Abstract
Many transcription factors and DNA binding proteins play essential roles in the development of organs in which they are highly and/or specifically expressed. Embryonic stem cell (ESC)-associated transcript 15-1 (ECAT15-1) and ECAT15-2, also known as developmental pluripotency-associated 4 (Dppa4) and Dppa2, respectively, are enriched in mouse ESCs and preimplantation embryos, and their genes encode homologous proteins with a common DNA binding domain known as the SAP motif. Previously, ECAT15-1 was shown to be important in lung development, while it is dispensable in early development. In this study, we generated ECAT15-2 single and ECAT15-1 ECAT15-2 double knockout (double KO) mice and found that almost all mutants, like ECAT15-1 mutants, died around birth with respiratory defects. Paradoxically, the expression of neither ECAT15-1 nor ECAT15-2 was detected in lung organogenesis. Several genes, such as Nkx2-5, Gata4, and Pitx2, were downregulated in the ECAT15-2-null lung. On the other hand, genomic DNA of these genes showed inactive chromatin statuses in ECAT15-2-null ESCs, but not in wild-type ESCs. The chromatin immunoprecipitation (ChIP) assay revealed that ECAT15-2 binds to the regulatory region of Nkx2-5 in ESCs. These data suggest that ECAT15-2 has important roles in lung development, where it is no longer expressed, by leaving epigenetic marks from earlier developmental stages.
INTRODUCTION
ECAT15-1 and ECAT15-2 are members of embryonic stem cell (ESC)-associated transcripts (ECATs), which were identified as genes enriched in mouse ESCs by in silico differential display screening (18). The two genes were also known as Dppa4 (developmental pluripotency-associated 4) and Dppa2, respectively, which were identified as novel markers of undifferentiated mouse ESCs with expression patterns similar to Oct3/4 (3).
ECAT15-1 and ECAT15-2 are tandemly located on the 16th chromosome in the mouse genome and have similar exon-intron structures, encoding polypeptides with 32% identity at the amino acid sequences (13). They contain a common putative DNA binding domain, the SAP motif, which consists of two amphipathic helices separated by an invariant glycine, and have DNA/RNA binding ability and function in chromatin modification (1). ECAT15-1 and ECAT15-2 show a weak homology to another SAP domain-containing protein, PGC7/Stella/Dppa3 (3), which binds DNA and protects the maternal genome from global demethylation in fertilized eggs (19). Therefore, ECAT15-1 and ECAT15-2 may regulate gene expression through modifying epigenetic status, like Dppa3. Indeed, ECAT15-1 has been shown to associate with chromatin and may therefore play a role in transcriptionally active regions (16).
The specific expressions suggest that the two genes play roles in pluripotency and early mouse development. However, ECAT15-1 single and ECAT15-1 ECAT15-2 double mutant ESCs showed no significant phenotypes (11). Furthermore, ECAT15-1 deletion in mice did not affect early embryogenesis. Thus, ECAT15-1 and ECAT15-2 are dispensable in early mouse development and derivation and maintenance of ESCs regardless of their specific expressions.
Unexpectedly, ECAT15-1 single mutant mice showed perinatal lethality, with deficiency in lung alveolar formation and rib abnormalities (11). Paradoxically, the authors were unable to detect the expression of ECAT15-1 in these organs in wild-type mice by reverse transcription-PCR (RT-PCR) (11). Thus, it remains elusive how ECAT15-1 contributes to functional organogenesis where it is no longer expressed. The functions of ECAT15-2 in vivo also remained to be determined.
In order to further clarify the functions of ECAT15-1 and ECAT15-2, as well as the relationship of the two related proteins, we generated mutant mice deficient in ECAT15-1, ECAT15-2, or both. Since ECAT15-1 and ECAT15-2 are tandemly located on the same chromosome, we established an ECAT15-1 ECAT15-2 double conditional targeting system using a bacterial artificial chromosome (BAC).
MATERIALS AND METHODS
Construction of ECAT15 double conditional targeting BAC vector.
A BAC DNA pool of a mouse BAC library (Research Genetics Co., catalog no. 0.96021) was screened according to the manufacturer's instructions. The yielded BAC DNA contains 140 kbp around the ECAT15-1 and ECAT15-2 locus. A loxP-pgk-Neor-loxP fragment (loxP-Neo cassette; Gene Bridge) was inserted into the ECAT15-1 locus, and an FLP recombination target (FRT)-pgk-Hygr-FRT fragment (FRT-Hyg cassette; Gene Bridge) was inserted into the ECAT15-2 locus by Red/ET recombination technology (Gene Bridge) according to the manufacturer's instructions. The BAC DNA modification of each locus was performed through six steps. First, the loxP-Neo cassette was inserted into the 3′ locus of ECAT15-1, and then the pgk-Neo part was excised with Escherichia coli 294-Cre (Gene Bridge). Finally, the loxP-Neo cassette was again inserted into the 1st intron of ECAT15-1 locus. The same steps were employed to insert the FRT-Hyg cassette into the modified ECAT15-2 locus with E. coli 294-FLPe (Gene Bridge).
Generation of the ECAT15 mutant mice and the 15-2KO ESCs.
The ECAT15 double conditional targeting BAC vector was digested with SalI and introduced into RF8 ESCs by electroporation. The primers for probe amplification of Southern blotting and real-time PCR based on TaqMan probe analysis are listed in Table S2 in the supplemental material. The two clones correctly recombined were injected into C57BL/6J blastocysts, thus yielding chimeric mice that transmitted the targeting allele through the germ line from both clones. The detailed modification steps are indicated in Fig. 1D. The ECAT15-2 knockout (15-2KO) ESCs were established from the 15-1+/CT 15-2+/− intercrossing (where CT represents “conditional targeting”), as described previously (24). The primers for genotype PCR are listed in Table S2 in the supplemental material.
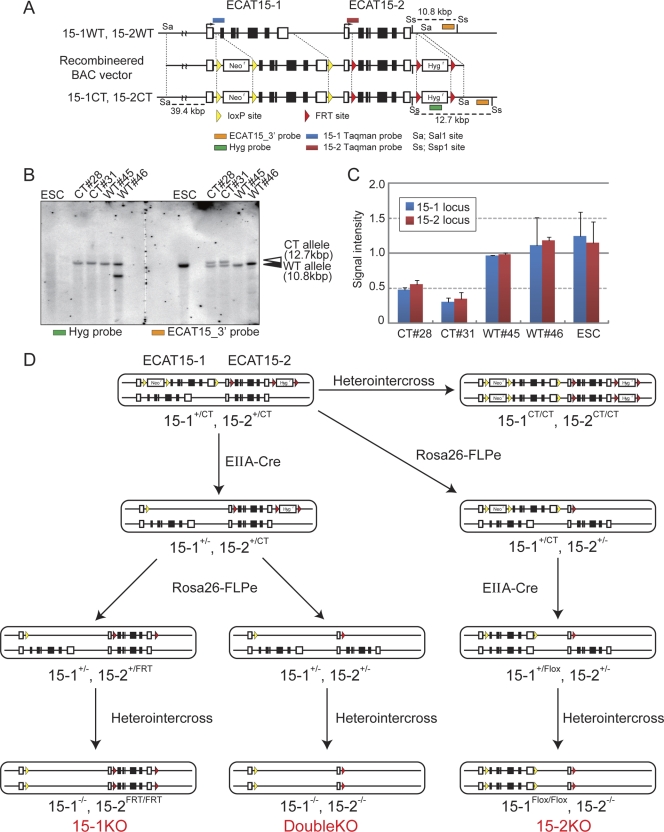
Fig. 1.
