Abstract
To examine the role of nucleosome occupancy in the evolution of gene expression, we measured the genome-wide nucleosome profiles of four yeast species, three belonging to the Saccharomyces sensu stricto lineage and the more distantly related Candida glabrata. Nucleosomes and associated promoter elements at C. glabrata genes are typically shifted upstream by ∼20 bp, compared to their orthologs from sensu stricto species. Nonetheless, all species display the same global organization features first described for Saccharomyces cerevisiae: a stereotypical nucleosome organization along genes and a division of promoters into those that contain or lack a pronounced nucleosome-depleted region (NDR), with the latter displaying a more dynamic pattern of gene expression. Despite this global similarity, however, nucleosome occupancy at specific genes diverged extensively between sensu stricto and C. glabrata orthologs (∼50 million years). Orthologs with dynamic expression patterns tend to maintain their lack of NDR, but apart from that, sensu stricto and C. glabrata orthologs are nearly as similar in nucleosome occupancy patterns as nonorthologous genes. This extensive divergence in nucleosome occupancy contrasts with a conserved pattern of gene expression. Thus, while some evolutionary changes in nucleosome occupancy contribute to gene expression divergence, nucleosome occupancy often diverges extensively with apparently little impact on gene expression.
INTRODUCTION
Nucleosomes restrict the access of regulatory proteins to the DNA and serve as the scaffold for various histone marks (22). The positioning of nucleosomes therefore is generally believed to play an important role in the regulation of DNA transcription, replication, recombination, and repair. We previously characterized the genome-wide nucleosome positioning of the budding yeast Saccharomyces cerevisiae using high-density tiling arrays (21). This and related studies uncovered general properties common to several model organisms. First, most promoters are characterized by a nucleosome-depleted region (NDR), a region that is relatively depleted of nucleosomes and enriched with transcription factor binding sites (TFBSs), just upstream of the transcription start site (TSS) (26, 44). Second, the NDR is established at least in part by AT-rich sequences as well as several trans factors that exclude nucleosome binding (2, 11, 13, 15, 19, 32, 43). Third, at coding regions, nucleosomes form an ordered array, with the +1 nucleosome bordering the TSS and an approximately constant distance (165 bp in S. cerevisiae) between adjacent nucleosomes. Recent work further suggested that this periodic pattern is governed by an ATP-dependent nucleosome packing mechanism (45).
The global patterns discussed above were observed for most S. cerevisiae genes. However, we also identified a significant subset of S. cerevisiae promoters that deviated from this organization and for which the TSS-proximal region was occupied by a nucleosome (33). Notably, these genes are enriched with TATA boxes and are characterized by a flexible expression pattern that is readily modulated upon changing conditions, displays a high cell-to-cell variability (noise), and diverges rapidly between related species or strains; we refer to this collection of behaviors as reflecting a dynamic pattern of gene expression. Together, these observations have led to the suggestion that the different nucleosome organizations at promoters code for distinct strategies of gene regulation (5, 11, 34).
A comparative genomics approach can discern the significance of nucleosome organization by examining whether it is conserved during evolution and whether changes in nucleosome patterns are associated with gene expression divergence (36). Two recent studies compared distantly related yeast species and concluded that changes in nucleosome positioning are associated with divergence of gene expression, thus implicating nucleosome positioning as a major driving force in the evolution of gene expression (10, 39). In contrast, we recently compared the genome-wide profiles of nucleosome occupancy in S. cerevisiae to its closest sequenced relative, Saccharomyces paradoxus (∼5 million years), and found that evolutionary changes in nucleosome positioning were, overall, not associated with changes in gene expression (37). To attempt to reconcile these apparently opposing conclusions, we carried out an analysis of nucleosome occupancy in S. cerevisiae and three additional yeast species that cover a range of evolutionary distances yet still allow comparison of orthologous promoters. We found that the global organization of nucleosomes is largely similar among different yeast species. However, this global conservation contrasts with an extensive divergence of nucleosome patterns at individual genes, whose expression patterns are, nonetheless largely conserved. Our results support the idea that many different configurations of nucleosome positioning can encode the same regulatory function and therefore that most changes in nucleosome patterns did not influence gene expression and may have evolved primarily through neutral drift.
