing increased apoptosis (brown staining) in Redd1−/− cells following UV irradiation (30 J/m2). (F) Immunoblots showing increased activated (*) caspase 3 and cleaved PARP1 (bottom arrow at right) in Redd1−/− tissues compared to the results for wild-type tissues following irradiation (12 Gy) of newborn mice at the indicated times (h). β-Actin serves as a loading control. (G) Summary of cell counts from histological sections of the cerebellum of newborn mice irradiated as described for panel F and stained for activated caspase 3. Error bars show the standard error of the mean (SEM) for 5 representative fields; 2,500 cells were counted per genotype. (H) Cell counts following TUNEL staining of thymus in newborn mice irradiated as described for panel F. Error bars show the SEM for 7 representative fields; 2,000 cells were counted per genotype. **, P < 0.01; ***, P < 0.001.
We then validated these findings in vivo by generating Redd1−/− mice. These mice do not have obvious developmental defects and do not exhibit an increased incidence of spontaneous tumors (30). We tested tissue sensitivity to DNA damage by irradiating newborn mice and then examining apoptotic cell death in the brain and thymus, as these tissues are known to undergo apoptosis following irradiation (20, 34). Redd1−/− mice exhibited a substantial increase in radiation-induced apoptosis in both tissues compared with that in their wild-type littermates, demonstrated by enhanced caspase 3 activation, PARP1 cleavage, and TUNEL staining of tissues (Fig. 1F to H). Increased DNA damage-induced apoptosis was further confirmed in Redd1−/− primary thymocytes. Consistent with our results following ionizing radiation, Redd1−/− thymocytes exhibited an increase in cell death and activated caspase 3 following doxorubicin treatment in vitro (not shown), in keeping with the increased apoptosis observed in vivo. Together, these findings demonstrate that genetic loss of REDD1 confers an increase in DNA damage-induced apoptosis both in vitro and in vivo.
Deregulation of p53 protein and activity in Redd1−/− cells and tissues.
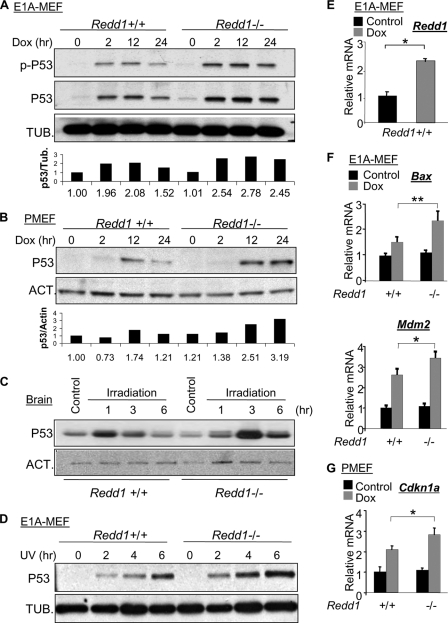
To uncover the explanation for increased DNA damage sensitivity, we first examined the activation of p53, which is known to be important for the apoptotic response to DNA damage in the brain and thymus (20, 34). We observed a consistent increase in p53 protein in Redd1−/− cells compared to the levels in wild-type MEFs following DNA damage. This increase was observed as early as 2 h following doxorubicin treatment, and the difference in p53 levels became even more apparent at later time points (Fig. 2A and B). This increase was also reflected in elevated phosphorylated (Ser-15) p53 (Fig. 2A) and was observed in both E1A-immortalized and primary Redd1−/− MEFs (Fig. 2B). In the brain, baseline levels of p53 protein were low in mice of both genotypes, but p53 protein was induced significantly more highly in irradiated Redd1−/− mice than in litter-matched wild-type controls (Fig. 2C). Similarly, UV-irradiated Redd1−/− E1A immortalized MEFs demonstrated an increase in p53 levels compared to the levels in wild-type cells (Fig. 2D). Thus, genetic loss of Redd1 is associated with elevated levels of p53 protein following DNA damage.
Fig. 2.
Increased p53 level and activity in Redd1−/− cells following DNA damage. (A, B) Immunoblots showing increased p53 and phosphorylated p53 (p-p53, S15) in Redd1−/− cells following doxorubicin treatment (0.2 μM). Bottom panels show corresponding densitometry values normalized to those of the respective loading controls. PMEF, primary MEF. (C) Increased p53 protein in Redd1−/− newborn brain following irradiation (12 Gy) in vivo. (D) Increased p53 in Redd1−/− cells following UV irradiation (20 J/m2). (E) Induction of Redd1 by doxorubicin (0.2 μM, 12 h). (F, G) Increased p53 activity in Redd1−/− cells treated as described for panel E, evidenced by hyperinduction of Bax and Mdm2 in E1A-immortalized MEFs (F) and of Cdkn1a (p21) in PMEFs (G), assessed by real-time QRT-PCR. Error bars show the SD for triplicate measurements from a representative experiment. **, P < 0.01; *, P < 0.05.
