Abstract
Several tumor necrosis factor receptor (TNFR) family members activate both the classical and the alternative NF-κB pathways. However, how a single receptor engages these two distinct pathways is still poorly understood. Using lymphotoxin β receptor (LTβR) as a prototype, we showed that activation of the alternative, but not the classical, NF-κB pathway relied on internalization of the receptor. Further molecular analyses revealed a specific cytosolic region of LTβR essential for its internalization, TRAF3 recruitment, and p100 processing. Interestingly, we found that dynamin-dependent, but clathrin-independent, internalization of LTβR appeared to be required for the activation of the alternative, but not the classical, NF-κB pathway. In vivo, ligand-induced internalization of LTβR in mesenteric lymph node stromal cells correlated with induction of alternative NF-κB target genes. Thus, our data shed light on LTβR cellular trafficking as a process required for specific biological functions of NF-κB.
INTRODUCTION
The tumor necrosis factor receptor (TNFR)/tumor necrosis factor ligand (TNFL) superfamily forms a complex network of cytokines and receptors that are important for biological functions ranging from cell homeostasis and inflammation to lymphoid organ development (20). This family is subdivided according to structural features within the cytosolic tail such as the presence of a death domain (DD) and/or tumor necrosis factor receptor-associated factor (TRAF) binding sites (6, 11). The DD is involved in the recruitment of other death domain-containing proteins like FADD and TRADD, which connect the receptor to downstream signaling pathways leading to cell death or cell survival. Likewise, the recruitment of adaptor protein TRAF triggers both mitogen-activated protein kinase (MAPK) and NF-κB signaling pathways. Two consensus binding sites for TRAF2, (P/S/A/T)X(Q/E)E and PXQXXD, and one for TRAF3, PXQX(S/T), have been defined based on alignment of TNFR family members (11, 57). However, noncanonical TRAF binding sites have also been identified for other substrates, such as NF-κB-inducing kinase (NIK) or TRADD (30, 38). TRAF proteins connect TNFR to at least two NF-κB signaling pathways. The first, named the classical (or canonical) NF-κB pathway, is activated by most TNFRs. Our current understanding is that TRAF proteins are recruited directly or indirectly (e.g., through TRADD for TNFR1) to TNFR, which then recruits the IκB kinase (IKK) complex that is targeted for posttranslational modifications impinging on its catalytic activity (51). Among those, autophosphorylation of IKKβ takes place shortly after TNFR engagement (10), while the regulatory subunit NEMO (or IKKγ) seems to be modified by both K63 and linear polyubiquitination (22). Then, the IKK complex phosphorylates IκB proteins to trigger their proteasomal degradation and the release of related NF-κB dimers. The latter control many of the proinflammatory and antiapoptotic roles associated with NF-κB through binding to a specific cis regulatory region called the κB site (37, 39).
An additional biological outcome accounts for TNFR members by the fact that a subclass (e.g., TNFRI and HVEM) activates solely the classical NF-κB pathway whereas other TNFRs (e.g., lymphotoxin β receptor [LTβR] and CD40) activate both the classical and the alternative (or noncanonical) NF-κB pathways (8, 51). The alternative pathway involves the activation of the NF-κB-inducing kinase (NIK), which activates IKKα, and both phosphorylate the inhibitor p100, leading to its subsequent polyubiquitination and partial proteasomal degradation into p52 (46, 56). In particular, it was demonstrated that neither LTβR nor BAFF-R required NEMO or IKKβ for inducing p100 processing (7–9). Ultimately, NIK and IKKα activate the dimer p52/RelB, which controls a set of genes involved in secondary lymphoid organ development, B cell survival, and osteoclastogenesis (45, 53). Indeed, NIK- and IKKα-deficient mice share a panel of developmental abnormalities reminiscent of mice deficient in LTβR, BAFF-R, or RANK (13, 15, 25, 46, 47, 49, 58). Deregulation of the alternative NF-κB pathway has also been associated with malignancy. For instance, transgenic mice expressing inducers of the alternative pathway such as BAFF or LTα1β2 display lymphoid malignancies and hepatocellular carcinoma development, respectively (2, 18, 28). On the other hand, elevated expression of NIK and/or loss of expression of its negative regulators is a signature found in multiple myeloma and B cell lymphoma (1, 26, 42).
Thus, NIK appears to play a central role in many biological functions, but the molecular determinants that dictate its activation are still poorly characterized. The current model depicts TRAF3 as a bridge between TRAF2-associated c-IAP1/2 E3 ligase complex and the N-terminal domain of NIK promoting its constitutive K48-linked polyubiquitination and proteasomal degradation. Upon stimulation of CD40, TRAF3 is polyubiquitinated by c-IAP1/2 and degraded by the proteasome, allowing the stabilization and accumulation of NIK (30, 52, 60). Hence, TRAF3 recruitment has been proposed as a hallmark of the TNFR-induced alternative NF-κB pathway (17). However, HVEM, a TNFR that binds TRAF3, fails to activate the alternative pathway (5, 33). Thus, it is likely that the capacity to recruit TRAF3 is necessary but not sufficient for inducing the alternative NF-κB pathway.
Thus, the molecular mechanisms linking the outcome of TRAF-associated TNFR and the activation of p100 processing are far from being fully understood and need further biochemical and biological characterization. In this study, we have addressed how LTβR activates both the classical and the alternative NF-κB pathways. We found that activation of these two pathways is spatially and temporally regulated by LTβR trafficking.
MATERIALS AND METHODS
Plasmids, cloning, and mutagenesis.
Expression vectors and sequences of primers used for cloning and mutagenesis are available upon request. PCR amplification of cDNAs was performed with Goldstar DNA polymerase (Eurogentec), and mutagenesis of LTβR was performed using the QuikChange site-directed mutagenesis XL kit (Stratagene) according to the manufacturer's instructions.
Abs and reagents.
The following commercially available antibodies (Abs) were used for several applications: p100/p52 (05-361) and anti-ubiquitin Lys48-specific antibody (05-1307) from Millipore; phospho-p100 (4810), phospho-IκBα (9246), anti-NIK (4994), and Myc tag (2276) from Upstate Cell Signaling; antihemagglutinin (anti-HA) (sc-805), LTβR-N15 (sc-8375), TRAF2 H-249 (sc-7187), TRAF3 H-122 (sc-1828), TRAF3 H-20 (sc-948-G), TRAF5 H-257 (sc-7220), and clathrin heavy chain (CHC) (sc-12374) from Santa Cruz Biotechnology; actin (69100) from MP Biomedicals; HA (MMS-101R) from Covance; Flag M2 (F3165) and Beads Red anti-Flag M2 (F2426) from Sigma; h-LTβR (AF629) from R&D Systems; glutathione S-transferase (GST) tag (71097-3) from Novagen; EEA1 (610456); AP50 (612620) from BD Bioscience; anti-dynamin-2 (PA1-661) from Thermo Scientific; and anti-NIK (ab8111) from Abcam.
Anti-IκBα monoclonal antibody (MAb) 10 was a gift from Ron Hay (University of St. Andrews, St. Andrews, Scotland). Anti-murine LTβR, mICAM-1, mVCAM-1, mMAdCAM-1, CD45, and mp100/p52 were previously described (3, 9). The following reagents were purchased from the indicated companies: chloroquine (C6628), bafilomycin A1 (B1793), N-ethylmaleimide (E1271), and n-octyl-β-d-glucopyranoside (29836-26-8) from Sigma; dithiobis[succinimidyl propionate] (DSP) (22585) from Thermo Scientific; recombinant LTα1β2 (678-LY) from R&D Systems; digitonin (300410) from Calbiochem; and Topro-3 iodide from Invitrogen.
Mice and lymph node explants.
C57BL/6 (H-2b) and RelB−/− (H-2b) mice (backcrossed for more than 10 generations in a C57BL/6J background) (54) were bred and maintained under specific-pathogen-free conditions in the Biomedical Service Unit at the University of Birmingham according to Home Office and local ethics committee regulations. The day of vaginal plug detection was designated E0. Freshly isolated fetal lymph nodes were isolated at the indicated time and either analyzed directly or explanted in organ cultures and incubated for 72 h in the presence of an agonistic antibody to LTβR at 2 μg/ml (3).
Cell lines, transfection, and retroviral infection.
HEK 293, HEK 293T, HeLa, and HeLa Tet/On Advanced (Clontech) cells and LTβR knockout (KO) MEFs were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% antibiotics, and 1% l-glutamine. For transient transfections, cells were seeded at 70 to 80% confluence and transfected using Lipofectamine 2000 (Invitrogen) or Fugene HD (Roche Applied Sciences). For analyzing LTβR-mediated p100 processing into HEK 293T cells, 6-well plates were used and 100 ng of LTβR expression vector was transfected per well. Retrovirus was collected from the supernatant of Phoenix Ampho packaging cells transfected with pRetroX-Tight-HA-μ2-D176A/W421A or pBabe-puro-LTβR encoding wild type (wt) or ΔI 389–395, ΔI 345–358, or ΔI 359–368 mutant and used to infect HeLa Tet/On and HEK 293T cells or LTβR KO MEFs, respectively. Single clones were obtained after antibiotic selection, and expression of AP2μ2-DN (dominant negative) was achieved by adding 2 μg/ml of doxycycline for the indicated period.
siRNA knockdown experiments.