MATERIALS AND METHODS
Strains and growth conditions.
S. cerevisiae BY4741, S. kudriavzevii IFO 1802, S. bayanus NRRL Y-11845, and C. glabrata CBS138 were grown in YPD (2% peptone, 1% yeast extract, 2% dextrose) starting from an overnight culture until they reached mid-log phase (optical density, 0.8) at 27°C. Cells were then cross-linked with formaldehyde, and mononucleosomes were isolated according to protocols modified from those described by Lee et al. (21).
Illumina sequencing of mononucleosomal DNA.
Gel-extracted mononucleosome fragments from digestions that resulted in mainly mononucleosomes were end repaired, subjected to amplification-free adaptor ligation, and then size selected on a 2% agarose gel. Clusters were generated on a single-read flow cell using Illumina's cBot and sequenced to 36 to 40 bases using Illumina's GAIIx instrument.
Processing of Illumina sequence data.
The genomic sequences of all species were retrieved from the Saccharomyces Genome Database and Genolevures. Reads of each species were mapped to their respective genome using Novoalign (Novocraft), and uniquely mapped reads were retained. As reads covered both ends of the ∼150-bp DNA fragments that correspond to mononucleosomes, we converted the mapped positions into the estimated center position by adding or removing (depending on the strand) half the length of a typical DNA fragment. This typical length was evaluated in each sample by the distances between consecutive peaks of reads from the forward and backward strands and was 107 to 126 bp in all samples (see Table S3 in the supplemental material). Importantly, the fragment length of C. glabrata samples was intermediate between those of the sensu stricto species, indicating that the differential nucleosome patterns observed in C. glabrata cannot be accounted for by differences in the degree of MNase digestion (42). Highly similar DNA fragment lengths among the different species were also estimated with an Agilent Bioanalyzer high-sensitivity chip (see Table S3). For S. bayanus and C. glabrata, the reads from two biological repeats were combined into a single data set. The raw and processed data are available at the GEO database (GSE23577) and can be visualized at http://barkai-serv.weizmann.ac.il/nuc_species/.
Estimation of nucleosome occupancy and positions.
Nucleosome occupancy was computed by extending each mapped position to the surrounding 147-bp region. A Gaussian filter was used to define nucleosome positioning scores, and nucleosome calls were defined as peaks of this score, with a minimal distance of 100 bp between two consecutive peaks. The 10% calls with either the lowest score or the lowest occupancy were removed. Nucleosome numbers (−1, +1 etc.) were assigned first in S. cerevisiae such that the +1 nucleosome is the first nucleosome whose center is downstream of the TSS of each gene, and the +1 nucleosome in the other species was defined as the closest one to the S. cerevisiae +1, as long as it was closer than 75 bp to the S. cerevisiae +1 position (otherwise, nucleosome numbers were not defined).
Promoter classification as DPN and OPN.
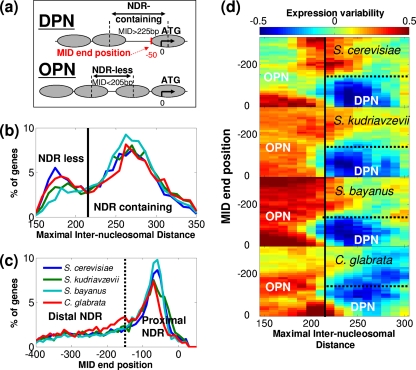
The maximal distance between promoter nucleosomes (maximal internucleosomal distance [MID]) was computed as the maximal distance between consecutive nucleosome peaks in the window of −600 to +100 (with respect to the ATG), and MID positions were defined as the distance of the NDR from the ATG, which was calculated as position of the nucleosome peak at the 3′ end of the MID minus 75 bp. Genes with a MID above 350 or below 150 or a MID position below −400 (relative to ATG) were excluded (see Table S1 in the supplemental material). Promoters with a MID shorter than 205 bp were defined as OPN (occupied by nucleosomes), while promoters with a MID larger than 225 bp whose position was more than 150 bp upstream of the TSS were defined as DPN (depleted promoter nucleosome) (see Fig. 2a and b).