Generation of ECAT15-1 and ECAT15-2 single or double mutant mice using ECAT15 double conditional targeting system. (A) Schematic illustration of ECAT15 double conditional targeting. An ∼70-kbp fragment digested from BAC targeting vector with SalI was introduced into RF8 ESCs. (B) Screening of homologous recombineered ESCs and detection of nonhomologous insertion site by Southern blotting. All genomic DNAs which were digested by SspI were loaded in one gel, and the membrane was separated after blotting. The left membrane was detected using a Hyg probe (green box in panel A), and the right membrane was detected by the ECAT15_3′ probe (orange boxes in panel A). The upper band indicates the targeted allele (CT allele), and the lower band indicates the wild-type (WT) allele. (C) Confirmation of homologous recombination using TaqMan probe-based quantitative genomic PCR. Each TaqMan probe for wild-type ECAT15-1 or ECAT15-2 loci was designed as described in panel A (ECAT15-1, blue boxes; ECAT15-2; red boxes). The CT clones indicate that the correctly recombineered and WT clones were not correctly recombineered when examined by Southern blotting. Error bars represent the standard deviations (SD) of three experiments. (D) Strategy of ECAT15-1 and/or ECAT15-2 deletion. 15-1+/CT 15-2+/CT mice were mated with EIIA-Cre or Rosa26-FLPe transgenic mice step by step as shown.
Gene expression analysis.
RT-PCR and Western blotting were performed as described previously (12). The primers for RT-PCR are listed in Table S2 in the supplemental material. For calculation of expression level by RT-PCR, each value was normalized by Nat1 signal values as an internal control. The antibodies used for immunoblotting were anti-ECAT15-1 antibody (TMD-PB-DP4; Cosmo Bio), rabbit anti-ECAT15-2 antiserum (generated against 158 amino acids of mouse ECAT15-2), anti-Oct3/4 (sc-5279, Santa Cruz), anti-Sox2 (14), anti-Sall4 (24), anti-β-actin (A5441; Sigma), anti-mouse IgG-horseradish peroxidase (HRP) (7076; Cell Signaling), and anti-rabbit IgG-HRP (7074; Cell Signaling).
Histological analysis.
Extirpated lung samples were fixed with 4% paraformaldehyde–phosphate-buffered saline (PBS) at 4°C overnight. They were embedded in paraffin and cut into 3-μm sections with a microtome. Sections were deparaffinized and stained with hematoxylin and eosin (HE) solution. For immunohistochemistory, sections were stained with Vectastain Universal Elite ABC kit (Vector Laboratories, catalog no. PK-6200) according to the manufacturer's instructions. Anti-Scgb1b1 antibody (sc-9772; Santa Cruz) and anti-surfactant protein type C (anti-SP-C) antibody (AB3786; Millipore) were used at 1/100 and 1/2,000 dilutions. Paraffin-embedded blocks and sections of mouse tissues for in situ hybridization (ISH) were obtained from Genostaff Co., Ltd. The following procedures for ISH were performed according to the manufacturer's instructions (Genostaff Co., Ltd.). Probes for ECAT15-1 were generated with RF8 ESC cDNA and primers listed in Table S2 in the supplemental material.
ESC culture and immunocytochemistry analysis.
ESCs were harvested on gelatin-coated dishes as described previously (18). Immunocytochemistry was performed as described previously (22). The primary antibodies used for immunocytochemistry were anti-ECAT15-1 antibody or anti-ECAT15-2 antiserum and anti-HP1α antibody (17). Secondary antibodies were Cy3-conjugated anti-rabbit IgG antibody (Invitrogen) and Alexa 488-conjugated anti-mouse IgG antibody (Invitrogen). The nuclei were stained with Hoechst 33342 (1/10,000; Invitrogen). Fluorescent signals were observed using confocal microscopy (LSM710; Carl Zeiss).
Generation of the 15-2-EGFP transgenic ESCs and mice.
For construction of the 15-2-enhanced green fluorescent protein (EGFP) vector, the EGFP-FRT-PGK-Hygr-FRT cassette was inserted into the start codon of ECAT15-2 loci of ECAT15 BAC DNA. The 15-2-EGFP reporter ESCs were generated by introduction of the BAC vector into RF8 ESCs. The 15-2-EGFP reporter mice were generated by introduction of the BAC vector into a C57BL/6J mouse one-cell embryo. Then the 15-2-EGFP-Hygr transgenic mice were mated with Rosa26-FLPe transgenic mice in order to delete hygromycin-resistant gene, and the 15-2-EGFP transgenic mice were obtained. The 15-2-EGFP transgenic mice were genotyped by detection of EGFP cassette.
Flow cytometry.
Whole testis and lung of a 15-2-EGFP reporter embryo at embryonic day 14.5 (E14.5) were digested with 0.25% trypsin–1 mM EDTA for 20 min at 37°C, and the EGFP signals were analyzed by JSAN flow cytometry (Bay Bioscience) as described previously (8). The rhodamine filter was used for detection of autofluorescence.
DNA microarray.
Total RNA from each sample was labeled with Cy3. Samples were hybridized with a whole-mouse genome microarray (Agilent, catalog no. G4122F) as described previously (20). Data were analyzed with GeneSpring GX version 11. Quantile normalization was performed. The gene ontology analysis was performed with the program NEXTBIO using an entity list (106 entities; see Table S1B in the supplemental material).
Bisulfate sequencing.
Extracted genomic DNA from wild-type ESCs and the15-2KO counterparts were analyzed as described previously (12). The primers for bisulfate sequencing analysis are listed in Table S2 in the supplemental material.
Immunoprecipitation.
Protein lysate was extracted from mouse ESCs by CelLytic NuCLEAR extraction kit (NXTRACT-1KT; Sigma) according to the manufacturer's instructions with a small modification. The nuclei were lysed with high-salt extraction buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 500 mM NaCl, 0.5 mM dithiothreitol [DTT], supplemented with Complete protease inhibitor cocktail [Roche, catalog no. 11 697 498 001]) by sonication with a Biorupter (CosmoBio). The lysates were dialyzed in dialysis buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 100 mM KCl, 0.2 mM EDTA, and 0.5 mM DTT, supplemented with Complete protease inhibitor cocktail). Then the protein samples were incubated with magnetic beads (Invitrogen, catalog no. 112.03D) conjugated with anti-ECAT15-2 antibody at 4°C overnight. The beads were washed six times with dialysis buffer and then boiled in SDS sample buffer (50 mM Tris-HCl [pH 6.8], 12.5% glycerol, 1% sodium dodecyl sulfate, 0.01% bromophenol blue, 5% β-mercaptoethanol) and separated by SDS-PAGE followed by immunoblotting as described above.
ChIP assay.