In order to specifically test p53 activity in this context, we examined p53 target gene expression following DNA damage. Redd1 was induced as expected (Fig. 2E), and more importantly, the p53 transcriptional target genes Bax and Mdm2 (Fig. 2F) and Cdkn1a (p21) (Fig. 2G) were both significantly more highly induced following doxorubicin treatment in Redd1−/− compared to their induction in wild-type E1A-immortalized and primary MEFs, respectively. These findings therefore agree with the increased p53 levels and DNA damage sensitivity observed in Redd1−/− cells in this setting.
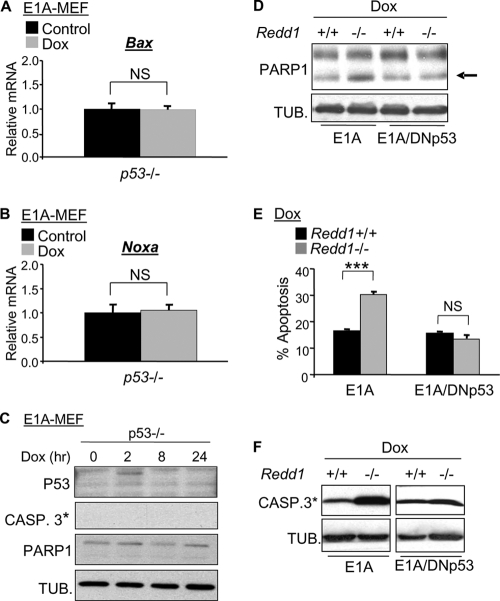
To confirm that the observed DNA damage-induced effects on cell viability and gene induction were mediated by p53, we prepared p53−/− MEFs expressing E1A. As predicted, the induction of proapoptotic genes (Fig. 3A and B) and apoptosis (Fig. 3C) following DNA damage was abolished in the absence of p53. Furthermore, blocking p53 activity in wild-type and Redd1−/− cells with a C-terminal p53 protein (dominant negative p53 [DNp53]), which is well established as functioning in a dominant negative manner (28), abolished the increase in cell death observed in Redd1−/− cells following doxorubicin treatment (Fig. 3D to F). Taken together, these data demonstrate that the p53 protein level and activity are deregulated in the absence of Redd1, leading to an increase in DNA damage-induced cell death.
Fig. 3.
Loss of p53 abolishes proapoptotic gene induction and apoptosis following DNA damage. (A, B) Absence of induction of Bax (A) and Noxa (B) following doxorubicin treatment (Dox, 0.2 μM, 12 h) in p53−/− cells, assessed by real-time QRT-PCR. (C) Immunoblot showing absence of p53, caspase 3 cleavage, and PARP1 cleavage following treatment of cells with doxorubicin. (D) Dominant negative p53 (DNp53) abrogates the increase in apoptosis observed in Redd1−/− MEFs, assessed by PARP1 cleavage (arrow at right). Redd1−/− E1A-expressing MEFs (E1A-MEFs) were infected with DNp53 or the control vector, followed by doxorubicin treatment as described for panel A. (E) DNp53 abrogates increased annexin V-PI staining of Redd1−/− E1A-MEFs following doxorubicin treatment as described above, assessed by FACS analysis. Error bars show the SD for triplicate plates in a representative experiment. (F) DNp53 abolishes increased caspase 3 activation in Redd1−/− E1A-MEFs following doxorubicin treatment, shown by immunoblot for cleaved (*) caspase 3. ***, P < 0.001; NS, nonsignificant P value.
Unaltered DNA damage and p53 stability in Redd1−/− cells.
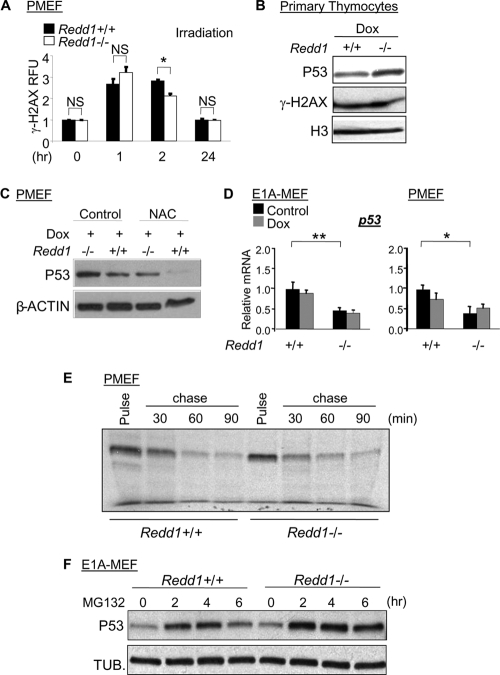
One possible explanation for increased p53 and apoptosis in the absence of REDD1 is an increase in DNA damage itself or a failure of DNA repair in these cells. To explore this possibility, we first tested phosphorylation of the variant histone H2AX (γ-H2AX), a sensitive indicator of DNA breaks induced by a variety of genotoxins (9). We therefore treated MEFs with a single dose of ionizing radiation and followed γ-H2AX using a quantitative fluorescence-activated cell sorter (FACS) assay. We observed little or no significant difference between wild-type and Redd1−/− cells in basal γ-H2AX or in its induction or resolution following ionizing radiation (Fig. 4A). Similarly, in primary thymocytes, no difference in γ-H2AX was observed between wild-type and Redd1−/− cells following DNA damage (Fig. 4B). Importantly, using the same lysates, we observed elevated p53 in Redd1−/− cells under the same conditions in which no difference in γ-H2AX was observed (Fig. 4B).