Fig. 2.
Classification of promoter nucleosome patterns. (a) Scheme showing a DPN gene, whose promoter contains a large NDR which is proximal to the ATG, and an OPN gene, whose promoter does not contain a large NDR, as defined by the MID. (b) For each promoter, we calculated the maximal distance between consecutive nucleosomes (MID). The distribution of these distances is shown for each of the species (the color key is given in panel c). The vertical bar shows the distinction between NDR-containing and NDR-less promoters. (c) Distribution of MID end positions (i.e., the distance between the 3′ border of NDRs and the ATG; see Fig. 2a) among the NDR-containing genes. Genes falling to the right of the vertical line were redefined here as DPN. (d) Heat maps of expression variability in the four species, as a function of MID and MID end position. Expression variability was calculated from a large expression compendium of S. cerevisiae (17), and each value in the heat maps was calculated by averaging the variability of 150 genes with closest values in both MID (horizontal axis) and NDR position (vertical axis).
Comparison of nucleosome occupancy.
One-to-one orthologs were defined based on the work of Cliften et al. (8) and Wapinski et al. (41). The promoter depletion score was calculated as the 100-bp window with the lowest average occupancy within −300 to 0 relative to the ATG (11). Nucleosome occupancy patterns from −600 to +1000 (relative to the ATG) were compared among orthologs by Pearson correlation after they had been aligned by the position of the +1 nucleosome. For genes shorter than 1 kb, we examined the pattern only until the end of the gene. As a control, Pearson correlations were calculated for randomly selected genes (instead of orthologs) from the same species pairs.
Analysis of gene expression.
Expression levels of all species (39) were averaged over the different replicates of each species. Response to stress in C. glabrata was defined as the average log2 ratio of expression levels under 4 stress conditions (oxidative stress, osmotic stress, glucose starvation, and heat shock) (28). Similarly, the S. cerevisiae response to stress was calculated as the average log2 ratio of expression levels under the corresponding conditions (14). Expression variability in S. cerevisiae was defined as the absolute value of the average log2 ratio of expression levels under more than a thousand conditions (17). Expression variability of S. kudriavzevii, S. bayanus, and C. glabrata was defined like that of the S. cerevisiae orthologs, as there are not sufficient data for each of these species. Expression variability has probably diverged to some extent among the different species, and therefore these estimates are not entirely accurate. Despite the divergence of nucleosome patterns, OPN genes in all species are associated with high expression variability (as defined for S. cerevisiae), suggesting that this association is even stronger and is decreased by the use of inaccurate measures of expression variability.
Identification of TSS positions in S. cerevisiae and C. glabrata.
Cells were grown in YPD to mid-log phase followed by snap-freezing in liquid nitrogen. Total RNA was isolated using acid phenol, and then mRNA was purified using NucleoTrap mRNA from Macherey-Nagel. The mRNA was then processed using NEBNext mRNA sample preparation reagent set 1. The resulting cDNA was prepared for sequencing using the Illumina HiSeq protocols.
To predict TSS positions, we searched for genes in which 100-bp segments in the coding region have at least 0.5 reads per base (expressed), while their upstream sequences contain a 100-bp segment with less than 0.05 read per base (nonexpressed). The TSS was defined as the transition point from the nonexpressed to the expressed segment, as predicted with a two-state hidden Markov model.
RESULTS
Patterns of nucleosome occupancy of four yeast species.