We performed the chromatin immunoprecipitation (ChIP) assay for ESCs as previously described (12). The antibodies used in this experiment were anti-ECAT15-2 antiserum (also used for Western blotting), anti-H3K4me3 antibody (ab1012; Abcam), anti-H3K27me3 antibody (07-449; Upstate), and anti-H3K9me2 antibody (ab1220; Abcam). We also performed a ChIP assay with E18.5 whole lung tissue (P-2008-24; Epigentek) according to the manufacturer's instructions.
Microarray data accession number.
All reported microarray data have been deposited in the public database Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE31584.
RESULTS
Generation of ECAT15-1 and ECAT15-2 single and double heterozygous mutant mice using the double conditional BAC vector.
We generated ECAT15-1 and ECAT15-2 single mutant mice and ECAT15-1 ECAT15-2 double mutant mice in order to investigate the functions and relationships between ECAT15-1 and ECAT15-2 in vivo. It would be difficult to generate ECAT15-1 ECAT15-2 double mutant mice by mating single mutant mice of each allele because the two genes are located on the same chromosome and are separated by only ∼17 kbp (13). Therefore, we constructed an ECAT15-1 ECAT15-2 double conditional targeting vector using BAC (Fig. 1A). This BAC vector has three loxP sites and a pgk-neomycin-resistant gene cassette in the ECAT15-1 locus and three FRT sites and a pgk-hygromycin-resistant gene cassette in the ECAT15-2 locus. The vector was introduced into RF8 ESCs (18) by electroporation, and the cells were selected using both G418 and hygromycin. Southern blot analyses with an external probe (ECAT15_3′ probe) showed that two clones (CT 28 and CT 31), out of 48 screened, possessed bands corresponding to the correctly targeted alleles (where “CT allele” represents the “conditional targeting allele”) at both ECAT15-1 and ECAT15-2 (Fig. 1B; see Fig. S1B and C in the supplemental material). Real-time PCR with TaqMan probes showed that these two clones have only one copy of the wild-type alleles of ECAT15-1 and ECAT15-2, further confirming the homologous recombination (Fig. 1C). Southern blot analyses with an internal probe (Hyg probe) did not detect extra bands, indicating that these clones are free random integrations of the BAC targeting vector (Fig. 1B; see Fig. S1B and C in the supplemental material). These ESCs were injected into blastocysts to generate 15-1+/CT 15-2+/CT mice and subsequently 15-1CT/CT 15-2CT/CT mice (see Fig. S1A in the supplemental material), which were normal in gross appearance and fertile. Therefore, the conditional alleles do not disturb the development of somatic organs and germ cells.
ECAT15-1 and ECAT15-2 were singly or doubly deleted by mating the 15-1+/CT 15-2+/CT mice with EIIA-Cre transgenic mice (10) and Rosa26-FLPe transgenic mice (5) (Fig. 1D). To disrupt ECAT15-1, the 15-1+/− 15-2+/CT mice were generated by mating the 15-1+/CT 15-2+/CT mice with EIIA-Cre mice. Then the 15-1+/− 15-2+/CT mice were mated with Rosa26-FLPe transgenic mice to obtain the 15-1+/− 15-2+/FRT mice (see Fig. S2A in the supplemental material). To delete ECAT15-2, the 15-1+/CT 15-2+/− mice were generated by mating the 15-1+/CT 15-2+/CT mice with Rosa26-FLPe mice. Then the 15-1+/CT 15-2+/− mice were mated with EIIA-Cre mice to obtain the 15-1+/Flox 15-2+/− mice (see Fig. S2B in the supplemental material). The ECAT15-1 ECAT15-2 double heterozygous mutant (15-1+/− 15-2+/−) mice were generated from the mating of the 15-1+/− 15-2+/CT mice and Rosa26-FLPe mice (see Fig. S2C in the supplemental material).
Although there were no obvious abnormalities in the 15-1CT/CT 15-2CT/CT mice, the residual loxP and FRT sites may affect the expression of ECAT15-1 or ECAT15-2. Thus, we selected single heterozygous mutant mice in which these cassettes had been removed by Cre- or FLPe-mediated recombination (the 15-1+/− 15-2+/FRT mice and 15-1+/Flox 15-2+/− mice). Nevertheless, quantitative RT-PCR (qRT-PCR) analysis detected abnormal expression from the 15-2FRT allele in testis (more than 1,000-fold increase in comparison to wild type) (see Fig. 2D and E in the supplemental material). However, the ECAT15-2 protein was not detected by Western blotting in testis of the 15-1+/− 15-2+/FRT mice (see Fig. S2F in the supplemental material). The aberrant ECAT15-2 transcript was not detected in other organs or tissues examined. All three types of heterozygous mutant mice were normal in gross appearance and fertile. Therefore, we concluded that the aberrant ECAT15-2 transcript did not disturb normal development.
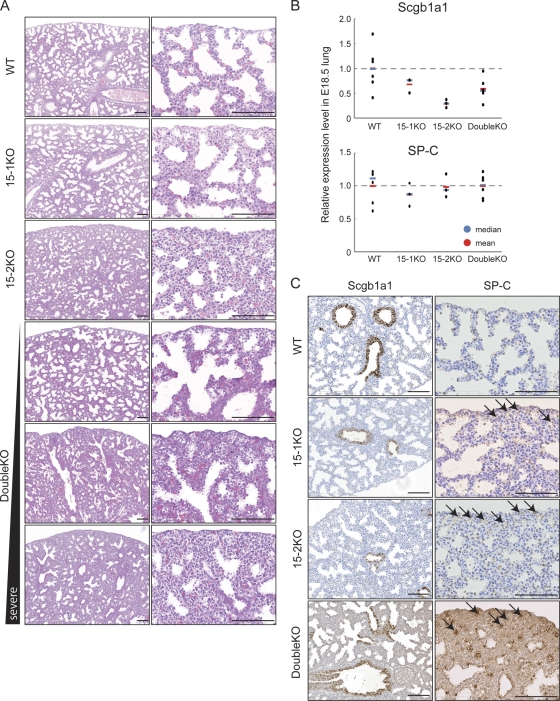
Fig. 2.
Respiratory disorder in ECAT15-1 ECAT15-2 mutant embryos. (A) Representative images of HE staining of lung sections at E18.5. (B) Expression levels of Scgb1a1 and SP-C in the E18.5 lung were examined by qRT-PCR. Blue and red bars in the graph indicate the median and mean. (C) Expression levels of Scgb1a1 and SP-C in the E18.5 lung were examined by immunohistochemistry with anti-Scgb1a1 and anti-SP-C antibody. Arrows indicate alveoli that have SP-C-positive cells and disrupted architecture. All scale bars = 100 μm.
Analyses of ECAT15 single and double homozygous mutant mice.
Heterointercross analysis using each of the ECAT15 heterozygous mutant mice was performed. The 15-1+/− 15-2+/FRT intercross generated a slightly smaller number of the 15-1KO (15-1−/− 15-2FRT/FRT) embryos than that expected from Mendelian law at E18.5 (Table 1). The mortality of the 15-1KO pups was increasing at birth (Table 1), and most of the 15-1KO neonates died within 3 days (see Fig. S3A in the supplemental material) and around weaning age (∼5 weeks old), we found that only 14 of 423 (3.3%) were the 15-1KO neonates (Table 1). The few surviving 15-1KO neonates showed growth retardation (see Fig. S3B in the supplemental material), but they caught up with wild-type and the 15-1+/− 15-2+/FRT mice by 20 weeks of age (see Fig. S3C in the supplemental material). These results indicated that ECAT15-1 is dispensable for peri-implantation development but important for the growth and survival during neonatal period.