Knockdown experiments for targeting CHC or Dyn2 in HeLa cells were carried out with On Target Plus Smart pools M-004001 and L-004007, respectively (Dharmacon). The experiments were performed according to the protocol described by Motley et al. with minor modifications (36). HeLa cells were transfected with either small interfering RNA (siRNA) green fluorescent protein (GFP) as negative control or siRNA CHC. For the first round of transfection, cells were plated in 6-cm dishes and incubated for 4 h in serum/antibiotic-free medium in the presence of 12 μl Oligofectamine (Invitrogen) and 120 pmol of siRNA. Cells were trypsinized 24 h posttransfection and seeded into a 10-cm dish. After the second round of transfection on day 3, the cells were split into 6-well plates for further analyses on day 5. Knockdown experiments for targeting human AP2μ2 in HeLa Tet/On rat AP2μ2-DN were performed with two siRNAs (GCGAGAGGGUAUCAAGUAU and AGUUUGAGCUUAUGAGGUA) designed to efficiently target human AP2μ2 but not rat AP2μ2-DN (J-008170; Dharmacon). The cells were transfected twice, on day 1 and day 3, with siRNA AP2μ2 or with GFP siRNA as negative control (Dharmacon). Doxycycline was added at 2 μg/ml in the medium over the 5 days of the experiment.
Immunoblotting, IP, and cell extract fractionation.
Whole-cell extracts were prepared by lysing cells in 0.5% SDS containing 5 mM sodium fluoride (NaF), 1 mM sodium orthovanadate (Na3VO4), 20 mM β-glycerolphosphate (βGP), and Complete protease inhibitor (Roche). Cell extracts were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Millipore) followed by incubation with specific primary antibodies and secondary horseradish peroxidase (HRP)-conjugated antibodies (Dako). For coimmunoprecipitation (co-IP) assays, cells were analyzed after stimulation with LTβR antibody or 40 h posttransfection. Cells were lysed in TNT buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 0.5 to 1% Triton X-100, 5 mM NaF, 1 mM Na3VO4, 20 mM βGP, and Complete protease inhibitor). After a preclearance of 1 h with protein A agarose beads, cell lysates were incubated overnight with the indicated antibodies and with protein A or G agarose beads for an additional hour. For double immunoprecipitation (IP), an excess of 3× Flag peptide was used overnight to release the Flag-immunoprecipitated material and a second immunoprecipitation was carried out with either an anti-HA or an anti-Myc antibody. The agarose beads were washed four times with TNT buffer before addition of 4× loading buffer. Samples were then subjected to SDS-PAGE. In order to determine the presence of LTβR mutants in detergent-insoluble fraction, HEK 293T cells were lysed 40 h posttransfection in 0.5% Triton-TNT buffer for 15 min on ice. Supernatants corresponding to detergent-soluble fraction were isolated after centrifugation at 13,200 rpm for 15 min. The pellets corresponding to the detergent-insoluble fraction were resolubilized by sonication in octylglucoside-containing buffer (60 mM n-octyl-β-d-glucopyranoside, 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.1% SDS, 5 mM NaF, 1 mM Na3VO4, 20 mM βGP, and Complete protease inhibitor). After incubation for 30 min on ice, the detergent-insoluble lysates were clarified by centrifugation at 13,200 rpm for 15 min at 4°C. Samples were analyzed by immunoblotting for LTβR expression.
For studying the formation of K48-linked polyubiquitination, we transfected 293T cells. Thirty-six hours posttransfection, cells were incubated for 6 h with the proteasome inhibitor MG132 prior to lysis under denaturing conditions with 0.5% SDS. Following clearance of cell extracts and dilution up to 0.1% (final concentration) SDS, we performed two sets of immunoprecipitations for assessing the K48-linked polyubiquitination of both TRAF3 and NIK. Cells transfected with NIK, TRAF2, TRAF3, and HA-ubiquitin or the same combination in addition to LTβR ΔS were lysed, and equal amounts of cell extracts were used for immunoprecipitation with either control IgG or anti-TRAF3 antibodies (1st IP). Following incubation with protein A agarose beads, the supernatants were kept for a second round of immunoprecipitation (2nd IP) whereas the immunoprecipitates were loaded on a Western blot for the detection of TRAF3 and K48-linked polyubiquitinated TRAF3. On the other hand, the second set of immunoprecipitations (2nd IP) was carried out with supernatants from the first immunoprecipitation experiment (1st IP) by incubation with either control beads or Flag-M2 beads. The immunoprecipitates were then used for detecting Flag-NIK and K48-linked polyubiquitinated NIK. Quantification of signals was performed with the Image J software.
Oligomerization of LTβR and GST pulldown.
For studying the ability of LTβR to form trimers, we performed transient transfections of HEK 293T cells with a combination of HA-, Flag-, and Myc-tagged LTβR expression vectors. A first immunoprecipitation was performed using Flag-M2 agarose beads. The immunoprecipitated material was then selectively released overnight by competition with an excess of 3× Flag peptide (Sigma). The supernatants were subjected to a second immunoprecipitation with either an anti-HA or an anti-Myc antibody for 2 h at 4°C. The immunoprecipitated materials were analyzed by immunoblotting with an anti-Myc or an anti-HA antibody, respectively. To analyze aggregation of ectopic LTβR, HEK 293T cells were collected 40 h posttransfection, washed twice with phosphate-buffered saline (PBS), and then incubated for 30 min at room temperature in PBS containing 1 mM dithiobis[succinimidyl propionate] (DSP; Pierce/Thermo Fisher Scientific). After cross-linking, the cells were washed once in PBS and incubated for 15 min in 20 mM Tris-HCl (pH 7.4)–PBS to stop the reaction. The cells were scraped in SDS buffer, and protein extracts were analyzed by immunoblotting for LTβR expression. pGEX-4T1-wt LTβR or mutant expression vectors were transformed into Escherichia coli BL21. Bacterial cultures were grown to an A600 of 0.6 and were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. After washing in PBS, the bacterial pellets were sonicated in NENT buffer (250 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8, 1.5% Nonidet P-40). The lysates were cleared by centrifugation and incubated with glutathione-Sepharose 4B beads for 1 h at 4°C. After three washes in NENT buffer, the beads were collected in TWB buffer (20 mM HEPES, pH 7.9, 60 mM NaCl, 1 mM dithiothreitol [DTT], 6 mM MgCl2, 8.2% glycerol, 0.1 mM EDTA). Expression of GST fusion proteins was estimated by SDS-PAGE and Coomassie blue staining. 35S labeling of TRAF proteins was generated in vitro using the TNT kit from Promega. For GST fusion protein interactions with 35S-TRAF proteins, an aliquot of glutathione-Sepharose beads containing 1 μg of GST fusion protein was incubated with 7.5 μl of in vitro-translated TRAF for 1 h at 4°C in NENT buffer. Beads were washed five times in NENT buffer and then boiled in SDS loading buffer for 5 min. Bound proteins were analyzed by SDS-PAGE and then by autoradiography. For analyzing the binding of overexpressed mammalian TRAF proteins, cell lysates were prepared from transfected 293T cells in TGH buffer (50 mM HEPES, pH 7.4, 1% Triton X-100, 10% glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitors). One microgram of GST fusion protein was incubated with 150 μg of cell lysates for 3 h at 4°C. Beads were washed five times in TGH buffer and then boiled in SDS loading buffer for 5 min. Bound proteins were analyzed by SDS-PAGE and immunoblotting.
Flow cytometry.
LTβR-expressing HEK 293 cells were analyzed 2 days posttransfection. Cells were fixed in 1% paraformaldehyde, permeabilized or not with 0.5% saponin, and incubated for 30 min with an anti-LTβR antibody. After washing in PBS-bovine serum albumin (BSA), cells were stained with a goat anti-LTβR and a donkey anti-goat antibody–phycoerythrin (PE). To study the internalization of LTβR in MEFs or in HT29 cells, the cells were stimulated with 2 μg/ml of agonistic antibody to murine LTβR or 0.2 μg/ml of agonistic antibody to human LTβR and were incubated for the indicated period of time. After collection of the cell, the staining of cell surface and internalized LTβR was achieved with the same agonistic antibody followed by incubation with PE-labeled goat anti-rat secondary antibody or PE-labeled donkey anti-goat antibody. Samples were analyzed on a FACSCanto II flow cytometer (BD Biosciences), and at least 10,000 events were acquired for each sample using FACSDiva6 software (BD Biosciences). Staining of mesenteric lymph node (mLN) stromal cells was performed as previously described (3).
Immunofluorescence microscopy.
Transfected HEK 293T or HeLa cells were grown on glass coverslips. Two days posttransfection, cells were fixed in 4% paraformaldehyde for 30 min and permeabilized with 0.5% saponin-PBS for 20 min. 293T cells were then incubated for 1 h with anti-human LTβR antibody and subsequently incubated for 50 min with a donkey anti-goat antibody–Alexa 546 (Invitrogen). Nuclei were stained with Topro-3 iodide (Invitrogen).
Transfected HeLa cells with HA-tagged wt LTβR, ΔS-LTβR, and ΔS ΔI 345–358 LTβR were stained with anti-HA sc-805 at a 1/50 dilution and an anti-rabbit antibody–Alexa 546, and nuclei were visualized with 4′,6-diamidinophenylindole (DAPI). For the colocalization of LTβR and endosomes, staining was performed on HT29 cells treated with the agonist antibody to LTβR. Following the indicated kinetic of stimulation, cells were fixed for 20 min with 2% of p-formaldehyde and permeabilized for 15 min at room temperature with 50 μM digitonin. LTβR was detected with agonist primary antibody and a donkey anti-goat secondary antibody–Alexa 546. Early endosomes were detected with a mouse monoclonal anti-EEA1 antibody and a donkey anti-mouse-Alexa 488 secondary antibody.