Mononucleosomal DNA fragments were isolated following MNase digestion using standard protocols from four yeast species grown to mid-log phase in the same media. The length of mononucleosome fragments was comparable among the different species (see Materials and Methods), and these fragments were subjected to Illumina high-throughput sequencing, producing ∼10 to 40 million reads per species that were mapped to each of the four genomes (Fig. 1 a). The species analyzed were the model budding yeast Saccharomyces cerevisiae, two additional species of the Saccharomyces sensu stricto group (S. kudriavzevii and S. bayanus), which are highly similar to S. cerevisiae in morphology, gene repertoire, and genomic sequences (∼78% coding sequence identity, ∼10 to 20 million years) (8), and a more distantly related yeast (Candida glabrata), which is a major human pathogen that still retains considerable similarity to S. cerevisiae (∼57% coding sequence identity, ∼50 million years; note that in spite of its name, C. glabrata is much more closely related to S. cerevisiae than it is to C. albicans) (9). Figure 1a shows the phylogenetic tree of these species along with read density and nucleosome positioning scores at two orthologous loci.
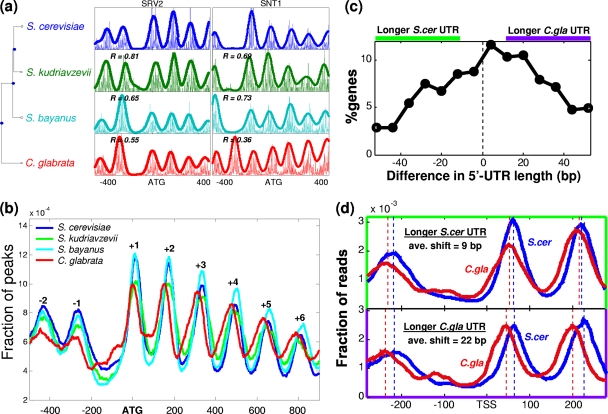
Fig. 1.
Patterns of nucleosome positioning of four yeast species. (a) Phylogenetic tree and examples of two orthologous loci. At each locus, the density of reads is shown along with a smoothed pattern (nucleosome positioning score). The correlation between nucleosome score patterns of S. cerevisiae and each of the other species is also indicated for each gene. (b) Distribution of peaks in nucleosome positioning scores relative to their ATG, over all genes analyzed in each species (see Table S1 in the supplemental material). (c) Distribution of differences in 5′ UTR lengths between S. cerevisiae and C. glabrata orthologs. The regions with larger 5′ UTRs in C. glabrata and S. cerevisiae (by at least 10 bp) are marked above the graph, and the corresponding genes are used in panel d. (d) Normalized read density relative to the TSS of S. cerevisiae and C. glabrata orthologs. Genes with longer UTRs (by at least 10 bp, as shown in panel c) in S. cerevisiae and C. glabrata are shown, along with the corresponding peaks of read densities (dotted lines) and the average shift of the peaks.
To examine whether the typical nucleosome organization described previously for S. cerevisiae is also seen in these other species, we examined the distribution of predicted nucleosome center positions (peaks) relative to the ATG. These distributions were highly similar among the four species and were consistent with the typical organization described for S. cerevisiae (Fig. 1b). While the general pattern was conserved and was in fact indistinguishable among the Saccharomyces sensu stricto species, the pattern appears to have slightly shifted upstream in C. glabrata (relative to ATG) (Fig. 1b). This shift in nucleosome positioning was accompanied by a similar shift in the positions of predicted transcription factor binding sites (TFBSs) (see Fig. S1 in the supplemental material) and is consistent with a shift in promoter DNA bendability patterns that we reported previously (35). The shift is observed both for promoter and for coding-region nucleosomes, although coding-region sequences are not shifted among the different species, suggesting that small changes in the positions of promoter elements have influenced the coding regions, causing multiple nucleosomes within an array to shift upstream (37).