Table 1.
Genotyping analysis of offspring from 15-1+/− 15-2+/FRT intercross of heterozygous mutant mice
| Time | No. of offspring |
No. of offspring with abnormalitya | ||
|---|---|---|---|---|
| WT (+/+ +/+) | 15-1 heterozygous (+/− +/FRT) | 15-1KO (−/− FRT/FRT) | ||
| E18.5 | 16 | 27 | 11 | 6 |
| P0 | ||||
| Alive | 26 | 45 | 13 | |
| Dead | 2 | 3 | 7 | |
| 1 wk | 27 | 66 | 0 | |
| Weaningb | 128 | 281 | 14 | |
“Abnormality” indicates offspring whose genomic DNA could not be extracted.
Time of weaning, 1 to ∼5 weeks of age.
Next, the 15-1+/Flox 15-2+/− mice were intercrossed to study the functions of ECAT15-2. The 15-2 homozygous mutant (15-2KO 15-1Flox/Flox 15-2−/−) embryos were identified in accordance with the Mendelian ratio by E16.5. However, at E18.5 and postnatal day 0 (P0), there were fewer 15-2KO embryos than expected, as they started to show mortality (Table 2). None of these 15-2KO neonates survived by weaning age (Table 2). These data indicated that ECAT15-2 is dispensable for the peri-implantation embryo but plays important roles during late embryogenesis and is essential for the survival of neonates.
Table 2.
Genotyping analysis of offspring from 15-1+/Flox 15-2+/− intercross of heterozygous mutant mice
| Time | No. of offspring |
No. of offspring with abnormalitya | ||
|---|---|---|---|---|
| WT (+/+ +/+) | 15-2 heterozygous (+/Flox +/−) | 15-2KO (Flox/Flox −/−) | ||
| E10.5 | 6 | 7 | 9 | 4 |
| E12.5 | 10 | 12 | 4 | 7 |
| E15.5 | 9 | 24 | 8 | 8 |
| E16.5 | 4 | 15 | 6 | 7 |
| E18.5 | 36 | 60 | 10 | 28 |
| P0 | ||||
| Alive | 10 | 29 | 2 | |
| Dead | 1 | 2 | 3 | |
| Weaningb | 153 | 290 | 0 | |
“Abnormality” indicates offspring whose genomic DNA could not be extracted.
Time of weaning, 1 to ∼5 weeks of age.
Finally, the 15-1+/− 15-2+/− mice were intercrossed to determine whether ECAT15-1 and ECAT15-2 function complementarily in early embryogenesis, where both proteins are expressed. Unexpectedly, at E18.5, the double KO embryos (15-1−/− 15-2−/−) were found in accordance with the Mendelian ratio (Table 3). The double KO neonates had small birth weights (see Fig. S3D in the supplemental material), and their numbers also appeared to be significantly small compared to those of sibs (Table 3). At the weaning age, only 13 out of 451 mice (2.9%) were found to be double KO mice (Table 3). These data suggested that both ECAT15-1 and ECAT15-2 are dispensable for early embryogenesis; however, they are important for survival at the perinatal stage. Interestingly, the deletion of both ECAT15-1 and ECAT15-2 resulted in a phenotype less severe than that of the 15-2KO mice (Tables 1 to 3).
Table 3.
Genotyping analysis of offspring from 15-1+/− 15-2+/− intercross of heterozygous mutant mice
| Time | No. of offspring |
No. of offspring with abnormalitya | ||
|---|---|---|---|---|
| WT (+/+ +/+) | Double heterozygous (+/− +/−) | Double KO (−/− −/−) | ||
| E16.5 | 9 | 30 | 10 | 5 |
| E18.5 | 36 | 62 | 37 | 12 |
| P0 | ||||
| Alive | 20 | 33 | 7 | |
| Dead | 0 | 1 | 2 | |
| Weaningb | 152 | 286 | 13 | |
“Abnormality” indicates offspring whose genomic DNA could not be extracted.
Time of weaning, 1 to ∼5 weeks of age.
Next, we examined whether ECAT15-1 and ECAT15-2 play roles in normal fertility, since they are expressed in gonads (13). We bred the 15-1KO mice and the double KO mice that were more than 8 weeks of age with wild-type mice for a long period (more than 5 months). Both the male and female 15-1KO mice were fertile enough to result in births several times (data not shown). Furthermore, the double KO male and female mice were also fertile; their fertility also resulted in birth several times (data not shown). It was not possible to perform sufficient mating to detect minor abnormalities related to fertility, since most of the 15-1KO or the double KO mice died prior to maturation. Nevertheless, these facts showed that the functional germ cells are generated in the absence of ECAT15-1 or both ECAT15-1 and ECAT15-2.
Respiratory defects in ECAT15-1/15-2 single and double mutant embryos.
Further analyses were performed to make clear the reason why the three types of ECAT15 mutants die around birth. In the previous report, the ECAT15-1 mutant mice had respiratory defects (11). This suggested that the 15-2KO mice and the double KO mice also have breathing abnormalities. E18.5 embryos were collected from the 15-1+/CT 15-2+/− intercrosses by caesarian section. Six of 50 neonates rapidly became cyanotic and died within 2 h (data not shown). Five of the six dead neonates were found to be the 15-2KO (15-1CT/CT 15-2−/−). Whereas the lungs from surviving mice floated in water, the lungs from dead mice did not, thus indicating respiratory failure (data not shown).
Histological analyses of the lungs from the 15-1KO, 15-2KO, and double KO E18.5 embryos revealed that 15-1KO lungs were similar to the wild type, except for their slightly thicker mesenchyme (Fig. 2A). The 15-2KO lungs also showed thicker mesenchyme. In addition, they showed smaller alveolar spaces; their defects were apparently more severe than those of the 15-1KO lungs (Fig. 2A). The double KO lungs also showed thicker mesenchyme and smaller alveolar spaces, but the severity was more diverse than those of the 15-1KOs and 15-2KOs (Fig. 2A). At E16.5, in contrast, no clear differences were observed between wild-type lung and the 15-2KO mutants (see Fig. S4 in the supplemental material). Thus, the ECAT15 KO lungs have a morphologically normal proximal epithelium, but an impaired alveolar architecture, and the lung deformities appear mainly during the saccular stage, which starts from ∼E17.5 in mouse development (21).
Next, the expression pattern of lung airway epithelium markers was assessed by qRT-PCR and immunohistochemistry. Scgb1a1 (secretoglobin, family 1A, member 1) is expressed in proximal lung epithelium (25), and SP-C (surfactant protein type C) is specific for alveolar cell type 2 and serves as a distal epithelium marker (6). The 15-2KO lungs (E18.5) expressed significantly small amounts of Scgb1a1, and the 15-1KO and the double KO lungs also expressed slightly smaller amounts of Scgb1a1 (Fig. 2B), despite the fact that the morphology of epithelial cells is normal (Fig. 2C). The lungs of three types of ECAT15 knockout mice had normal SP-C expression levels (Fig. 2B), but SP-C-positive cells were buried in the mesenchyme (arrows in Fig. 2C).