Fig. 4.
DNA damage and p53 stability are not altered in Redd1−/− cells. (A) No difference in basal level, induction, or resolution of γ-H2AX in Redd1−/− cells compared to the results for wild-type MEFs following irradiation (6 Gy), assessed by FACS analysis. Error bars represent the SD for three independent samples from a representative experiment. (B) Increased p53 in Redd1−/− cells coincides with no difference in γ-H2AX following doxorubicin treatment (Dox, 0.4 μM, 12 h) in isolated thymocytes. Histone H3 serves as a loading control. (C) Deregulation of ROS does not explain elevated p53 in Redd1−/− cells. MEFs were pretreated with NAC (10 mM, 2 h), followed by doxorubicin (0.4 μM, 12 h). (D) Modestly decreased p53 mRNA levels in Redd1−/− E1A-MEFs (left) and primary MEFs (PMEF, right) at baseline and following doxorubicin treatment (0.2 μM and 0.4 μM, respectively). (E) No difference in p53 stability in Redd1−/− versus wild-type MEFs following doxorubicin treatment (0.4 μM), as assessed by 35S pulse-chase and then immunoprecipitation. (F) Chemical stabilization with MG132 (5.0 μM) does not normalize p53 levels between Redd1−/− and wild-type E1A-MEFs. *, P < 0.05; ** P < 0.01.
Since increased damage does not appear to explain deregulated p53 in Redd1−/− cells, we next tested whether increased reactive oxygen species (ROS) may play a role, given that altered ROS have been linked to both Redd1 loss (13) and p53 stabilization (2). We therefore treated MEFs with the potent antioxidant N-acetyl cysteine (NAC) and examined the effect on p53 levels following DNA damage. NAC treatment decreased p53 levels in both wild-type and Redd1−/− cells, as expected (16); however, this treatment did not normalize p53 levels in Redd1−/− cells and instead may have further increased the relative difference in p53 levels between Redd1−/− and wild-type cells (Fig. 4C). These findings demonstrate that deregulation of ROS does not explain the enhanced p53-mediated damage response in the absence of Redd1.
To determine the mechanism of p53 deregulation, we then examined p53 mRNA levels and found that they were not elevated in Redd1−/− MEFs but instead were approximately half as high in these cells as in matched wild-type cells. This was true both in the presence or absence of E1A expression and in the presence or absence of DNA damage (Fig. 4D). We then turned to address the stability of p53 protein in Redd1−/− compared to its stability in wild-type cells. Initially, we treated MEFs with doxorubicin followed by cycloheximide in order to block new protein synthesis. In this setting, the half-life of p53 protein was not prolonged in Redd1−/− cells compared to its half-life in wild-type cells (not shown). In order to address the half-life of p53 more rigorously, we performed a pulse-chase analysis of p53 in Redd1−/− and wild-type MEFs. We treated cells with doxorubicin for 12 h, and then pulse-labeled cells with [35S]methionine for 30 min prior to chase with unlabeled methionine and then p53 immunoprecipitation. This experiment showed the expected stabilization of p53 following DNA damage (a half-life of approximately 30 min) (35) but, again, was unequivocal in demonstrating no difference in p53 stability between wild-type and Redd1−/− cells (Fig. 4E). Finally, we tested the effect of chemical p53 stabilization, which would be expected to normalize p53 levels if increased p53 stability accounted for its elevation in Redd1−/− cells. Consistent with our pulse-chase analysis, treatment with the proteasome inhibitor MG132 did not normalize p53 levels but instead yielded a clear elevation of p53 protein in Redd1−/− cells versus the level in wild-type MEFs (Fig. 4F). Together, these experiments show that increased p53 protein in Redd1−/− cells following DNA damage is not mediated by an increase in either p53 transcription or its protein stability. Notably, this conclusion is also consistent with our finding described above that p53 deregulation in Redd1−/− cells is not due to increased or persistent DNA damage, which would be expected to result in stabilized p53 (33).
Increased translation of p53 in Redd1−/− cells.