Localization of endogenous TRAF3/NIK complex was achieved using the Duolink II Orange kit (Eurogentec, Belgium). Anti-rabbit plus and anti-goat minus antibodies were used as secondary antibodies for recognition of anti-TRAF3 H-20 (sc-948-G; Santa Cruz Biotechnology) and anti-NIK (ab8111; Abcam), and polymerase reaction was performed for 150 min. Nuclei were visualized with DAPI. Confocal microscopy was performed with either a Leica TCS SP2 microscope or an Olympus FV1000 microscope.
RESULTS
Molecular determinants of LTβR involved in induction of p100 processing.
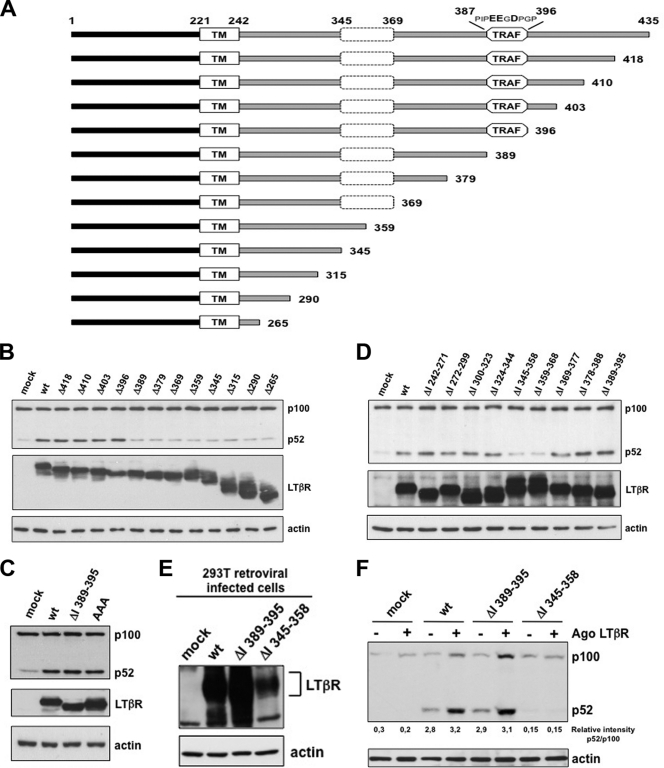
It has been previously shown that overexpression of LTβR is sufficient to activate NF-κB in a ligand-independent way (14). We constructed a panel of deletion mutants of the cytosolic tail of LTβR and evaluated their ability to induce the processing of p100 (Fig. 1 A). All deletion mutants expressing at least the first 395 amino acids induced the processing of p100, whereas the shorter mutants did not (Fig. 1B). Thus, removal of the six residues upstream of amino acid 389 was sufficient to prevent LTβR-induced p100 processing. This region was characterized as a TRAF binding region involved in NF-κB activation (14). We narrowed down the critical region up to amino acid 392 (see Fig. S1A in the supplemental material). Surprisingly, alteration of acidic amino acids crucial for the recruitment of TRAF proteins into alanine (29) or internal deletion from amino acid 389 to 395 did not abrogate LTβR-induced p100 processing (Fig. 1C). Titration of wt LTβR and LTβR ΔI 389–395 expression level indicated that this TRAF binding site was dispensable for the induction of p100 processing (see Fig. S1B). These results suggested the requirement of another region upstream of amino acid 392 for LTβR-mediated p100 processing. Bioinformatics analysis of the cytosolic tail revealed another putative TRAF3 binding site, PQQQS, at position 319 to 323 matching the consensus PXQX(S/T). However, the substitutions Q321A and S323A did not inhibit LTβR-induced p100 processing (see Fig. S1C). By generating additional internal deletions, we identified two mutants, named ΔI 345–358 and ΔI 359–368, missing critical regions for the processing of p100 (Fig. 1D). Additional shorter internal deletions along the region 345 to 368 displayed an alteration of LTβR-induced p100 processing to the same extent as ΔI 345–358 and ΔI 359–368 mutants (see Fig. S1D). In order to confirm our results under physiological conditions, we generated 293T clones stably expressing wt and mutant LTβR for which ligand inducibility was maintained (Fig. 1E). While agonistic anti-LTβR treatment allowed the induction of p100 processing in wt and ΔI 389–395 LTβR-expressing cells, cells expressing ΔI 345–358 LTβR failed to respond (Fig. 1F and data not shown). Similar results were obtained when we complemented LTβR KO MEFs with the same expression vectors (see Fig. S1E and F).
Fig. 1.
Identification of the cytoplasmic domain involved in LTβR-mediated p100 processing. (A) Schematic representation of human full-length and deletion mutant LTβR. The black bar, the gray bar, and TM represent the extracellular domain (amino acids 1 to 121), the cytosolic tail (amino acids 242 to 425), and the transmembrane domain (amino acids 222 to 241), respectively. (B to D) HEK 293 cells were transiently transfected with different sets of LTβR deletion mutants of the cytosolic tail and with the triple D390D391E393/AAA mutant. (E) 293T cells were stably infected with retrovirus encoding the indicated Myc-tagged LTβR, and expression was analyzed with anti-Myc immunoblotting. (F) Stable clones were stimulated for 6 h with an agonistic (Ago) anti-LTβR antibody prior to analysis by Western blotting of the processing of p100 into p52.
Conformation of LTβR dictates the avidity of its bipartite binding sites toward specific TRAF proteins.
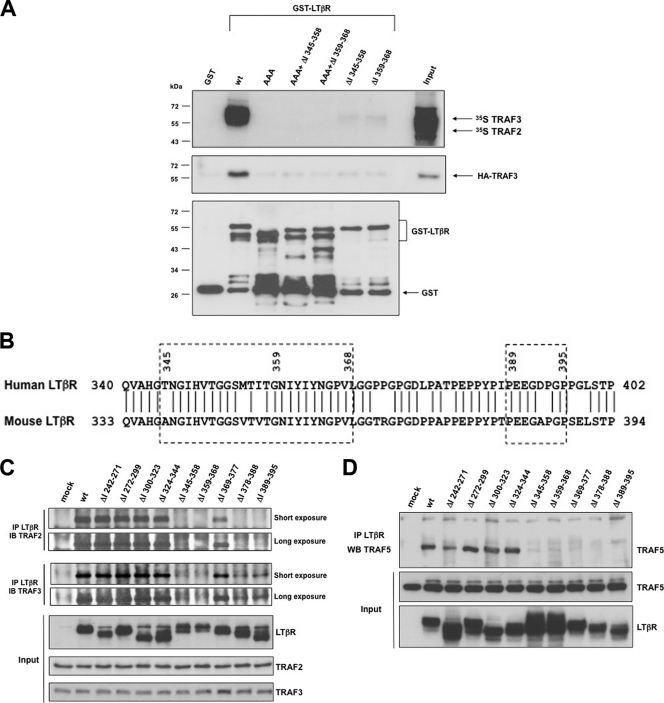
So far, a single TRAF binding site has been characterized and located at position 387 to 396 of human LTβR (14, 29). However, we demonstrated that this region was dispensable for the induction of the alternative NF-κB pathway (Fig. 1C). Therefore, we hypothesized that LTβR may contain another TRAF binding site required for the induction of p100 processing. We analyzed the recruitment of in vitro-translated 35S-TRAF2 and 35S-TRAF3 with recombinant GST-LTβR. We detected TRAF association with wt GST-LTβR but not with the mutant GST-LTβR-AAA, which contains point mutations disrupting TRAF recruitment within the region 387 to 396 (29) (Fig. 2 A). Similarly, both GST-ΔI 345–358 and GST-ΔI 359–368 failed to bind in vitro-translated TRAF proteins or HA-TRAF3-containing 293 cell extracts (and TRAF2) (data not shown). Thus, the amino acid stretch 345 to 368, similarly to the region 387 to 396 of human LTβR, is required for the binding of TRAF proteins when expressed as GST fusion proteins. Analysis of amino acid content of the region 345 to 368 did not reveal any conserved consensus TRAF binding site within either mouse or human LTβR (Fig. 2B). When we overexpressed wt and mutant LTβR into 293 cells, we observed that disruption of the region 389 to 395, 378 to 388, 359 to 368, or 345 to 358 completely abolished the recruitment of TRAF2 and TRAF5. However, removal of the regions 389 to 395 and 378 to 368 did affect the recruitment to TRAF3, but a significant residual binding was still observed despite this deletion (Fig. 2C and D). Hence, on one hand, when the cytosolic tail of LTβR is fused to GST, any mutation or deletion within one of the two TRAF binding sites is sufficient to abrogate the binding of TRAF2 or TRAF3. On the other hand, when full-length LTβR is expressed in 293 cells, the region 345 to 358 is sufficient for the recruitment of TRAF3 and the induction of p100 processing.
Fig. 2.
Conformation of LTβR dictates the mechanism of TRAF recruitment through an unconventional bipartite site. (A) GST pulldown of either in vitro-translated TRAF2 and TRAF3 or 293 cell extracts containing HA-TRAF3 with GST-LTβR and the indicated mutants. (B) Alignment of human and mouse LTβR. The two TRAF binding sites are boxed with dashed lines. (C and D) HEK 293T cells were transfected with internal deletion mutant LTβR, and immunoprecipitated LTβR was analyzed by Western blotting for the recruitment of TRAF2 and TRAF3 (C) or ectopic Flag-TRAF5 (D).
Internalization of LTβR is required to induce the processing of p100.