ECAT15-1 and ECAT15-2 expression in lung development.
The above-mentioned roles of ECAT15s led to the expectation that they are expressed during lung organogenesis. Lung and gonad RNA was prepared from C57/BL6 mouse embryos at several developmental stages and analyzed by qRT-PCR. Both ECAT15-1 and ECAT15-2 transcripts were detected in developing gonads at approximately ∼10% of the level in undifferentiated ESCs (Fig. 3A). However, no expression of ECAT15 proteins was detected in lungs (Fig. 3A).
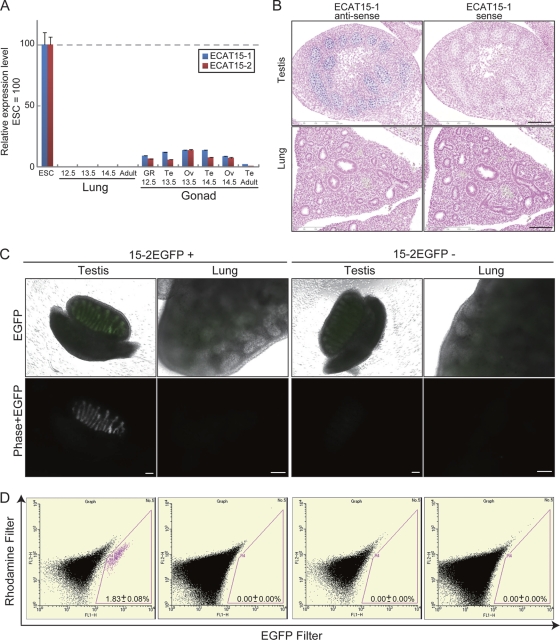
Fig. 3.
ECAT15 expression during lung organogenesis. (A) Relative expression levels of ECAT15-1 and ECAT15-2 in the undifferentiated ESCs, developing lungs, and gonads were examined by qRT-PCR. RNAs from developing lungs and gonads were mixtures of RNA samples from several individuals. The numbers under the graph indicates the embryonic day. Te, testis; Ov, ovary; GR, genital ridge (did not distinguish testis and ovary). Error bars represent the SD of three experiments. (B) Serial axial-proximal sections of the whole embryo at E14.5 were stained by in situ hybridization using antisense ECAT15-1 probe and sense ECAT15-1 probe. (C) Lung and testis of offspring at E14.5 of the 15-2-EGFP transgenic mice and C57BL/6J. Genotypes were confirmed by genomic PCR. (D) The EGFP-positive cells from the trypsinized 15-2-EGFP transgenic lungs and testis were examined by flow cytometry. Totals of 50,000 testis cells and 500,000 lung cells were examined. A rhodamine filter was used for detection of autofluorescence. All scale bars = 100 μm.
To further address whether ECAT15 proteins were expressed in specific cell populations such as somatic stem cells, the expression of ECAT15 transcripts was examined histologically either by in situ hybridization or by utilizing EGFP reporter mice. ECAT15-1 probes were generated for in situ hybridization and used to analyze serial axial-proximal axis sections of whole mouse embryos at E14.5. There were positive signals in the testis, but not in the lung (Fig. 3B). The 15-2-EGFP reporter mouse was generated to study the expression of ECAT15-2. The EGFP cDNA was introduced into the 1st exon of ECAT15-2 in the BAC clone, and the reporter BAC was inserted into RF8 ESCs. The reporter ESCs showed strong EGFP signals when undifferentiated, but the signal rapidly decreased upon differentiation (see Fig. S5A in the supplemental material). The reporter BAC was injected into fertilized eggs to generate the 15-2-EGFP reporter transgenic mice. The EGFP signals were detected in E3.5 blastocysts (see Fig. S5B in the supplemental material). During the later developmental period, the EGFP signal was detected only in the genital ridge of E14.5 embryos (Fig. 3C). No EGFP signals were detected in other organs or tissues, including the lung (Fig. 3C).
To further confirm the absence of ECAT15-2 expression in developing lung, we performed flow cytometry analysis using dissociated cells isolated from lung and testis of a 15-2-EGFP reporter embryo at E14.5. Approximately 1,000 EGFP-positive cells were detected out of 50,000 cells derived from testis (Fig. 3D). In great contrast, no EGFP-positive cells were identified out of 500,000 cells from lung (Fig. 3D). Taken together, these data demonstrated that neither ECAT15-1 nor ECAT15-2 is expressed during lung organogenesis.
Aberrant gene expression in ECAT15-2 mutant lungs.
To identify molecular mechanisms of lung disorders in three types of the ECAT15 KO mice, the global gene expression pattern of the 15-2KO lungs was examined at E18.5, which showed the most severe phenotype with the smallest divergence among the three types of ECAT15 KO mice. Comparisons between three wild-type and three 15-2KO lungs revealed 106 entities that showed >2-fold changes in expression (Fig. 4A; see Table S1A in the supplemental material). Gene ontology analyses using the NEXTBIO program revealed significant enrichment of genes involved in contraction and muscle function among the 106 entities (see Table S1B in the supplemental material). These changes in global gene expression are consistent with the thicker mesenchyme detected in mutant lungs histologically.
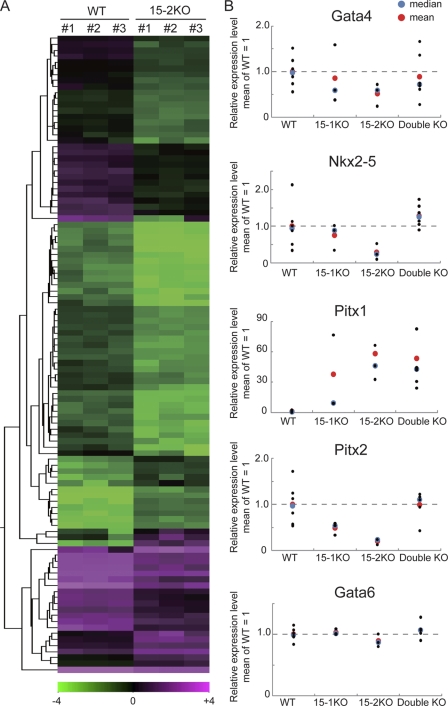
Fig. 4.
Global gene expression in the 15-2KO lung. (A) Transcriptome analysis by microarray using the total lung RNA of wild-type and the 15-2KO sibs at E18.5. The heat map shows the 106 probes with over 2-fold differences in expression between the wild-type and the 15-2KO lung. The color range indicates the log2 scale. (B) Relative levels of expression of Gata4, Nkx2-5, Pitx1, and Pitx2 in the lungs at E18.5 were examined by qRT-PCR. Gata6 was analyzed as an unchanged control that was not differentially expressed in the microarray analysis.
The microarray analyses also detected aberrant expression of several development-related transcription factors, Gata4, Nkx2-5, Pitx1, and Pitx2, in the 15-2KO lungs (see Table S1A in the supplemental material). The expression of the genes among three types of the ECAT15 mutant lungs was examined by qRT-PCR (Fig. 4B). For about all genes examined, the expression pattern in the 15-2KO lungs showed the biggest changes compared to the wild type among three types of ECAT15 mutant lungs (Fig. 4B). There were also the aberrant expressions of Pitx2 in the 15-1KO lungs and Pitx1 in both the 15-1KO and the double KO lungs (Fig. 4B). These data suggest that the abnormal expression of these transcription factors may contribute to the impaired lung architecture found in the three types of ECAT15 KO mice.