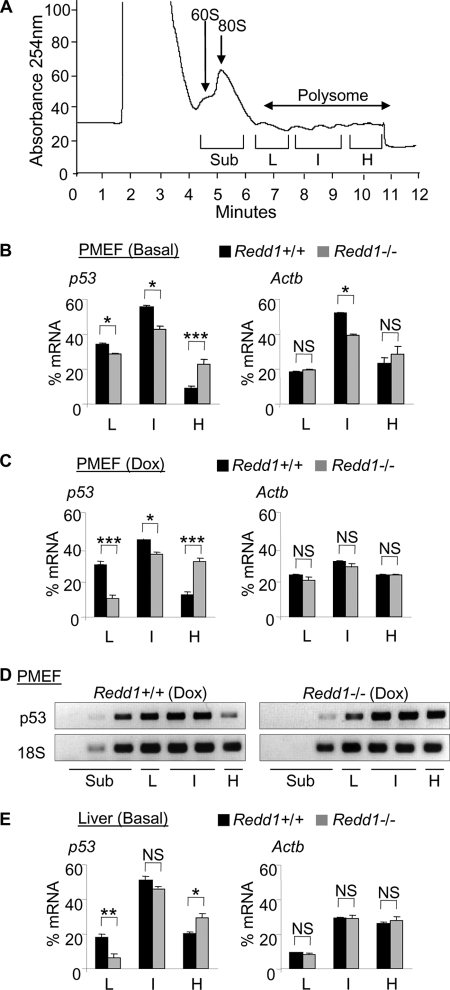
Based on these findings, we asked whether increased p53 synthesis might explain the accumulation of p53 protein observed following DNA damage in Redd1−/− cells. Increased p53 protein synthesis was initially suggested by the finding that brief pulse-labeling analysis demonstrated equal levels of radiolabeled p53 protein in Redd1−/− and wild-type MEFs (Fig. 4E, 1st and 5th lanes), despite the fact that Redd1−/− cells have substantially lower p53 mRNA levels (Fig. 4D). To address the rate of p53 translation directly, we performed polysome profiling of wild-type and Redd1−/− cells following doxorubicin treatment. This technique involves sucrose density gradient centrifugation of cytosolic lysates in order to separate mRNA molecules based on the number of associated ribosomes, a measure of the translation rate (Fig. 5A) (7).
Fig. 5.
Increased p53 translation in Redd1−/− cells. (A) Representative polysomal profile of cytosolic lysates from MEFs. Ribosomal subunit RNA peaks, polysome regions, and collection times are indicated. Subpolysome (Sub) and light (L), intermediate (I), and heavy (H) polysome regions are indicated as defined in Materials and Methods. (B) Quantitative RNA analysis from polysome fractions shows that p53 (left) but not Actb (β-actin, right) mRNA is overrepresented in the heaviest (H, most highly translated) polysome fraction and underrepresented in lighter (L, I) polysome fractions in Redd1−/− cells compared to the results for wild-type cells, as assessed by real-time QRT-PCR and normalized to rRNA. Error bars show the SD for triplicate samples from a representative experiment. (C) A similar selective increase in p53 but not Actb translation is observed in Redd1−/− cells compared to the results for wild-type cells following doxorubicin treatment (0.4 μM, 12 h), assessed from polysome profiles generated and analyzed as described for panel B. (D) Increase in absolute p53 mRNA levels in the heaviest (H) polysome fraction from Redd1−/− cells compared to the results for wild-type cells, assessed by RT-PCR. Also apparent is increased p53 mRNA in subpolysome (Sub) fractions in wild-type cells. Loading control 18S rRNA is shown. (E) Increased p53 but not Actb translation in Redd1−/− liver compared to the results for wild-type liver in vivo. RNA analysis of polysome profiles from livers of adult mice of the indicated genotypes, analyzed as described for panel B. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
We performed a series of profiling experiments in wild-type and Redd1−/− cells and tissues, collecting heavy, intermediate, and light polysome fractions and determining the proportions of p53 mRNA in each fraction (Fig. 5A). Remarkably, p53 mRNA was dramatically more abundant in the heaviest polysome fraction (representing the most highly translated mRNAs) in Redd1−/− cells than in wild-type MEFs, even in the absence of DNA damage (Fig. 5B). In contrast, the intermediate and light polysome fractions contained significantly more p53 mRNA in the wild-type cells. As a control, we examined the translation of Actb (β-actin), which showed little difference between Redd1−/− and wild-type cells (Fig. 5B). Increased p53 translation in Redd1−/− cells was consistently observed in multiple polysome-profiling experiments, both in the presence and absence of DNA damage (Fig. 5B and C). We next performed analysis of absolute RNA levels in different polysome fractions. These studies confirmed an increase in the absolute number of highly translated p53 mRNA molecules in Redd1−/− versus wild-type cells (Fig. 5D). Finally, we performed polysome profiling of the livers of Redd1−/− versus wild-type mice. These experiments showed a similar increase in p53 translation but not β-actin translation in Redd1−/− tissues in vivo (Fig. 5E). Collectively, these findings demonstrate that loss of REDD1 is associated with a constitutive increase in p53 translation both in vitro and in vivo. Subsequent stabilization of p53 by DNA damage (Fig. 2) or proteasome inhibition (Fig. 4F) leads to elevated p53 protein and activity, resulting in increased DNA damage sensitivity.
mTORC1-dependent regulation of p53 and cellular survival by REDD1.