Previous studies have shown that the position of the +1 nucleosome is tightly correlated with the TSS (18). We thus speculated that the widespread shift of nucleosome positions may be linked to shifts in 5′ untranslated-region (UTR) lengths (and thus TSS positions). To test this possibility, we used RNA-Seq to profile the transcriptomes and TSS positions of S. cerevisiae and C. glabrata. Comparison of orthologs showed a tendency toward longer 5′ UTRs in C. glabrata than in S. cerevisiae (Fig. 1c; the median and average shift are 7 and 6.7 bp, respectively). However, this small shift in TSS positions cannot completely account for the larger shift in nucleosome positioning (∼20 bp), and thus we still observed a shift in nucleosome positioning when aligning genes by the TSS (Fig. 1d). Interestingly, the remaining shift in nucleosome positioning (relative to the TSS) was larger for genes with a longer 5′ UTR in C. glabrata than for genes with a longer 5′ UTR in S. cerevisiae (Fig. 1d). Thus, larger C. glabrata 5′ UTRs may be associated with an upstream shift in nucleosome positioning both through their immediate effects on TSS positions and through additional, as-yet-unknown effects.
Promoters of all species are naturally divided into NDR-containing and NDR-lacking types.
A key insight that emerged from analysis of the nucleosome profiles in S. cerevisiae is that promoters can be classified into two types: those which contain a pronounced NDR just upstream of the TSS (so-called DPN [depleted promoter nucleosome] promoters) and those in which the TSS-proximal promoter region is occupied by nucleosomes (the so-called OPN [occupied promoter nucleosome] promoters) (33). We previously classified genes into these two types by analyzing the relative occupancy of TSS-proximal promoter regions as defined by tiling arrays. Our new data sets, generated by high-throughput sequencing at high coverage, allow a better estimation of the canonical nucleosome positions by directly examining whether each gene contains a promoter region that is depleted of nucleosomes.
We predicted the canonical positions of nucleosome centers as peaks of nucleosome reads and calculated the largest distance between two consecutive nucleosome centers at each promoter (Fig. 2 a; also, see Materials and Methods). This maximal distance between promoter nucleosomes displayed a bimodal distribution (Fig. 2b): the first mode centered around 150 to 200 bp, corresponding to a linker of 0 to 50 bp (NDR-less promoters), while the second mode, 220 to 300 bp, corresponds to the typical NDR, with a length of 70 to 150 bp (NDR-containing promoters). As expected, NDRs were highly enriched directly upstream of the ATG (Fig. 2c). We thus redefined the DPN class as comprising NDR-containing genes in which the NDR is proximal to the ATG (Fig. 2c), and the OPN genes as NDR-less genes (Fig. 2a). Approximately half of the genes were classified as DPN, a quarter were OPN, and the rest had an intermediate pattern and thus were not classified (see Table S1). Interestingly, the maximal distance between promoter nucleosomes is correlated with mRNA levels among DPN genes (R ∼ 0.25) but not among OPN genes (R < 0), further demonstrating the OPN/DPN dichotomy (see Fig. S2 in the supplemental material).
Notably, this bimodal distribution of the maximal internucleosomal distances and the biased distributions of NDR locations were observed for all four species examined, allowing us to define the respective DPN and OPN classes of each species (Fig. 2b and c). Importantly, in all species examined, OPN genes were associated with high expression variability (Fig. 2d) and enriched among TATA-containing genes (see Fig. S3 in the supplemental material), as defined for S. cerevisiae (3, 33).
Gene-specific nucleosome occupancy patterns are poorly conserved.
We asked whether the patterns of nucleosome occupancy are conserved among orthologous genes. Based on the results from the preceding section, we first asked whether classification of orthologs into OPN and DPN patterns was conserved across species. Using the maximal distances between promoter nucleosomes (MID) (Fig. 2a) as a proxy for this division, we found that conservation was high among sensu stricto orthologs but very low between the sensu stricto species and C. glabrata (Fig. 3 a; also, see Fig. S4 and Table S2 in the supplemental material). Importantly, the use of the maximal distance between nucleosomes as a measure of OPN/DPN conservation eliminates any potential for artifacts caused by use of ATG rather than TSS (which have not been defined for S. kudriavzevii and S. bayanus) to define the location of the NDR. Interestingly, while conservation of the OPN pattern between the sensu stricto species and C. glabrata was generally close to that expected by chance, conservation of this pattern was markedly higher for genes with high expression variability and a TATA box (Fig. 3b). These results suggest that the OPN pattern is functionally important, and thus conserved, for genes with high expression variability, while for other genes it readily diverges without functional consequences.