Aberrant epigenetic status in ECAT15-2 mutant ESCs.
Due to the DNA binding domain, the SAP motif, we hypothesized that ECAT15-1 and ECAT15-2 affect lung development by modifying the epigenetic status of critical genes, such as the four transcription factors (Fig. 4B), during the earlier developmental stage when they are expressed. To address this hypothesis, the 15-2KO ESCs were generated as an in vitro model of preimplantation embryos. The 15-2KO (15-1CT/CT 15-2−/−) ESCs were established from blastocysts of heterozygous intercross mice (see Fig. S6A in the supplemental material). The established 15-2KO ESCs showed morphology similar to that of their wild-type counterparts (see Fig. S6B in the supplemental material), but they proliferated slightly slower than the wild-type cells (see Fig. S6C in the supplemental material). The 15-2KO ESCs expressed pluripotency markers such as Oct3/4, Sox2, and Sall4 at levels comparable to those in wild-type ESCs (see Fig. S6D in the supplemental material). Thus, ECAT15-2 is dispensable in maintaining pluripotency of mouse ESCs.
However, a comparison of global gene expression between wild-type and the 15-2KO ESCs revealed that many genes were downregulated in ECAT15-2KO ESCs (see Fig. S6E in the supplemental material). Of note, many of the suppressed genes are involved in gonads and gametogenesis, such as Ddx4, Mael, and Syce1. We also established rescue cells by expressing Flag-tagged ECAT15-2 in ECAT15-2KO ESCs by means of the piggyBac vector (26) (see Fig. S7A in the supplemental material). In the established cells, the expression level of ECAT15-2 was approximately 5-fold higher than that in wild-type ESCs (see Fig. S7B and C in the supplemental material). Quantitative RT-PCR (see Fig. S7C) and DNA microarray analyses (see Fig. S7D) demonstrated that the altered levels of gene expression observed in ECAT15-2KO ESCs were reverted, albeit partially, by the forced expression of Flag-tagged ECAT15-2. The partial rescue may be attributable to the abnormally high expression levels of ECAT15-2. Nevertheless, these data demonstrated that ECAT15-2 is involved in the regulation of many genes in mouse ESCs.
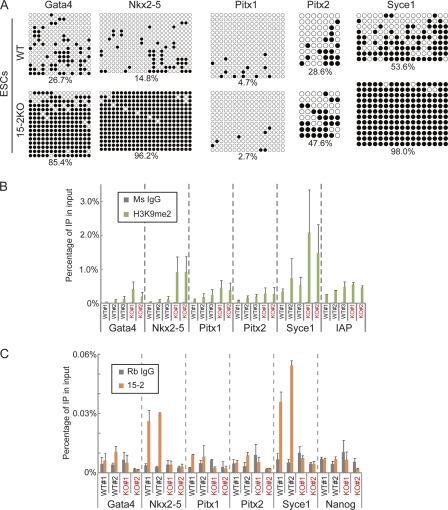
We next analyzed epigenetic status in ECAT15-2KO ESCs. DNA methylation status was analyzed by bisulfate sequencing using genomic DNA of the 15-2KO ESCs, and we found that the promoter regions of Syce1, Gata4, and Nkx2-5 were hypermethylated in the 15-2KO ESCs (Fig. 5A). The promoter region of Pitx2 in the 15-2KO ESCs was also methylated more than that of wild-type ESCs (Fig. 5A). Histone modification studies by ChIP assays revealed the enrichment of dimethylation at H3K9 in the Nkx2-5 and Syce1 promoter regions in the 15-2KO ESCs (Fig. 5B). In contrast, histone trimethylation at H3K4 and H3K27 was normal in the mutant ESCs (see Fig. S8 in the supplemental material). To address whether these aberrant epigenetic statuses were regulated by ECAT15-2 directly, we also performed ChIP analyses using anti-ECAT15-2 antibody and found that ECAT15-2 binds to the promoters of Nkx2-5 and Syce1, but not to that of Gata4 (Fig. 5C). These data demonstrated that ECAT15-2 directly or indirectly maintains the active DNA and histone modification statuses of the target genes in ESCs.
Fig. 5.
Aberrant epigenetic status in the 15-2KO ESCs. (A) Methylation status of five candidate gene promoters in wild-type and the 15-2KO ESCs. The numbers under the panels indicated the percentages of methylated CpG dinucleotides. (B) ChIP analysis using anti-H3K9me2 antibody. Precipitated DNA of the wild-type (WT) and the 15-2KO (KO) ESCs was examined by qPCR. Ms IgG, mouse IgG. (C) ChIP analysis using anti-ECAT15-2 antibody. Precipitated DNA of the wild-type (WT) and the 15-2KO (KO) ESCs was examined by qPCR. Rb IgG, rabbit IgG. Error bars represent the SD of three experiments.
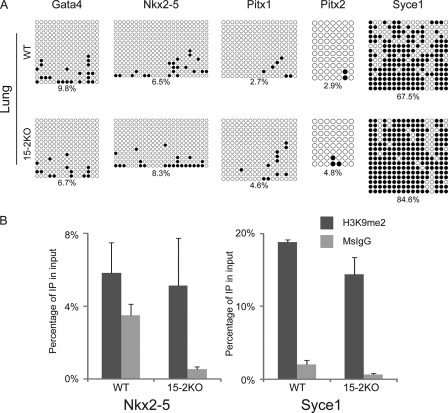
The epigenetic status was also examined in the 15-2KO lungs. Bisulfate genomic sequences showed that the promoter regions of five genes tested, except Syce1 in the 15-2KO lungs, had similar patterns of DNA methylation to the wild type (Fig. 6A). The promoter of Syce1 had a slightly methylated DNA pattern compared to the wild type (Fig. 6A). Similarly, ChIP analysis showed that histone dimethylation at H3K9 did not increase in the 15-2KO lungs in comparison to wild-type lungs (Fig. 6B). Taken together, these data indicated that ECAT15-2 may affect gene expression in developing lung through the regulation of epigenetic status in the early embryonic stage.
Fig. 6.
Epigenetic analysis of the 15-2KO lung. (A) DNA methylation status of five candidate gene promoters in wild-type and the 15-2KO lungs. Numbers under panels indicate the percentages of methylated CpG dinucleotides. (B) ChIP analysis using anti-H3K9me2 antibody. Precipitated DNAs from wild-type and the 15-2KO lungs were examined by quantitative PCR. IP, immunoprecipitate. Error bars represent the SD of three experiments.
Analyses of molecular moieties of ECAT15s.