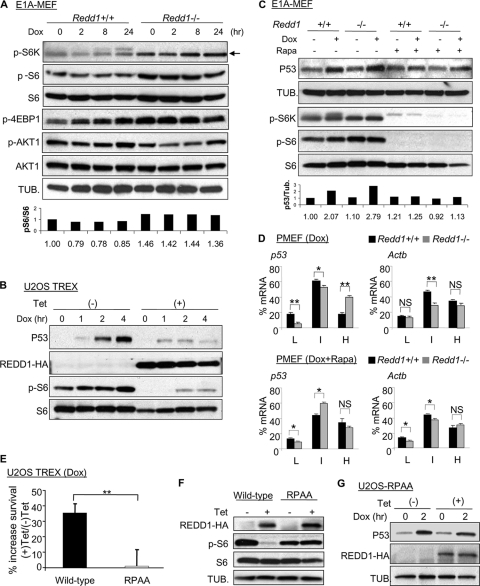
Given that REDD1 functions as a potent inhibitor of mTORC1, a key regulator of cellular translation, we tested the hypothesis that increased mTORC1 kinase activity might mediate p53 deregulation in this setting. Consistent with this hypothesis, we observed increased mTORC1 activity both at baseline and following doxorubicin treatment in Redd1−/− cells, evidenced by increased phosphorylation of the mTORC1 substrate p70 ribosomal S6 kinase 1 (p70-S6K1) and its downstream substrate ribosomal protein S6 (RPS6) (Fig. 6A). We then turned to address the specific contributions of REDD1 and mTORC1 to p53 levels following DNA damage. We first asked whether overexpression of REDD1 would lead to the inverse of the Redd1−/− phenotype and, therefore, result in a decrease in DNA damage-induced p53 levels. For these experiments, we used U2OS osteosarcoma cells engineered to express tetracycline-regulated REDD1, which results in inhibition of endogenous mTORC1 activity (30). These cells are a suitable model for our studies, as they express endogenous wild-type p53 which is induced following DNA damage (10). Indeed, REDD1 expression by tetracycline was sufficient to suppress DNA damage-induced p53 levels, and this suppression was coincident with inhibition of mTORC1 activity by REDD1 (Fig. 6B).
Fig. 6.
Deregulation of mTORC1 mediates increased translation, p53 levels, and damage sensitivity in Redd1−/− cells. (A) Increased mTORC1 activity in Redd1−/− cells at baseline and following doxorubicin treatment (0.2 μM), assessed by immunoblot for phosphorylated p70-ribosomal protein S6 kinase 1 (p-S6K, T389), ribosomal protein S6 (p-S6, S235/S236), and EIF-4E binding protein 4E-BP1 (p-4EBP1, T70) in E1A-MEFs. Phosphorylated AKT (p-AKT, S473) is modestly decreased in Redd1−/− cells, consistent with a Redd1-dependent feedback effect on mTORC1 activity. Densitometry values for pS6 normalized to total S6 protein are shown below. (B) Tetracycline (Tet)-induced REDD1 suppresses p53 levels and mTORC1 activity, assessed by pS6 (S235/S236) level, following doxorubicin treatment (2.0 μM). (C) mTORC1 suppression by rapamycin (Rapa) treatment (25 nM) normalizes DNA damage-induced p53 protein to wild-type levels in Redd1−/− E1A-MEFs following doxorubicin treatment (0.2 μM, 12 h). Compare the 2nd and 4th and the 6th and 8th lanes. Densitometry values are shown below. (D) Increased p53 translation in Redd1−/− MEFs following doxorubicin treatment (0.2 μM, top) reverts to wild-type levels with rapamycin treatment (25 nM, bottom). Cells were treated and profiled in parallel in the presence or absence of rapamycin, and RNA was prepared from the indicated polysome profile fractions and then assessed by real-time QRT-PCR and normalized to rRNA. (E) REDD1 promotes cell survival after DNA damage via mTORC1. REDD1 or the inactive mutant REDD1-RPAA was induced with tetracycline followed by doxorubicin treatment (0.1 μM, 72 h), and surviving cells were determined by crystal violet staining relative to that in controls without tetracycline. (F) Immunoblot demonstrates equal levels of induced wild-type and mutant REDD1 in cells used in the experiment whose results are shown in panel E, as well as failure of REDD1-RPAA to suppress mTORC1 activity (pS6, S235/S236). (G) Induction of the inactive REDD1-RPAA mutant by tetracycline fails to suppress p53 following doxorubicin treatment (Dox, 2.0 μM). **, P < 0.01; *, P < 0.05.
To assess the link between endogenous REDD1-regulated mTORC1 activity and p53 protein translation, we treated Redd1−/− MEFs or their wild-type counterparts with rapamycin, a potent mTORC1 inhibitor, followed by doxorubicin. In the absence of rapamycin, p53 was more highly induced in Redd1−/− cells than in wild-type cells, as shown previously. In contrast, suppression of mTORC1 activity with rapamycin completely eliminated this REDD1-dependent difference in p53 protein levels (Fig. 6C). We then performed parallel polysome profiles in wild-type and Redd1−/− cells following doxorubicin treatment in the presence or absence of rapamycin. Treatment with rapamycin abolished the previously seen preferential heavy polysome distribution of p53 mRNA in Redd1−/− cells in comparison to its distribution in wild-type cells (Fig. 6D). These findings argue for a direct link between REDD1-dependent increased mTORC1 activity and REDD1-dependent deregulation of p53 translation.