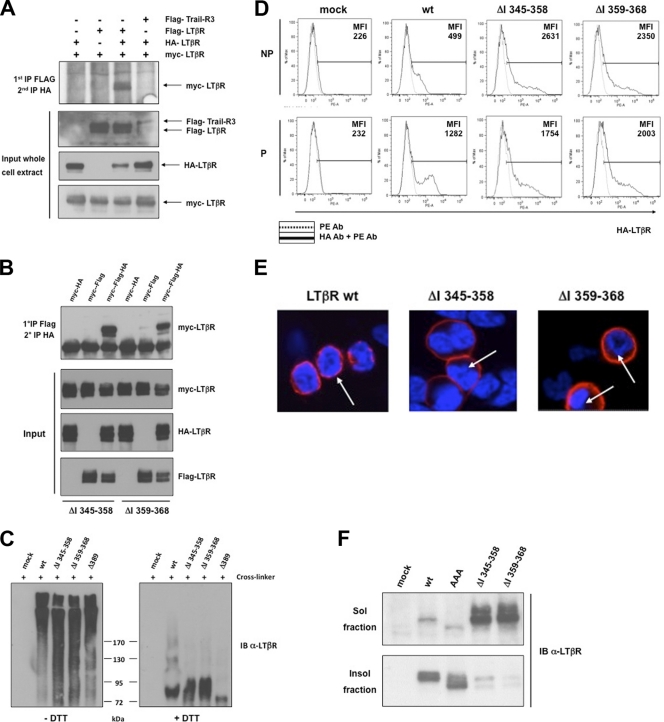
Trimerization of TNFR is a prerequisite for triggering their downstream pathways. Thus, we addressed whether the LTβR mutants deficient for the induction of p100 processing were able to trimerize. We transfected 293 cells with three differently tagged wt or mutant LTβR ΔI 345–358 and ΔI 359–368 constructs (Flag, HA, and Myc). The resulting cell lysates were subjected to a double immunoprecipitation (IP) procedure (see Materials and Methods), and the immunoprecipitated materials were analyzed by Western blotting for expression of the third tagged LTβR. Under those conditions, wt LTβR was able to trimerize (Fig. 3 A; see also Fig. S3A in the supplemental material). Interestingly, mutants LTβR ΔI 345–358 and ΔI 359–368 were still able to form trimeric complexes, while other TNFR-related proteins, such as Trail-R3 and TNFRII, did not coimmunoprecipitate (co-IP) in single-IP or double-IP procedures (Fig. 3B; see Fig. S3B). We also analyzed the ability of wt LTβR and mutants ΔI 345–358, ΔI 359–368, and Δ389 to form aggregates upon overexpression in 293 cells. Thirty-six hours posttransfection, cells were cross-linked with a membrane-permeable cross-linker (DSP) and LTβR aggregation was analyzed by Western blotting in the absence or presence of DTT (reversed cross-link). All mutants defective for p100 processing were as efficient as wt LTβR in forming high-molecular-weight aggregates (Fig. 3C). Overall, our data showed that deletions ΔI 345–358 and ΔI 359–368 did not affect the ability of LTβR to multimerize. Meanwhile, when we monitored by flow cytometry the outcome of cell surface expression of wt and mutants ΔI 345–358 and ΔI 359–368 in both nonpermeabilized (NP) and permeabilized (P) 293 transfected cells, we observed striking differences. As expected, a pool of wt LTβR was detected at the cell surface of nonpermeabilized 293 cells. However, the intensity of fluorescence was elevated in permeabilized cells, indicating that part of wt LTβR was present within an intracellular compartment (Fig. 3D). In contrast, both LTβR mutant ΔI 345–358 and mutant ΔI 359–368 were exclusively localized at the cell surface, since no further increase of fluorescence intensity was detected upon cell permeabilization. We confirmed by confocal microscopy that wt LTβR displayed a perinuclear staining whereas both mutant ΔI 345–358 and mutant ΔI 359–368 were exclusively seen in the plasma membrane (Fig. 3E). We also analyzed by Western blotting the cellular distribution into soluble and insoluble Triton X-100 fractions of active wt LTβR and LTβR-AAA and inactive ΔI 345–358 and ΔI 359–368 LTβR, for the processing of p100. Our results revealed that internalized wt LTβR and LTβR-AAA were mainly present in the insoluble fraction while the mutants ΔI 345–358 and ΔI 359–368 were mainly found in the soluble fraction (Fig. 3F).
Fig. 3.
LTβR defective for p100 processing is sequestered into the plasma membrane. (A) HEK 293T cells were transfected with three differently tagged LTβRs (HA, Flag, and Myc tagged), and double immunoprecipitations were performed to analyze the trimerization of wt LTβR (see the supplemental material for details). (B) The same procedure as in panel A was applied for internal deletion mutants ΔI 345–358 and ΔI 359–368. (C) HEK 293T cells were transfected with wt LTβR, ΔI 345–358, ΔI 359–368, and Δ389. The cross-linker DSP was used prior to immunoprecipitation and immunoblotting of LTβR under nonreduced (−DTT) and reduced (+DTT) conditions. (D) Flow cytometry analysis of HEK 293 cells mock transfected or transfected with expression vector for wt LTβR, LTβR ΔI 345–358, or LTβR ΔI 359–368 and stained for cell surface LTβR (nonpermeabilized [NP]) or cell surface and intracellular LTβR (permeabilized [P]). MFI (mean fluorescence intensity) represents the value of one measurement out of three independent experiments. (E) Localization of wt LTβR, LTβR ΔI 345–358, and LTβR ΔI 359–368 in HEK 293 cells. Arrows indicate the perinuclear compartment. (F) Cell fractionation of LTβR into Triton X-100-soluble and -insoluble fractions from HEK 293 cells transfected with the indicated LTβR constructs.
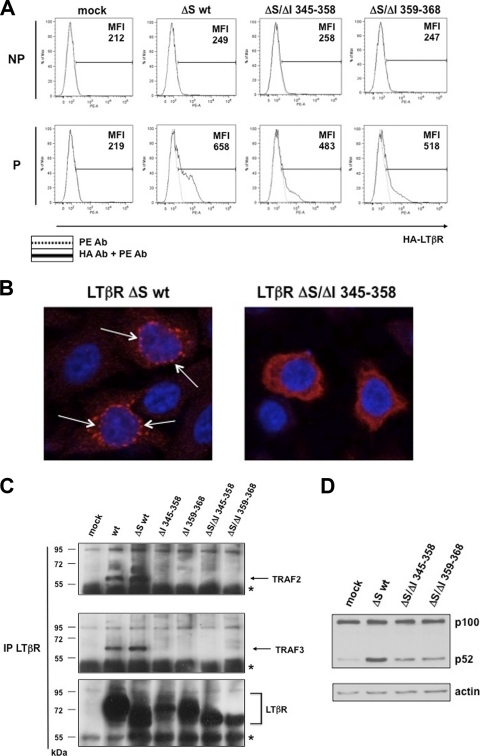
We next explored the ability of LTβR to recruit TRAF2 and TRAF3 from the intracellular compartment. We first constructed signal sequence-deficient (ΔS) expression vectors for wt LTβR, ΔI 345–358, and ΔI 359–368 and checked their cellular localization in nonpermeabilized (NP) and permeabilized (P) cells. In all cases, we detected LTβR expression only in permeabilized cells, confirming the inability of LTβR to migrate to the cell surface (Fig. 4 A). We then looked at the cellular location of these signal sequence-defective mutants by confocal microscopy. We observed a punctate staining of LTβR ΔS wt mainly localized in the perinuclear compartment. However, the staining was largely diffuse when the region 345 to 358 of LTβR was absent, indicating that this stretch of the receptor directs its specific intracellular location (Fig. 4B). We then showed that LTβR ΔS wt could recruit endogenous TRAF2 and TRAF3 as wt LTβR. However, the recruitment was impaired in the absence of the region 345 to 358 or 359 to 368 irrespective of the presence of the signal sequence (Fig. 4C). These results highlight a dual role of the region 345 to 368 of LTβR in mediating internalization from the plasma membrane (Fig. 3D and E) and TRAF recruitment from an internal cellular compartment. In addition, we observed that the ability of LTβR ΔS wt to induce the processing of p100 was severely impaired for the mutants ΔS/ΔI 345–358 and ΔS/ΔI 359–368 (Fig. 4D).
Fig. 4.
Perinuclear location of LTβR is a prerequisite for the recruitment of endogenous TRAF proteins and induction of p100 processing. (A) Flow cytometry analysis of HEK 293 cells mock transfected or transfected with expression vector for LTβR ΔS wt, LTβR ΔS/ΔI 345–358, or LTβR ΔS/ΔI 359–368 and stained for cell surface LTβR (nonpermeabilized) or cell surface and intracellular LTβR (permeabilized). MFI (mean fluorescence intensity) represents the value of one measurement out of three independent experiments. (B) HeLa cells transiently transfected with the indicated HA-tagged construct were stained for LTβR (in red) and nuclei (DAPI). Arrows indicate the punctate perinuclear staining of LTβR. (C) HEK 293 cells were mock transfected or transfected with LTβR expression vectors encoding either wt, ΔI 345–358, ΔI 359–368, or their signal sequence (ΔS)-defective counterpart. Immunoprecipitated LTβR was analyzed by Western blotting for the recruitment of endogenous TRAF2 and TRAF3. The asterisks represent the cross-reactivity with Ig heavy chains. (D) Extracts from cells transfected with signal sequence (ΔS)-defective mutants were used to analyze the processing of p100 by Western blotting.
Overall, our results reveal that internalization of LTβR and binding of intracellular TRAF2 and TRAF3 are a prerequisite for the induction of p100 processing.