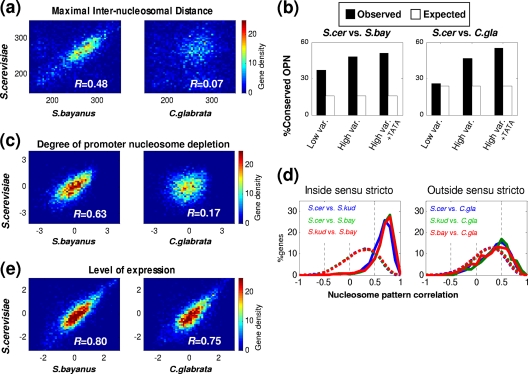
Fig. 3.
Low conservation of gene-specific nucleosome patterns and high conservation of gene expression between sensu stricto species and C. glabrata. (a, c, and e) Color-coded density plots comparing promoter maximal internucleosomal distance (a), the degree of promoter nucleosome depletion (c), and expression levels (e) at orthologous genes between S. cerevisiae and either S. bayanus or C. glabrata. Pearson correlations are also indicated. (b) Conservation of promoter classification to OPN between S. cerevisiae and either S. bayanus or C. glabrata for a third of the genes with lowest or highest expression variability and for those with both high expression variability and a TATA box. White bars represent the percentages of conservation expected by chance (assuming independent classifications to OPN/DPN in the different species). (d) We aligned nucleosome occupancy patterns (promoter plus 1 kb of the coding region) by the position of the +1 nucleosomes (see Fig. S4 in the supplemental material) and calculated the correlation between occupancy patterns of orthologs from the different species. Distribution of the correlation coefficients are shown for each pairwise species comparison, divided into comparisons within sensu stricto species (left) and those between sensu stricto species and C. glabrata (right). Dashed lines indicate the distributions for comparison of randomly selected genes (nonorthologs) from the same pairwise species comparisons.
To further examine the conservation of gene-specific nucleosome patterns, we compared the degree of promoter nucleosome depletion between orthologs, following the approach used by previous studies which used such measures for interspecies comparisons (10, 39). Strikingly, also for promoter depletion, we found a high degree of similarity among sensu stricto species yet very little similarity between the sensu stricto species and C. glabrata (Fig. 3c; also, see Fig. S4).
As an additional means of assessing the conservation of nucleosome patterns, we also calculated the correlations between the patterns of nucleosome occupancy throughout the promoters and coding regions of orthologs from the different species. Such correlations may be sensitive to local shifts in the positions of nucleosomes, which may be generated by local insertions/deletions or by differences in global chromatin organization, such as the shift we observed in C. glabrata (Fig. 1b). However, we reasoned that such a measure would be complementary to the previous promoter-centered measures and thus controlled for these issues by aligning orthologous nucleosome patterns to their corresponding +1 nucleosomes and by excluding genes with gaps in the interspecies sequence alignments (see Fig. S5 in the supplemental material). Once again, we found that nucleosome patterns were highly conserved among sensu stricto orthologs (R ∼ 0.75), while the similarity in nucleosome patterns between sensu stricto genes and their C. glabrata orthologs was almost as low as for randomly selected genes (Fig. 3d; some similarity is expected by chance even for randomly selected pairs of genes due to the relative nucleosome depletion of most promoters and the periodic positioning at coding regions).