Finally, we tested directly whether REDD1-associated effects on cell survival following DNA damage were mediated through its regulation of mTORC1. To do so, we took advantage of a REDD1 mutant, REDD1-RPAA, which we had previously shown to be inactive in regulating mTORC1 (6). We treated U2OS cells expressing tetracycline-regulated wild-type REDD1 or REDD1-RPAA with doxorubicin and then quantitated cell survival. The expression of wild-type REDD1 substantially increased cell survival following doxorubicin treatment (Fig. 6E), consistent with its ability to suppress p53 levels (Fig. 6B). In contrast, comparable levels of REDD1-RPAA induction conferred no protective effect whatsoever in response to doxorubicin compared to the results for uninduced cells (Fig. 6E), no suppression of mTORC1 activity (Fig. 6F), and no effect on p53 levels (Fig. 6G). Altogether, these experiments demonstrate that REDD1 modulates p53 protein levels and translation through its effect on mTORC1 activity. The absence of REDD1 is associated with increased mTORC1 activity and a consequent increase in p53 translation. Following DNA damage-induced p53 stabilization, abnormally elevated p53 translation leads to an inappropriate apoptotic response. These findings therefore define a feedback circuit for control of the p53 level and activity through REDD1 and mTORC1 (Fig. 7).
Fig. 7.
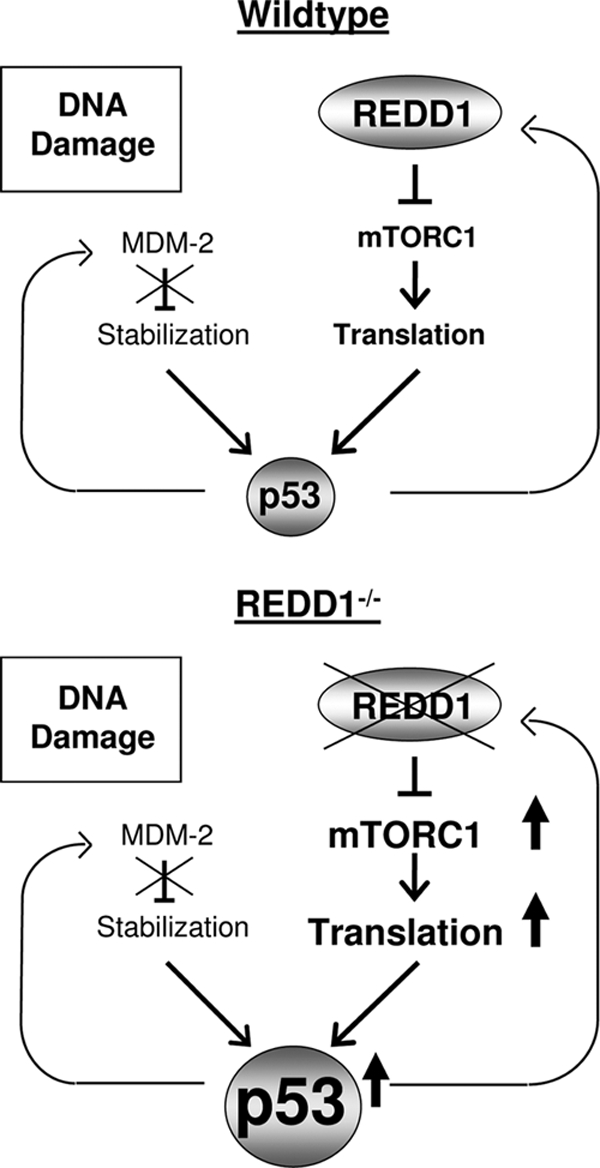
Model for inhibition of p53 through its transcriptional target gene REDD1. In normal cells, DNA damage-induced p53 stabilization leads to induction of REDD1, which exerts mTORC1-dependent feedback control to restrain p53 translation. In Redd1−/− cells, failed mTORC1 suppression contributes to increased p53 translation following DNA damage, leading to abnormally elevated p53 levels and enhanced apoptosis.
DISCUSSION
As a key nodal point in the response to DNA damage, p53 is subject to precise regulation in order to preserve cellular survival and organismal homeostasis. Here, we have uncovered a new and physiologically relevant mechanism which influences the p53 level and activity, involving the p53-induced gene Redd1. Redd1−/− cells and tissues show heightened DNA damage sensitivity (Fig. 1) that is associated with increased p53 protein levels and p53 target gene induction (Fig. 2). Deregulation of p53 in these mice is not attributable to enhanced DNA damage itself or failed DNA repair, a finding which is further supported by our observation that the stability of p53 is not affected in Redd1−/− cells (Fig. 4). Instead, we show directly that this phenotype results from increased p53 translation in both cultured cells and tissues of Redd1−/− mice (Fig. 5). In the absence of DNA damage, p53 protein levels remain low, yet following genotoxic stress and consequent p53 stabilization, increased p53 translation leads to its accumulation in Redd1−/− cells (Fig. 7). This work therefore suggests that p53-dependent induction of REDD1 serves to limit the p53 response following DNA damage, presumably as a mechanism to avoid the unnecessary elimination of cells that might otherwise undergo DNA repair and ultimately survive.