Internalization of LTβR in vivo correlates with induction of the alternative NF-κB pathway.
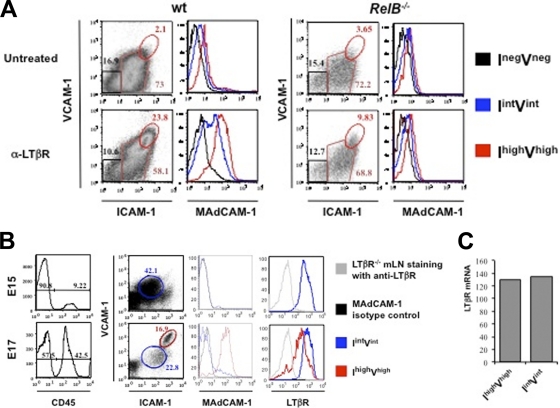
We next addressed the physiological relevance of our findings in biological settings for which an LTβR-induced alternative pathway is known to play a role through production of p52/RelB. Therefore, we isolated mesenteric lymph nodes (mLNs) from embryonic day 14 (E14) from wt and RelB-deficient mice and explanted them ex vivo in fetal organ culture. At that stage, mLN stromal cells had not yet fully matured and most CD45− cells were ICAM-1int VCAM-1int (3). However, treatment of wt explants with an agonistic antibody to LTβR for 3 days allowed ICAM-1int VCAM-1int cells to commit into ICAM-1high VCAM-1high mature stromal “organizer” cells (Fig. 5 A). During the maturation process, these cells expressed MAdCAM-1 at a high level. Interestingly, this transition was abrogated in mLNs from RelB−/− embryos. Thus, the absence of LTβR-mediated MAdCAM-1 upregulation in RelB−/− stromal cells revealed that MAdCAM-1 expression was a reliable readout for the activation of the alternative pathway ex vivo. Therefore, we extended our analyses in vivo by collecting mLNs from wt embryos at day E15 and day E17. At day E15, we detected ICAM-1int VCAM-1int cells but barely any ICAM-1high VCAM-1high cells (Fig. 5B). Nevertheless, at day E17, part of the pool of ICAM-1int VCAM-1int cells committed to the ICAM-1high VCAM-1high phenotype. Despite the fact that both populations expressed LTβR, only ICAM-1high VCAM-1high cells were positive for MAdCAM-1. In particular, LTβR cell surface expression in the ICAM-1high VCAM-1high MAdCAM-1+ cell population was reduced compared to that in ICAM-1int VCAM-1int MAdCAM-1− cells. However, this decrease was not due to a down-modulation of LTβR transcript since the mRNA levels of expression were similar in ICAM-1int VCAM-1int and ICAM-1high VCAM-1high cells (Fig. 5C). We conclude from these experiments that the activa tion of the alternative NF-κB pathway in vivo correlates with a downregulation of LTβR cell surface expression.
Fig. 5.
Internalization of stromal LTβR correlates with induction of MAdCAM-1. (A) RelB is necessary for a sustained induction of MAdCAM-1 expression in E14 mLN organ culture treated with an agonistic antibody to LTβR. Flow cytometry analysis of single cell suspensions of wt (left panel) and RelB−/− (right panel) E14 mLNs in organ cultures for 3 days. Percentages shown in histograms correspond to CD45− stromal cells. Three different stromal cell populations were gated: ICAM-1neg VCAM-1neg (Ineg Vneg), Iint Vint, and Ihigh Vhigh. Expression levels of MAdCAM-1 for each cell population are shown in histograms. (B) Emergence of the Ihigh Vhigh stromal organizer cells in mLNs. Flow cytometry analysis of single cell suspensions of mLNs at E15 and E17 showing the recruitment of CD45+ hematopoietic cells and the concomitant phenotypic changes in the CD45− stromal cells. Percentages shown in the histogram correspond to CD45− stromal cells and CD45+ hematopoietic cells. At E15, the stromal cell population expressed low levels of ICAM-1 and VCAM-1 (Iint Vint cell population). These cells expressed LTβR but were MAdCAM-1 negative. At E17, the stromal cells have matured and some cells express high levels of ICAM-1 and VCAM-1 (Ihigh Vhigh cell population, in red) and MAdCAM-1 but downregulated LTβR cell surface expression. (C) The Iint Vint and Ihigh Vhigh cell populations expressed the same levels of LTβR mRNA. Iint Vint and Ihigh Vhigh stromal cell populations of mLNs at E18 were sorted and analyzed for LTβR mRNA expression by real-time PCR. Ratios of the gene of interest to the β-actin gene are shown. Results are representative of at least 3 independent experiments.
LTβR-induced stabilization of intracellular NIK and activation of the alternative NF-κB pathway rely on dynamin-2 GTPase activity.
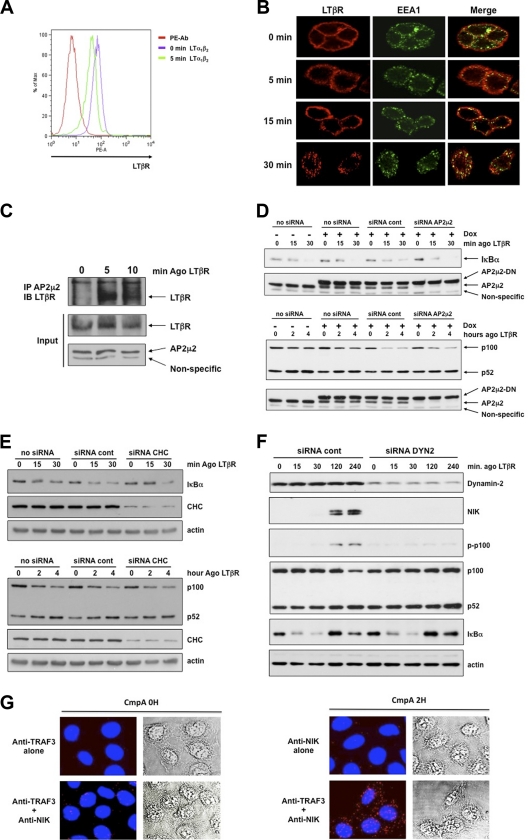
We further analyzed the mechanisms that control the internalization of endogenous LTβR after binding of its natural ligands. First, we performed flow cytometry analyses on human epithelial colon carcinoma HT29 cells and observed that the level of cell surface LTβR dropped as soon as 5 min after treatment with LTα1β2 or LIGHT (Fig. 6 A and data not shown). This early internalization of ligand-bound LTβR was conserved across species, since treatment of MEFs with an agonistic antibody resulted in a similar pattern of LTβR internalization (see Fig. S6A in the supplemental material). We confirmed the internalization of activated LTβR by performing confocal analyses of HT29 cells treated for different periods of time with an agonistic antibody to LTβR. LTβR was associated with a shift from a cell membrane staining (untreated cells) to a punctate pattern (LTβR stimulated), while the early endosome-associated protein EEA1 staining showed the typical punctate profile (Fig. 6B). We observed that, over time, activated LTβR accumulated in early endosomes and colocalized with EEA1. Several routes of internalization converge to early endosomes, and a role for AP2/clathrin complexes was highlighted for TNFR members such as TNFR1 or Fas (44). Interestingly, we found that LTβR is a bona fide partner of the adaptor complex AP2. Indeed, we observed that activation of LTβR led to a fast recruitment of the AP2μ2 subunit, which was almost concomitant with TRAF3 recruitment (Fig. 6C; see also Fig. S6B). This result prompted us to analyze the connection between the AP2μ2 and LTβR-mediated NF-κB activation. Therefore, we analyzed the responsiveness of LTβR-activated HeLa cells transfected with either siRNA control or siRNA AP2μ2. Despite a potent inhibition of endogenous AP2μ2 expression combined with expression of a dominant negative rat AP2μ2, neither IκBα degradation nor p100 processing was altered following LTβR stimulation (Fig. 6D). Similar results were obtained with siRNA CHC (Fig. 6E). Hence, LTβR-mediated p100 processing appeared to be AP2/clathrin independent.
Fig. 6.
Dynamin-dependent internalization of LTβR is required for the activation of the alternative but not the classical NF-κB pathway. (A) Flow cytometry analysis of cell surface LTβR expression in untreated and LTα1β2-treated HT29 cells. (B) Colocalization of LTβR and EEA1 to early endosomes. HT29 cells were stimulated with an agonistic antibody to LTβR for the indicated time prior to immunostaining. (C) Recruitment of endogenous AP2μ2 subunit to immunoprecipitated LTβR. (D) Inducible (+ Dox [doxycycline]) rat dominant negative (DN) AP2μ2-expressing HeLa cells transfected with control or siRNA AP2μ2 and stimulated with an agonistic anti-LTβR antibody. (E) HeLa cells transfected with siRNA clathrin heavy chain (CHC) and treated as indicated for the assessment of IκBα degradation and p100 processing. (F) The same cells were transfected with siRNA dynamin-2 and treated as in panel E for the analysis of classical and alternative NF-κB pathways. (G) HeLa cells were treated with the Smac mimetic CmpA prior to staining with the Duolink technology and the indicated antibodies. The endogenous NIK/TRAF3 complex appears in red, and nuclei appear in blue (DAPI).