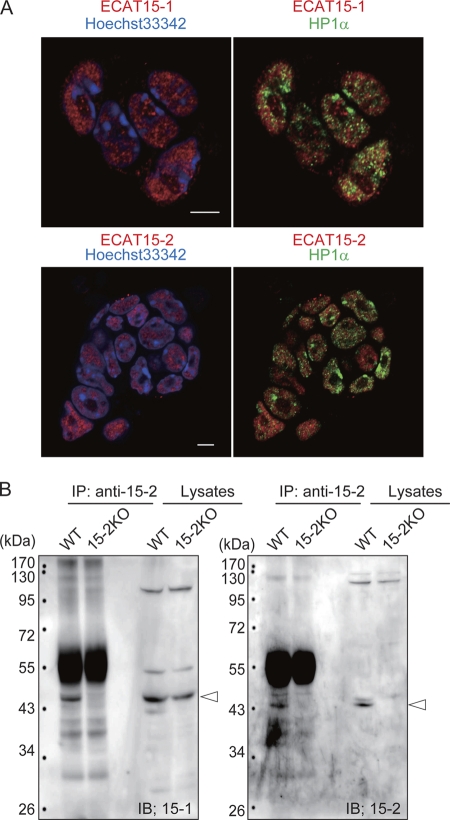
Finally, to address the reasons why the 15-2KO mice showed more severe phenotypes than the double KO mice did, molecular relationships between ECAT15-1 and ECAT15-2 were analyzed. The subcellular localization and protein-protein interactions of ECAT15-1 and ECAT15-2 were examined in the ESCs. Immunostaining with anti-ECAT15-1, anti-ECAT15-2, and anti-HP1α antibodies revealed that ECAT15-1 and ECAT15-2 were localized exclusively with HP1α in the nucleus (Fig. 7A). HP1α is known to localize in the heterochromatin region (2); thus, the result suggests that both ECAT15-1 and ECAT15-2 are located in euchromatin regions. Immunoprecipitation with anti-ECAT15-2 antibody using lysates from wild-type and 15-2KO ESCs showed that ECAT15-1 and ECAT15-2 interact with each other (Fig. 7B). These data indicated that ECAT15-1 and ECAT15-2 form a complex, and the balance of these genes may affect the gene expression and developmental process in lung.
Fig. 7.
Molecular moieties of ECAT15-1 and ECAT15-2 in ESCs. (A) Sublocalizations of ECAT15-1 and ECAT15-2 in ESCs were examined by immunocytochemistry using anti-ECAT15-1 or anti-ECAT15-2 antibody with anti-HP1α antibody. Nucleus was counterstained with Hoechst 33342. White bars = 5 μm. (B) Molecular interactions between ECAT15-1 and ECAT15-2 were examined by immunoprecipitation (IP) assay using anti-ECAT15-2 antibody with a nuclear protein lysate of wild-type and the 15-2KO ESCs. ECAT15-1 and ECAT15-2 were detected by immunoblotting (IB) with anti-ECAT15-1 and anti-ECAT15-2 antibody. Arrowheads indicate ECAT15-1 protein (left panel) and ECAT15-2 protein (right panel).
DISCUSSION
Our results demonstrated essential roles of ECAT15-1 and ECAT15-2 in functional lung development. Our results are consistent with the previous report that ECAT15-1 is important in normal lung function, indicating that the phenotypes we observed are attributable to loss of ECAT15 functions, but not to off-target effects. Despite these important roles, we were unable to detect the expression of ECAT15 proteins during lung organogenesis. In ESCs where ECAT15 proteins are expressed, we found that ECAT15-1 and ECAT15-2 bind to target genes as a complex and maintain active epigenetic statuses. We thus propose a model in which ECAT15 complex affects the expression of target genes at later developmental stages where ECAT15 proteins are no longer expressed, by leaving epigenetic memories from earlier developmental stages.
How ECAT15 proteins maintain active epigenetic statuses (i.e., protecting DNA and histone H3K9 from hypermethylation) in ESCs remains to be determined. We analyzed the amino acid sequences of ECAT15-1 and ECAT15-2 and compared them with those of Dnmt family members (Dnmt1, NP_001186360; Dnmt3a, NP_031898; Dnmt3b, NP_001003961; and Dnmt3l, NP_062321) or histone methyltransferases (HMTs) on histone H3K9 (Eset, NP_001157113.1; G9a, NP_665829.1; Glp, NP_001012536.2; Riz1, NP_001074824.3; Suv39h1, NP_035644.1; and Suv39h2, NP_073561.2), but did not identify any similarities to ECAT15 proteins. Recently, it has been indicated that Tet family members (Tet1, NP_081660.1; Tet2, NP_001035490.2; and Tet3, NP_898961.2) might have DNA demethylation ability through methylcytosine oxidization (9). Thus, the amino acid sequences of SAP domains and C-terminal regions of ECAT15-1 and ECAT15-2 were compared with the oxygenase domain of the Tet family, but again, there are no significant similarities. There have been no reports that SAP motif-containing proteins have DNA or histone demethylation ability. Thus, ECAT15 proteins per se might not regulate epigenetic statuses.
Alternatively ECAT15 proteins may bind and regulate other proteins involved in DNA methylation and histone modification. BioGRID (version 3.1, 12 December 2010) predicts that human ECAT15-2 protein interacts with SETD5 (Set domain containing 5). The SET domain is known as a methyltransferase domain, mainly for histone. Further studies are required to determine whether ECAT15 proteins associate with SETD5 or other epigenetic modifying proteins and regulate their functions.
We detected aberrant epigenetic status in ECAT15-2 mutant ESCs, but not in mutant lungs. The promoter regions of Gata4, Nkx2-5, and Syce1 seemed active, judging from DNA hypomethylation and H3K9me2 hypomethylation in ECAT15-2 mutant lungs. Considering the suppressed expression of these genes in ECAT15-2 mutant lungs, the low DNA histone and DNA methylation statuses of these genes are paradoxical. One possibility is that these genes are expressed in a small cell population within lungs, and thus analyses using whole-lung lysates failed to detect epigenetic abnormalities in minor cell types. Another possibility is that other types of epigenetic marks, such as other histone tail modifications, are involved. Alternatively, ECAT15 proteins might regulate the expression of these genes through regulatory elements such as enhancers and suppressors, which we did not analyze in the current study. Further experiments are required to determine the precise mechanisms of how ECAT15 proteins affect gene expression in organs in which they are no longer expressed.
Another unanswered question involves the functional interactions of the two ECAT15 proteins. They have 32% identities in amino acid sequences and have the common SAP domain. They physically interact with each other. Their expression patters are also similar. These characteristics suggest that the two proteins may have overlapping and compensatory functions. Contrary to this prediction, either ECAT15-1 or ECAT15-2 single deletion resulted in respiratory failure, suggesting that each protein has noncompensating roles in normal lung functions. We also found that deletion of both genes did not worsen the lung phenotypes. Rather, the double mutant resulted in a similar phenotype to that in ECAT15-1 single mutant lungs, which is milder than that observed in ECAT15-2 single mutant lungs. One model to explain this result is that ECAT15-1 has both supportive and detrimental effects on lung functions. The model predicts that the supportive effect also depends on ECAT15-2 protein, whereas the detrimental effect is antagonized by ECAT15-2. In this model, ECAT15-2 mutant lungs would suffer not only from the loss of the supportive effect of ECAT15s, but also from unmasking of the detrimental effect of ECAT15-1, which would become apparent due to loss of protection by ECAT15-2. In contrast, double mutant lungs would suffer only from the loss of the supportive effect of ECAT15s.