Our second key finding is that p53 translational deregulation in the setting of Redd1 nullizygosity is mediated via mTORC1, in keeping with the established role of REDD1 as an endogenous inhibitor of this kinase complex. We find that Redd1−/− cells demonstrate deregulated mTORC1 activity and that treatment with the mTORC1 inhibitor rapamycin normalizes p53 translation and protein expression to wild-type levels (Fig. 6). Perhaps most importantly, we link p53 regulation directly to mTORC1 via REDD1 by showing that wild-type REDD1 expression is sufficient to suppress p53 levels and to rescue cell viability following DNA damage, while a REDD1 point mutant that is inactive for mTORC1 regulation lacks both of these properties (Fig. 6). These findings therefore provide important new insight into p53 regulation, demonstrating not only that p53 controls its own translation but that it does so through a key pathway (TOR) which integrates numerous external environmental cues in order to control cellular metabolism. And although other pathways influenced by mTORC1 could contribute to altered cell survival in the setting of REDD1 loss (e.g., PI3 kinase/AKT), we demonstrate directly a p53-dependent mechanism (Fig. 3) resulting from enhanced p53 translation (Fig. 4), which together supports our central model (Fig. 7).
How mTORC1 regulates the translation of selective mRNA species, including p53, is an important question and a focus of intensive investigation. The mTORC1 complex is clearly associated with increased 5′-Cap-dependent translation through phosphorylation of the elongation initiation factor 4E (eIF-4E) binding protein 4E-BP1, which promotes its release from eIF-4E. However, many mRNAs selectively regulated by mTORC1 have highly structured 5′ untranslated regions (5′UTRs), suggesting a contribution, as yet poorly understood, of the UTR itself to this selectivity (37). A contribution of the p53 5′UTR to mTORC1-mediated translational regulation is in keeping with prior work showing p53 translational regulation through its 5′UTR (31, 36). Unraveling the details of this specificity will be an important future goal.
Previously described pathways for p53 regulation converge on the p53 target gene Mdm2, an essential negative regulator of p53 protein stability (12). Unlike Mdm2−/− mice, which die early in embryogenesis due to p53-dependent apoptosis, we find that Redd1−/− mice are viable and fertile (6, 23). This phenotypic difference is in accord with our findings, discussed above, that p53 stability is not affected in Redd1−/− mice and, consequently, in the absence of DNA damage, p53 protein levels remain low. Nevertheless, it is possible that the deregulation of baseline p53 translation we observe in the absence of Redd1 may give rise to additional p53-associated phenotypes over time. For example, with time, these mice may demonstrate a premature aging phenotype, as has been reported in some genetic models of increased p53 activity (32). It is also conceivable that increased p53 synthesis in Redd1−/− mice could contribute to enhanced tumor suppression, consistent with our observation that these mice appear not to be tumor predisposed despite having deregulated mTORC1 signaling. Notably, cultured Redd1−/− cells in which p53 function has been abrogated are highly tumorigenic in some genetic contexts compared to their wild-type counterparts (6, 13), in keeping with a prominent role for p53-mediated tumor suppression in the setting of Redd1 loss. Additional genetic studies will be needed to confirm whether these findings apply equally in the setting of endogenous tumor suppression.
REDD1 was initially reported as a pleiotropic regulator of cell survival, given that ectopic overexpression studies demonstrated both pro- and antiapoptotic properties depending on the target cell and experimental conditions (29). Subsequent work has confirmed that in different cellular models, REDD1 may either contribute to or protect from cell death, depending on the cell type and stress stimulus (5, 22). As discussed above, our data explain why the effects of Redd1 loss may differ dramatically depending on the p53 status of the cell. Our findings also predict that the effects of Redd1 loss on damage sensitivity may be tissue-specific, given that tissues differ significantly in their sensitivity to p53-dependent apoptosis following DNA damage in vivo. Consistent with our proposed model for feedback inhibition of p53 by REDD1, the tissues of Redd1−/− mice that show the most significant differences in DNA damage sensitivity are those that are most susceptible to p53-dependent apoptosis (20, 34).
Our finding that the REDD1/mTORC1 pathway feeds back to control p53 may explain in part the significant cooperation between p53 loss and mTORC1 activation in promoting tumorigenesis in some genetic contexts (24). This feedback pathway to p53 also has potentially important implications for the wide variety of therapeutic applications of mTORC1 inhibition. In particular, it is clear that in some advanced tumors, many of which have inactivated p53 function, rapamycin derivatives can contribute to the therapeutic response (15). However, mTORC1 inhibition is also being investigated for tumors arising in the setting of genetic diseases involving loss of function of mTORC1 pathway regulators, including the tuberous sclerosis complex genes TSC1 and TSC2 and the Peutz-Jeghers tumor suppressor gene STK11 (encoding the LKB1 kinase). The benign tumors (hamartomas) that arise in mutation carriers appear to have wild-type p53, and indeed, it has been proposed that p53 may serve as a barrier to malignant transformation in such tumors (18). Therefore, it seems plausible that therapeutic suppression of mTORC1 activity in patients harboring such tumors might hamper p53 function through a translational effect. Such therapy could ultimately be detrimental if it contributed to p53-dependent tumor progression. Furthermore, it is also established that mTORC1 exerts negative feedback to the phosphoinositol-3 kinase (PI3K) pathway (14). Thus, mTORC1 inhibition in these patients might ultimately fail to control tumor growth and might in some cases exert a paradoxical effect due to potential cooperation between p53 suppression and PI3K activation. Our findings highlight the importance of in vivo models in uncovering and validating such physiologically relevant feedback pathways.
ACKNOWLEDGMENTS
We thank Mary Lynch for help with polysome profiling, Zachary Nash and Michael Dennis for expert technical work, and Shyamala Maheswaran for critical review of the manuscript.