Since dynamin participates in clathrin-dependent as well as clathrin-independent endocytosis, we extended our analysis by using siRNA dynamin-2. Under these conditions, LTβR-induced NIK stabilization, phosphorylation, and depletion of p100 were completely abrogated (Fig. 6F). Conversely, the activation of the classical pathway remained intact since the pattern of IκBα degradation was similar. Thus, although depletion of dynamin-2 does not prevent recruitment of signaling proteins to the cytosolic tail of LTβR for the induction of the classical NF-κB pathway, the receptor is not able to alleviate the constitutive degradation of NIK. Thus, we speculated that NIK must be kept in check within an intracellular compartment. To address this question, we used a method that allowed us to observe endogenous NIK/TRAF3 within the cell. Since NIK is constantly degraded by c-IAP1/2, we first pretreated or not HeLa cells with the Smac mimetic compound A (CmpA) for 2 h. This approach allowed us to stabilize NIK without disrupting its binding to TRAF3. Then, using the Duolink technology, we observed that endogenous NIK/TRAF3 complex was physically localized within punctate cytosolic bodies (Fig. 6G). Thus, it is likely that dynamin-2 participates in the transport of activated LTβR in close proximity to NIK/TRAF3 bodies to allow TRAF3 recruitment and activation of the alternative NF-κB pathway.
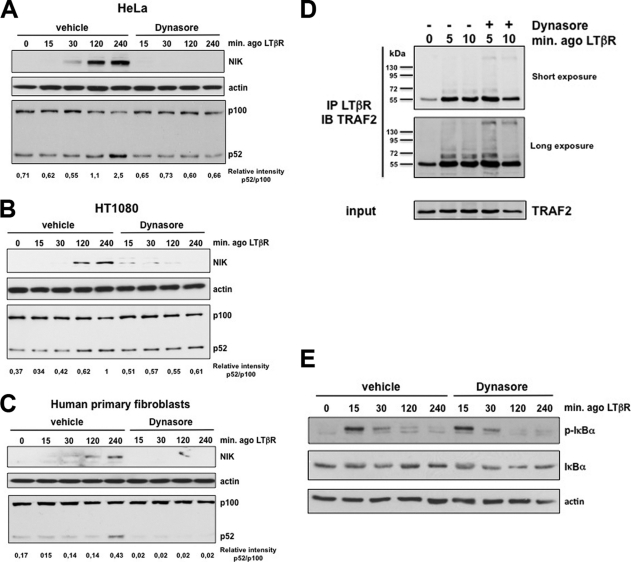
We next extended our analyses by using a noncompetitive inhibitor of the GTPase activity of dynamin, named Dynasore (32). We observed that preincubation of HeLa cells with Dynasore fully abrogated the stabilization of NIK and the processing of p100 in response to LTβR stimulation (Fig. 7A). These observations could be extended to other cell lines as well as to human primary fibroblasts (Fig. 7B and C).
Fig. 7.
The GTPase activity of dynamin-2 is involved in the induction of the processing of p100 in response to activated LTβR. (A) HeLa cells were incubated with DMSO (vehicle) or Dynasore (80 μM) 1 h before stimulation with the agonist (ago) to LTβR for the indicated time period. Total cell extracts were analyzed by immunoblotting for the indicated proteins. (B and C) Experiments were conducted as for panel A with HT1080 and human primary fibroblasts, respectively. (D) HeLa cells were pretreated or not with Dynasore for 1 h followed by a brief stimulation with the ago to LTβR. Cell extracts were immunoprecipitated with an anti-LTβR antibody prior to immunoblotting with anti-TRAF2. (E) The same extracts as in panel A were analyzed for the phosphorylation and degradation of IκBα.
Conversely, Dynasore inhibited neither the early phase of TRAF2 recruitment to activated LTβR nor the activation of the classical pathway since IκBα phosphorylation and degradation were similar in control and Dynasore-treated cells (Fig. 7D and E).
Altogether, these results confirmed our findings using the genetic approach with siRNA and revealed that the GTPase activity of dynamin-2 is crucial for the induction of the alternative NF-κB pathway.
TRAF3 degradation is secondary to LTβR-mediated p100 processing.
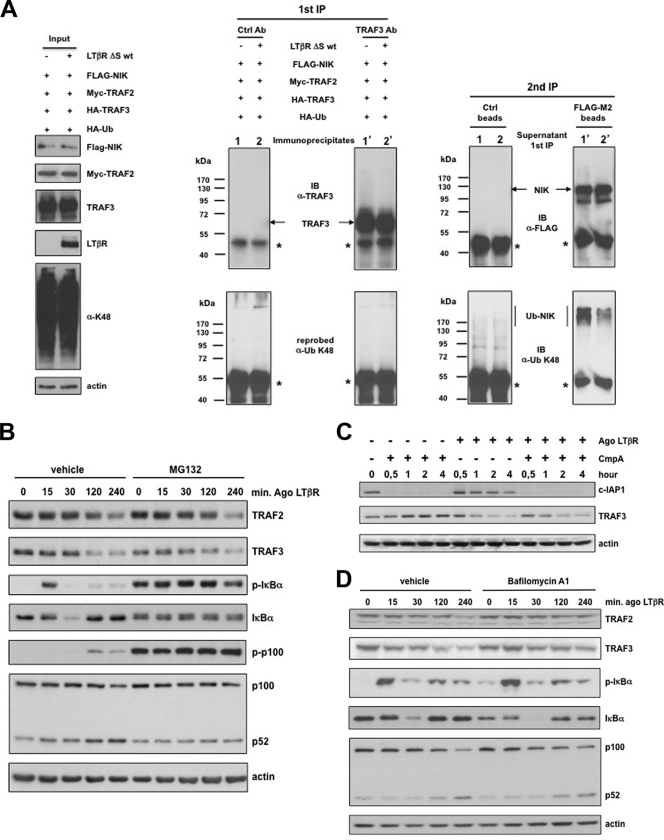
The current model is that TRAF3 recruitment and polyubiquitination by c-IAP1/2 occur at the cytoplasmic membrane-anchored receptor, leading to subsequent proteasomal TRAF3 degradation and NIK-induced p100 processing (52, 60). However, we observed that LTβR internalization is absolutely required for the activation of the alternative NF-κB pathway. Thus, we hypothesized that TRAF3 might be targeted for K48 polyubiquitination within the intracellular compartment. We established a system in which TRAF2, TRAF3, NIK, and HA-tagged ubiquitin were transiently coexpressed into 293T cells in the absence or presence of LTβR ΔS wt. Following blockade of the proteasome, we were able to efficiently immunoprecipitate, from SDS-denatured and renatured protein extracts, equal amounts of TRAF3 and NIK, in the absence and presence of LTβR ΔS wt (Fig. 8A). However, when we probed the same membranes with an anti-ubiquitin K48 antibody, we did not detect any K48-linked polyubiquitinated TRAF3 adducts. Conversely, in the absence of LTβR ΔS wt, we observed that NIK was constitutively polyubiquitinated. Nevertheless, in the presence of LTβR ΔS wt, the level of K48-linked polyubiquitinated NIK was drastically reduced without a concomitant TRAF3 K48-linked polyubiquitination. These results suggested that proteasomal degradation of TRAF3 would not be the only mechanism regulating its steady state. Indeed, blockade of the proteasome only marginally prevented TRAF2 and TRAF3 degradation upon LTβR stimulation, despite a potent inhibition of IκBα degradation and p100 processing (Fig. 8B). Moreover, inactivation of c-IAP1 by Smac mimetic (CmpA) did not prevent further TRAF3 degradation upon costimulation of LTβR (Fig. 8C). These results indicated that other mechanisms might account for LTβR-mediated TRAF3 degradation. We next analyzed the role of lysosomes for the degradation of TRAF2 and TRAF3 by using two different inhibitors of vacuolar ATPase activity, such as bafilomycin A1 and chloroquine. HeLa cells were treated for different periods of time with an agonistic antibody to LTβR in the absence or presence of bafilomycin A1 or chloroquine, and the phosphorylation and degradation of IκBα, as well as the processing of p100, were analyzed by Western blotting. Under both conditions, phospho-IκBα appeared as soon as 15 min after stimulation followed by an almost complete degradation of IκBα within 30 min. The efficiencies of p100 processing were also comparable between untreated and bafilomycin A1- or chloroquine-treated cells (Fig. 8D and data not shown). These results indicate that LTβR-mediated lysosomal degradation of TRAF2 and TRAF3 is dispensable for the activation of both the classical and the alternative NF-κB pathway. Overall, our results strongly suggest that internalized LTβR displaces intracellular TRAF3 from NIK, allowing its stabilization as previously proposed (41).
Fig. 8.
Intracellular LTβR activates the alternative NF-κB pathway independently of TRAF degradation. (A) HEK 293 cells were transiently transfected (+) or not (−) with the indicated expression vectors. Cells were lysed in 1% SDS and diluted up to 0.1% prior to the first round of immunoprecipitation (1st IP) with a control antibody (Ctrl Ab) or an anti-TRAF3 antibody. The immunoprecipitated material was analyzed by immunoblotting for TRAF3 and K48-linked TRAF3. The supernatants from the 1st IP were incubated with control beads or anti-Flag-coated beads. The immunoprecipitated materials were analyzed by immunoblotting for NIK and K48-linked ubiquitinated NIK. The asterisks represent the cross-reactivity with Ig heavy chains. (B) HeLa cells were stimulated with the agonist (Ago) to LTβR in the presence of DMSO (vehicle) or the proteasome inhibitor MG132, and cell extracts were analyzed by immunoblotting for the indicated proteins. (C) HeLa cells were treated with compound A (CmpA) or the Ago to LTβR alone or in combination as indicated, and the indicated proteins were analyzed by immunoblotting. (D) HeLa cells were stimulated as indicated with the Ago to LTβR in the absence (vehicle) or presence of bafilomycin A1. Total cell extracts were analyzed by Western blotting for the indicated proteins.