This putative detrimental effect of ECAT15-1, which is antagonized by ECAT15-2, may also exist in ESCs. We found that the deletion of ECAT15-2 showed abnormality in proliferation (see Fig. 6C in the supplemental material). In addition, overexpression of ECAT15-1 results in cell death during differentiation (15). In contrast, ECAT15-1 single and ECAT15-1 ECAT15-2 double mutant ESCs are apparently normal (11). Thus, like in lung, the stoichiometric balance of the two ECAT15 proteins might be important for proper functions of ESCs. When ECAT15-2 is suppressed or when ECAT15-1 is overexpressed, this balance might be destroyed and detrimental effects of ECAT15-1 become apparent.
Except for the slower proliferation of ECAT15-2 mutant ESCs, we found that ECAT15 proteins are dispensable in mouse ESCs and preimplantation embryos despite ECAT15's specific expression: even ECAT15-1 ECAT15-2 double mutant peri-implantation embryos are apparently normal. This result is consistent with the previous report that ECAT15-1 single mutant ESCs and ECAT15-1 ECAT15-2 double mutant ESCs were normal (11). However, ECAT15 proteins do regulate gene expression in ESCs, since DNA microarray analyses detected many genes, including those involved in germ cell development, which are downregulated in either ECAT15-1mutant ESCs (11) or ECAT15-2 mutant ESCs (see Fig. S6E in the supplemental material). Mutant ESCs may have developed compensation mechanisms, by which they can maintain pluripotency regardless of the altered gene expression. In contrast to our results, short hairpin RNA (shRNA)-mediated knockdown of ECAT15-1 or ECAT15-2 induced differentiation of mouse ESCs (4, 7, 15). The reasons for this discrepancy remain to be determined.
The suppression of germ cell-associated genes in mutant ESCs and significant expression in genital ridges in embryos, as well as in adult testes and ovaries suggest that ECAT15 proteins have important roles in germ cell development and gametogenesis. However, with the systemic gene targeting systems we used in this study, we obtained only a few ECAT15 homozygous mutant adult mice and were unable to perform detailed analyses of gametogenesis. Germ cell-specific gene disruption of ECAT15 proteins would answer this important question.
We found that the number of the 15-2KO embryos decreased even before birth, during E16.5 to E18.5 (Table 1). This suggests that the 15-2KO embryo had defects in organs that are indispensable for this developmental stage, in addition to the lung. In mutant lungs, we detected suppression of genes such as those for Nkx2-5 and GATA4, which are important for functional heart development (23). Thus, we postulated that the decrease in the number of homozygous mutant embryos during E16.5 to E18.5 may be attributable to abnormal heart development. However, we found that the hearts of the ECAT15-2 mutant E15.5 embryos beat normally and showed normal expression levels of Nkx2-5 and Gata4 (data not shown). It is still possible that ECAT15-2 mutant hearts have abnormalities, such as malformation of valves. However, it is more likely that other organs are affected in mutant embryos. Importantly, these results also demonstrated that several genes, such as Nkx2-5 and Gata4, were regulated by ECAT15-2 in lung, but not in heart.
In conclusion, our results demonstrated that the nuclear protein ECAT15-2 is essential for normal development of lung, in which the gene is not expressed during organogenesis. We propose that this represents a novel mechanism of gene regulation through epigenetic memories from earlier developmental stages when ECAT15-2 is expressed. Many important questions remained to be solved: What is the precise nature of epigenetic marks? Why is lung specifically affected? What is the relationship between ECAT15-1 and ECAT15-2? The ECAT15 double conditional targeting system developed in this study should be useful to answer these important questions.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Yamamoto, Y. Yamada, Y. Toda, and the members of the Yamanaka laboratory for valuable scientific discussions and administrative support. We thank T. Konishi, K. Iizuka, A. Okada, and M. Narita for technical assistance.
This work was supported in part by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (JSPS), the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Japan Science and Technology Agency (Yamanaka iPS Cell Special Project); a grant from the Leading Project of MEXT; and a grant from the Funding Program for World-Leading Innovative R&D on Science and Technology (First Program) of JSPS. T.N. was a Research Fellow of the Japan Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Aravind L., Koonin E. V. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112–114 [DOI] [PubMed] [Google Scholar]
- 2. Bannister A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- 3. Bortvin A., et al. 2003. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130:1673–1680 [DOI] [PubMed] [Google Scholar]
- 4. Du J., Chen T., Zou X., Xiong B., Lu G. 2010. Dppa2 knockdown-induced differentiation and repressed proliferation of mouse embryonic stem cells. J. Biochem. 147:265–271 [DOI] [PubMed] [Google Scholar]
- 5. Farley F. W., Soriano P., Steffen L. S., Dymecki S. M. 2000. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28:106–110 [PubMed] [Google Scholar]
- 6. Glasser S. W., et al. 2001. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 98:6366–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ivanova N., et al. 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442:533–538 [DOI] [PubMed] [Google Scholar]
- 8. Iwabuchi K. A., et al. 2011. ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency. PLoS One 6:e20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koh K. P., et al. 2011. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8:200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leneuve P., et al. 2003. Cre-mediated germline mosaicism: a new transgenic mouse for the selective removal of residual markers from tri-lox conditional alleles. Nucleic Acids Res. 31:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Madan B., et al. 2009. The pluripotency-associated gene Dppa4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol. Cell. Biol. 29:3186–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maekawa M., et al. 2011. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474:225–229 [DOI] [PubMed] [Google Scholar]
- 13. Maldonado-Saldivia J., et al. 2007. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells 25:19–28 [DOI] [PubMed] [Google Scholar]
- 14. Maruyama M., Ichisaka T., Nakagawa M., Yamanaka S. 2005. Differential roles for sox15 and sox2 in transcriptional control in mouse embryonic stem cells. J. Biol. Chem. 280:24371–24379 [DOI] [PubMed] [Google Scholar]
- 15. Masaki H., Nishida T., Kitajima S., Asahina K., Teraoka H. 2007. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J. Biol. Chem. 282:33034–33042 [DOI] [PubMed] [Google Scholar]
- 16. Masaki H., Nishida T., Sakasai R., Teraoka H. 2010. DPPA4 modulates chromatin structure via association with DNA and core histone H3 in mouse embryonic stem cells. Genes Cells 15:327–337 [DOI] [PubMed] [Google Scholar]
- 17. Meshorer E., et al. 2006. Hyperdynamic plasticity in pluripotent embryonic of chromatin proteins stem cells. Dev. Cell 10:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitsui K., et al. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631–642 [DOI] [PubMed] [Google Scholar]
- 19. Nakamura T., et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9:64–71 [DOI] [PubMed] [Google Scholar]
- 20. Okita K., Ichisaka T., Yamanaka S. 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317 [DOI] [PubMed] [Google Scholar]
- 21. Shi W., Bellusci S., Warburton D. 2007. Lung development and adult lung diseases. Chest 132:651–656 [DOI] [PubMed] [Google Scholar]
- 22. Takahashi K., et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 23. Takeuchi J. K., Bruneau B. G. 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459:708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsubooka N., et al. 2009. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 14:683–694 [DOI] [PubMed] [Google Scholar]
- 25. Weaver M., Yingling J. M., Dunn N. R., Bellusci S., Hogan B. L. 1999. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development 126:4005–4015 [DOI] [PubMed] [Google Scholar]
- 26. Woltjen K., et al. 2009. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.