This work was supported by RO1 CA-122589 to L.W.E. and by NRSA postdoctoral fellowship award F32 CA150633 to D.D.V.
Footnotes
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Brugarolas J., et al. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18:2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen K., Albano A., Ho A., Keaney J. F., Jr 2003. Activation of p53 by oxidative stress involves platelet-derived growth factor-beta receptor-mediated ataxia telangiectasia mutated (ATM) kinase activation. J. Biol. Chem. 278:39527–39533 [DOI] [PubMed] [Google Scholar]
- 3. Chen X., Ko L. J., Jayaraman L., Prives C. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10:2438–2451 [DOI] [PubMed] [Google Scholar]
- 4. Corradetti M. N., Inoki K., Guan K. L. 2005. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 280:9769–9772 [DOI] [PubMed] [Google Scholar]
- 5. Decaux O., et al. 2010. Inhibition of mTORC1 activity by REDD1 induction in myeloma cells resistant to bortezomib cytotoxicity. Cancer Sci. 101:889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeYoung M. P., Horak P., Sofer A., Sgroi D., Ellisen L. W. 2008. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 22:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dickson A. J., Pogson C. I. 1980. Polyribosomes in isolated liver cells. Preparative procedures, effects of incubation and correlation with protein synthesis. Biochem. J. 186:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellisen L. W., et al. 2002. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10:995–1005 [DOI] [PubMed] [Google Scholar]
- 9. Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amst.) 3:959–967 [DOI] [PubMed] [Google Scholar]
- 10. Florenes V. A., et al. 1994. MDM2 gene amplification and transcript levels in human sarcomas: relationship to TP53 gene status. J. Natl. Cancer Inst. 86:1297–1302 [DOI] [PubMed] [Google Scholar]
- 11. Fu L., Benchimol S. 1997. Participation of the human p53 3′UTR in translational repression and activation following gamma-irradiation. EMBO J. 16:4117–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris S. L., Levine A. J. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899–2908 [DOI] [PubMed] [Google Scholar]
- 13. Horak P., et al. 2010. Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 107:4675–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang J., Manning B. D. 2009. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 37:217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang S., Houghton P. J. 2003. Targeting mTOR signaling for cancer therapy. Curr. Opin. Pharmacol. 3:371–377 [DOI] [PubMed] [Google Scholar]
- 16. Karawajew L., Rhein P., Czerwony G., Ludwig W. D. 2005. Stress-induced activation of the p53 tumor suppressor in leukemia cells and normal lymphocytes requires mitochondrial activity and reactive oxygen species. Blood 105:4767–4775 [DOI] [PubMed] [Google Scholar]
- 17. Kenzelmann Broz D., Attardi L. D. 2010. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis 31:1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee C. H., et al. 2007. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 26:4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lowe S. W., Ruley H. E. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535–545 [DOI] [PubMed] [Google Scholar]
- 20. Lowe S. W., Schmitt E. M., Smith S. W., Osborne B. A., Jacks T. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847–849 [DOI] [PubMed] [Google Scholar]
- 21. Lu C., El-Deiry W. S. 2009. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis 14:597–606 [DOI] [PubMed] [Google Scholar]
- 22. Malagelada C., Jin Z. H., Greene L. A. 2008. RTP801 is induced in Parkinson's disease and mediates neuron death by inhibiting Akt phosphorylation/activation. J. Neurosci. 28:14363–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montes de Oca Luna R., Wagner D. S., Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206 [DOI] [PubMed] [Google Scholar]
- 24. Puzio-Kuter A. M., et al. 2009. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 23:675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reiling J. H., Hafen E. 2004. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18:2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Riley T., Sontag E., Chen P., Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell. Biol. 9:402–412 [DOI] [PubMed] [Google Scholar]
- 27. Schwarzer R., et al. 2005. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene 24:1138–1149 [DOI] [PubMed] [Google Scholar]
- 28. Shaulian E., Zauberman A., Ginsberg D., Oren M. 1992. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol. Cell. Biol. 12:5581–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shoshani T., et al. 2002. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22:2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sofer A., Lei K., Johannessen C. M., Ellisen L. W. 2005. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell. Biol. 25:5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123:49–63 [DOI] [PubMed] [Google Scholar]
- 32. Tyner S. D., et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415:45–53 [DOI] [PubMed] [Google Scholar]
- 33. Vousden K. H., Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137:413–431 [DOI] [PubMed] [Google Scholar]
- 34. Wood K. A., Youle R. J. 1995. The role of free radicals and p53 in neuron apoptosis in vivo. J. Neurosci. 15:5851–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wulf G. M., Liou Y. C., Ryo A., Lee S. W., Lu K. P. 2002. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 277:47976–47979 [DOI] [PubMed] [Google Scholar]
- 36. Yang D. Q., Halaby M. J., Zhang Y. 2006. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25:4613–4619 [DOI] [PubMed] [Google Scholar]
- 37. Yecies J. L., Manning B. D. 2011. mTOR links oncogenic signaling to tumor cell metabolism. J. Mol. Med. 89:221–228 [DOI] [PubMed] [Google Scholar]