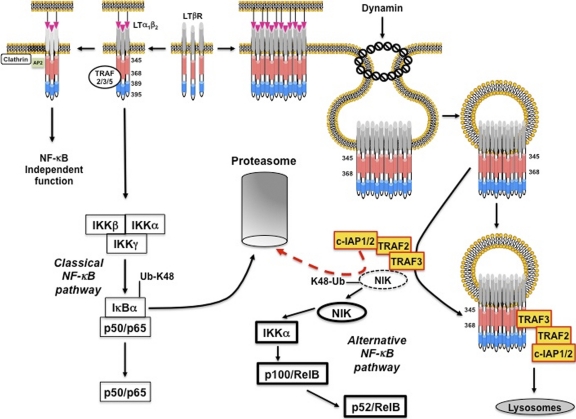
Altogether, we propose a model depicting ligand-bound LTβR complex outcomes and in which dynamin-dependent internalization uncouples the activation of the classical and the alternative NF-κB pathways (Fig. 9).
Fig. 9.
Model of LTβR trafficking and NF-κB activation. The binding of LTα1β2 to LTβR leads to its trimerization. This event allows a fast recruitment of TRAF proteins through the bipartite TRAF binding site (amino acids 389 to 395 in blue and 345 to 368 in red). This process then connects the receptor to the induction of IκBα degradation by the proteasome (classical NF-κB pathway). In the meantime, the complex AP2 in association with clathrin regulates an NF-κB-independent function of LTβR. While LTα1β2 accumulates at the cell surface of inducer cells, LTβR trimers form clusters on targeted cells. This process likely triggers the internalization of LTβR, which relies on the cytosolic region 345 to 368 and the presence of dynamin-2. Endocytic vesicles released from the plasma membrane expose the tail of LTβR toward the cytosol. This event allows LTβR to compete with intracellular NIK for the binding of its inhibitory complex TRAF3/TRAF2/c-IAP1/2. As a consequence, the constitutive proteasomal degradation of NIK (dashed line) is alleviated. Thus, NIK accumulates (solid line) and activates IKKα, and both events trigger the processing of p100 and the generation of p52/RelB. TRAF3/TRAF2/c-IAP1/2 complex is then degraded into lysosomes.
DISCUSSION
The biological functions fulfilled by members of the TNFR family rely on distinct signaling pathways for which recruitment of different TRAF proteins plays key roles. In this study, we identified an uncharacterized TRAF binding site spanning amino acid 345 to 368 of human LTβR. We showed that this region was as important as the triad D390/D391/E393 of LTβR for the recruitment of TRAF2 or TRAF3 in GST pulldown experiments (29). Of note, the primary sequence of amino acids 345 to 368 did not reveal any features of canonical TRAF2 or TRAF3 binding sites. Interestingly, other unconventional TRAF binding sites have also been characterized for TNFR2 (PLGVPDAGMKPS) and NIK (ISIIAQA), for which the recruitment of TRAF2 and TRAF3 appeared to be indirect and direct, respectively (16, 19, 30). Our data revealed that the affinity of the two TRAF binding sites of LTβR might fluctuate according to the oligomerization status of LTβR. Indeed, deletion of one of the two TRAF binding sites was sufficient to disrupt the recruitment of TRAF2 and TRAF3 to LTβR if expressed as a GST fusion protein. However, when LTβR was expressed as a native protein in eukaryotic cells, the region 345 to 368 retained some TRAF3 binding activity despite the deletion of the other TRAF binding site. It is reasonable to suggest that aggregation of LTβR increases the local concentration of LTβR, allowing it to increase its avidity toward TRAF3. The outcome of TRAF proteins following recruitment to TNFR varies from one receptor to another, involving degradation through either the proteasome or into lysosomes, as well as cellular relocalization to restricted cellular compartments (48). Recently, it was proposed that LTβR-mediated TRAF3 proteasomal degradation was required for stabilizing and accumulating NIK (41). However, under conditions for which overexpressed LTβR solely induced p100 processing, we did not observe an accumulation of K48-linked polyubiquitinated TRAF3, while the pool of K48-linked polyubiquitinated NIK was strongly reduced. Furthermore, it was shown that LTβR-mediated depletion of TRAF3 was required not only for the induction of the alternative pathway but also for the classical NF-κB pathway in MEFs, as well as in some colon epithelial cell lines (4, 59). Thus, proteasomal degradation of TRAF3 is associated not only with NIK stabilization. This statement can be also extended to signaling pathways downstream of CD40. Indeed, CD40-induced K48-linked polyubiquitination and proteasomal degradation of TRAF3 are strictly dependent on TRAF2 and c-IAP1/2 (21, 35, 52, 60). Based on these findings, a model had been proposed in which activated CD40 recruits TRAF2/TRAF3–c-IAP1/2 at the cell surface for promoting TRAF3 proteasomal degradation and NIK stabilization (52, 60). However, CD40-mediated K48-linked polyubiquitination and proteasomal degradation of TRAF3 are also required prior to cell membrane release of a MEKK1-containing complex that activates Jun N-terminal protein kinase (JNK) (34). Again, c-IAP1/2-mediated TRAF3 polyubiquitination is engaged in two distinct pathways involving MEKK1 and NIK. Therefore, assessing K48-linked TRAF3 polyubiquitination is not a readout strictly associated with an activation of NIK. Overall, TRAF3 appears to be a multitask protein that acts mainly as an inhibitor. It is likely that different pools of TRAF3-containing complexes exist, and according to the cell type and the duration of stimulation, TRAF3 is recruited and degraded at different locations to activate distinct pathways. We further observed that lymphotoxin-induced TRAF2 and TRAF3 degradation also occurred in the lysosomal compartment. However, potent inhibition of TNFR-mediated lysosomal TRAF degradation did not alter the extent of p100 processing, suggesting that this type of degradation is likely secondary to NIK stabilization.
Internalization of TNFR has been mainly considered a mechanism participating in recycling and/or degradation. In this study, we identified a new function assigned to LTβR trafficking, that is, the activation of the alternative NF-κB pathway. We found that toward lymphotoxin α1β2-induced mesenteric lymph node stromal cell maturation, internalization of LTβR correlated with RelB-induced MAdCAM-1 expression.
Similarly, ligand-induced down-modulation of cell surface LTβR has been observed in other settings, such as in myeloid dendritic cell (DC) homeostasis, which is strictly dependent on the alternative pathway, or SCS (subcapsular sinus) macrophage differentiation (23, 40, 55). Interestingly, we found that internalization of LTβR uncouples the activation of the alternative NF-κB pathway from the classical NF-κB pathway, probably reflecting the requirement of different adaptor proteins. This is reminiscent of the signaling pathways emerging from TLR4 for which TIRAP/MyD88 and TRAM/TRIF complexes regulate the proinflammatory and type I interferon responses from the plasma membrane and endosomes, respectively (24).
Other biological functions have been assigned to internalized TNFR family members. Indeed, TNFR1 also activates the classical NF-κB from the cell surface, but its internalization is rather associated with TNF-α-induced cell death (43). This process relies on a cytosolic region named the TRID domain, which contains a consensus YXXΦ motif that is targeted by the adaptor complex AP2 for sorting activated TNFR1 to endosomes (43). We have also detected an interaction between AP2 and activated LTβR. Unlike TNFR1, our bioinformatics analyses of LTβR did not reveal any consensus YXXΦ motif. Nevertheless, we found a dileucine motif at position 299 of human LTβR, which probably recruits AP2 through the binding of the σ2 subunit (27). However, we showed that deletion of the region encompassing this AP2 site did not alter the induction of the processing of p100, which is consistent with the fact that clathrin is dispensable for the activation of the alternative NF-κB pathway. Thus, AP2 may control LTβR internalization for the control of its half-life at the cell surface, its recycling, or a yet-undefined function.
The signaling pathways linked to TNFR internalization have been probably overlooked. Indeed, internalization of CD40 was reported to regulate the transcription of BAFF but also to allow the interaction with c-Rel for promoting B cell lymphoma proliferation (31, 61). Whether LTβR internalization controls nuclear functions is a possible scenario and is under investigation. Interestingly, ligand-dependent or -independent LTβR internalization might be linked to cancer progression. Indeed, we showed that dynamin-dependent internalization of LTβR is required for NIK stabilization, and exacerbated accumulation of NIK has been reported in hematopoietic as well as nonhematopoietic cancer cells (50). For instance, liver-specific LTα/LTβ expression leads to hepatitis-induced hepatocellular carcinoma development and this phenotype can be reversed when mice are treated with Fc-LTβR recombinant decoy receptors (18). Conversely, in LTβR-expressing melanoma cells, activation of NIK is driven in a ligand-independent way (12). In this particular case, the use of decoy LTβR would be useless and other strategies should be envisioned for preventing LTβR-mediated cell proliferation. The development of molecules that specifically block LTβR internalization, or other receptors, might be a promising research avenue for inflammatory disorders and cancer treatment.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Walloon Region (516056 PRALTER), the Inter-University Attraction Pole 6/18 (Federal Ministry of Science, Belgium), the FNRS (grant 1.5.047.07.F), and the Télévie, the Centre Anti-Cancéreux, and Foundation Jean Gol from the University of Liège and by a BBSRC project grant.
We thank Carl Ware (LIAI, San Diego, CA) for providing human wt LTβR cDNA and Tom Kirchhausen (Harvard Medical School/IDI) for the generous gift of Dynasore. We thank Alexander Sorkin (University of Pittsburgh School of Medicine) for the gift of AP2μ2-DN vector encoding HA-rat μ2D176A/W421A. We are greatly thankful to Srinivas Chunduru (TetraLogic Pharmaceuticals) for providing compound A. We thank Margaret Robinson for helpful discussions on AP2.
E.D., A.C., and J.P. are research associate, senior research associate, and research director at the FNRS, respectively.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Annunziata C. M., et al. 2007. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12:115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batten M., et al. 2004. TNF deficiency fails to protect BAFF transgenic mice against autoimmunity and reveals a predisposition to B cell lymphoma. J. Immunol. 172:812–822 [DOI] [PubMed] [Google Scholar]
- 3. Benezech C., et al. 2010. Ontogeny of stromal organizer cells during lymph node development. J. Immunol. 184:4521–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bista P., et al. 2010. TRAF3 controls activation of the canonical and alternative NFkappaB by the lymphotoxin beta receptor. J. Biol. Chem. 285:12971–12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheung T. C., et al. 2009. Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc. Natl. Acad. Sci. U. S. A. 106:6244–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chung J. Y., Park Y. C., Ye H., Wu H. 2002. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115:679–688 [DOI] [PubMed] [Google Scholar]
- 7. Claudio E., Brown K., Park S., Wang H., Siebenlist U. 2002. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat. Immunol. 3:958–965 [DOI] [PubMed] [Google Scholar]
- 8. Dejardin E. 2006. The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem. Pharmacol. 72:1161–1179 [DOI] [PubMed] [Google Scholar]
- 9. Dejardin E., et al. 2002. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 17:525–535 [DOI] [PubMed] [Google Scholar]
- 10. Delhase M., Hayakawa M., Chen Y., Karin M. 1999. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science 284:309–313 [DOI] [PubMed] [Google Scholar]
- 11. Dempsey P. W., Doyle S. E., He J. Q., Cheng G. 2003. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 14:193–209 [DOI] [PubMed] [Google Scholar]
- 12. Dhawan P., et al. 2008. The lymphotoxin-beta receptor is an upstream activator of NF-kappaB-mediated transcription in melanoma cells. J. Biol. Chem. 283:15399–15408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dougall W. C., et al. 1999. RANK is essential for osteoclast and lymph node development. Genes Dev. 13:2412–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Force W. R., et al. 2000. Discrete signaling regions in the lymphotoxin-beta receptor for tumor necrosis factor receptor-associated factor binding, subcellular localization, and activation of cell death and NF-kappaB pathways. J. Biol. Chem. 275:11121–11129 [DOI] [PubMed] [Google Scholar]
- 15. Futterer A., Mink K., Luz A., Kosco-Vilbois M. H., Pfeffer K. 1998. The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity 9:59–70 [DOI] [PubMed] [Google Scholar]
- 16. Grech A. P., et al. 2005. Tumor necrosis factor receptor 2 (TNFR2) signaling is negatively regulated by a novel, carboxyl-terminal TNFR-associated factor 2 (TRAF2)-binding site. J. Biol. Chem. 280:31572–31581 [DOI] [PubMed] [Google Scholar]
- 17. Hauer J., et al. 2005. TNF receptor (TNFR)-associated factor (TRAF) 3 serves as an inhibitor of TRAF2/5-mediated activation of the noncanonical NF-kappaB pathway by TRAF-binding TNFRs. Proc. Natl. Acad. Sci. U. S. A. 102:2874–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haybaeck J., et al. 2009. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell 16:295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He J. Q., Saha S. K., Kang J. R., Zarnegar B., Cheng G. 2007. Specificity of TRAF3 in its negative regulation of the noncanonical NF-kappa B pathway. J. Biol. Chem. 282:3688–3694 [DOI] [PubMed] [Google Scholar]
- 20. Hehlgans T., Pfeffer K. 2005. The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology 115:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hostager B. S., Haxhinasto S. A., Rowland S. L., Bishop G. A. 2003. Tumor necrosis factor receptor-associated factor 2 (TRAF2)-deficient B lymphocytes reveal novel roles for TRAF2 in CD40 signaling. J. Biol. Chem. 278:45382–45390 [DOI] [PubMed] [Google Scholar]
- 22. Iwai K., Tokunaga F. 2009. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 10:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kabashima K., et al. 2005. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity 22:439–450 [DOI] [PubMed] [Google Scholar]
- 24. Kagan J. C., et al. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaisho T., et al. 2001. IkappaB kinase alpha is essential for mature B cell development and function. J. Exp. Med. 193:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keats J. J., et al. 2007. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 12:131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelly B. T., et al. 2008. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456:976–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khare S. D., et al. 2000. Severe B cell hyperplasia and autoimmune disease in TALL-1 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 97:3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li C., et al. 2003. Structurally distinct recognition motifs in lymphotoxin-beta receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling. J. Biol. Chem. 278:50523–50529 [DOI] [PubMed] [Google Scholar]
- 30. Liao G., Zhang M., Harhaj E. W., Sun S. C. 2004. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279:26243–26250 [DOI] [PubMed] [Google Scholar]
- 31. Lin-Lee Y. C., et al. 2006. Nuclear localization in the biology of the CD40 receptor in normal and neoplastic human B lymphocytes. J. Biol. Chem. 281:18878–18887 [DOI] [PubMed] [Google Scholar]
- 32. Macia E., et al. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10:839–850 [DOI] [PubMed] [Google Scholar]
- 33. Marsters S. A., et al. 1997. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-kappaB and AP-1. J. Biol. Chem. 272:14029–14032 [DOI] [PubMed] [Google Scholar]
- 34. Matsuzawa A., et al. 2008. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 321:663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore C. R., Bishop G. A. 2005. Differential regulation of CD40-mediated TNF receptor-associated factor degradation in B lymphocytes. J. Immunol. 175:3780–3789 [DOI] [PubMed] [Google Scholar]
- 36. Motley A., Bright N. A., Seaman M. N., Robinson M. S. 2003. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162:909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Natoli G., Saccani S., Bosisio D., Marazzi I. 2005. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 6:439–445 [DOI] [PubMed] [Google Scholar]
- 38. Park Y. C., et al. 2000. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell 101:777–787 [DOI] [PubMed] [Google Scholar]
- 39. Pasparakis M. 2009. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat. Rev. Immunol. 9:778–788 [DOI] [PubMed] [Google Scholar]
- 40. Phan T. G., Green J. A., Gray E. E., Xu Y., Cyster J. G. 2009. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 10:786–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanjo H., Zajonc D. M., Braden R., Norris P. S., Ware C. F. 2010. Allosteric regulation of the ubiquitin:NIK and ubiquitin:TRAF3 E3 ligases by the lymphotoxin-beta receptor. J. Biol. Chem. 285:17148–17155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sasaki Y., et al. 2008. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF: BAFF-R-mediated survival signals in B cells. Proc. Natl. Acad. Sci. U. S. A. 105:10883–10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider-Brachert W., et al. 2004. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity 21:415–428 [DOI] [PubMed] [Google Scholar]
- 44. Schutze S., Tchikov V., Schneider-Brachert W. 2008. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat. Rev. Mol. Cell Bol. 9:655–662 [DOI] [PubMed] [Google Scholar]
- 45. Sen R. 2006. Control of B lymphocyte apoptosis by the transcription factor NF-kappaB. Immunity 25:871–883 [DOI] [PubMed] [Google Scholar]
- 46. Senftleben U., et al. 2001. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293:1495–1499 [DOI] [PubMed] [Google Scholar]
- 47. Shinkura R., et al. 1999. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat. Genet. 22:74–77 [DOI] [PubMed] [Google Scholar]
- 48. Silke J., Brink R. 2010. Regulation of TNFRSF and innate immune signalling complexes by TRAFs and cIAPs. Cell Death Differ. 17:35–45 [DOI] [PubMed] [Google Scholar]
- 49. Thompson J. S., et al. 2001. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science 293:2108–2111 [DOI] [PubMed] [Google Scholar]
- 50. Thu Y. M., Richmond A. 2010. NF-kappaB inducing kinase: a key regulator in the immune system and in cancer. Cytokine Growth Factor Rev. 21:213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vallabhapurapu S., Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 [DOI] [PubMed] [Google Scholar]
- 52. Vallabhapurapu S., et al. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 9:1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weih F., Caamano J. 2003. Regulation of secondary lymphoid organ development by the nuclear factor-kappaB signal transduction pathway. Immunol. Rev. 195:91–105 [DOI] [PubMed] [Google Scholar]
- 54. Weih F., et al. 1995. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell 80:331–340 [DOI] [PubMed] [Google Scholar]
- 55. Wu L., et al. 1998. RelB is essential for the development of myeloid-related CD8alpha− dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity 9:839–847 [DOI] [PubMed] [Google Scholar]
- 56. Xiao G., Harhaj E. W., Sun S. C. 2001. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 7:401–409 [DOI] [PubMed] [Google Scholar]
- 57. Ye H., Park Y. C., Kreishman M., Kieff E., Wu H. 1999. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell 4:321–330 [DOI] [PubMed] [Google Scholar]
- 58. Yin L., et al. 2001. Defective lymphotoxin-beta receptor-induced NF- kappaB transcriptional activity in NIK-deficient mice. Science 291:2162–2165 [DOI] [PubMed] [Google Scholar]
- 59. Zarnegar B., Yamazaki S., He J. Q., Cheng G. 2008. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. U. S. A. 105:3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zarnegar B. J., et al. 2008. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9:1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou H. J., et al. 2007. Nuclear CD40 interacts with c-Rel and enhances proliferation in aggressive B-cell lymphoma. Blood 110:2121–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.