Taken together, these analyses demonstrate that gene-specific nucleosome patterns are conserved among closely related species but have almost completely diverged between the sensu stricto species and C. glabrata, with the exception of OPN patterns at genes with high expression variability. In sharp contrast, we found that gene expression was largely conserved between orthologous S. cerevisiae and C. glabrata genes, both in absolute levels (Fig. 3e) (R = 0.75) and in the response to different stresses (see Fig. S6 in the supplemental material) (R ∼ 0.6). Furthermore, genes with differential expression among S. cerevisiae and C. glabrata are typically not more different in their nucleosome patterns or promoter nucleosome depletion than genes with similar expression patterns (see Fig. S7 in the supplemental material), suggesting that divergence of nucleosome patterns did not play a significant role in the divergence of gene expression. Similar results were obtained in our previous comparison of S. cerevisiae and S. paradoxus (37) and in comparisons of other pairs of yeast species. Importantly, these findings suggest that even for evolutionary distances considerably greater than that between S. cerevisiae and S. paradoxus, increased divergence of nucleosome occupancy generally does not result in divergence of gene expression.
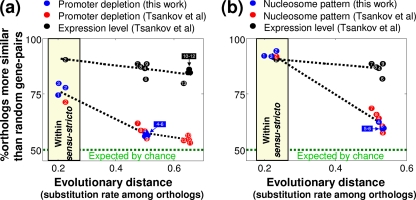
These results indicate that the evolutionary dynamics of nucleosome occupancy differ from those of gene expression. To further examine this phenomenon, we asked how well we can distinguish orthologs from random gene pairs based either on similarity in their nucleosome patterns or on similarity in their expression levels. Expression levels (39) were able to distinguish orthologs for all pairwise species comparisons, and this capacity decreased only slightly with evolutionary distance and remained high even for relatively distant species, such as S. cerevisiae and C. albicans (∼50% coding sequence identity) (Fig. 4 a). In contrast, orthology was distinguished by the patterns of nucleosomes only for the closely related species, and this distinction dropped sharply for more distant comparisons (Fig. 4). In fact, for all species comparisons, except those within the sensu stricto group (yellow section in Fig. 4a and b), the predictive power of nucleosome patterns was close to that expected by chance. Similar results were obtained when we used either promoter nucleosome depletion (Fig. 4a) or correlation in nucleosome patterns (Fig. 4b), and we used nucleosome data sets either from this work (blue circles) or from work by Tsankov et al. (39) (red circles). Notably, in the analysis of the data reported by Tsankov et al., we also included additional species (S. castellii and C. albicans) and still obtained similar results (Fig. 4). This consistency among analyses of different data sets, species, and measures of nucleosome similarity strongly supports the generality of our conclusions.
Fig. 4.
Evolution of nucleosome positioning versus gene expression. For each species pair, we estimated the rate of substitutions among aligned ortholog sequences using the Jukes-Cantor distance, as a measure of evolutionary distance (6). This was compared to the percentage of orthologs that are more similar than expected by chance (corresponding to the percentage of true positives at a false-positive rate of 50%), for either expression levels (black), promoter nucleosome depletion (a), or correlation of nucleosome occupancy patterns across the promoter and coding region (b). Note that comparison of nucleosome occupancy patterns was not performed for C. albicans (versus all other species), as C. albicans has higher nucleosome spacing than all other species examined (39), and thus the nucleosome patterns could not be aligned properly in these comparisons. Blue circles represent analysis of nucleosome data sets from this work, while red circles represent analysis of nucleosome data sets from the work of Tsankov et al. (39). Dashed black lines indicate linear regression, the green line indicates complete divergence (orthologs are as similar as random gene pairs), and the yellow section indicates comparisons only among the Saccharomyces sensu stricto species. Pairwise species comparisons are numbered as follows: 1, S. cerevisiae versus S. kudriavsevii; 2, S. cerevisiae versus S. bayanus; 3, S. kudriavsevii versus S. bayanus; 4, S. cerevisiae versus C. glabrata; 5, S. kudriavsevii versus C. glabrata; 6, S. bayanus versus C. glabrata; 7, S. cerevisiae versus S. castellii; 8, S. bayanus versus S. castellii; 9, C. glabrata versus S. castellii; 10, S. cerevisiae versus C. albicans; 11, S. bayanus versus C. albicans; 12, C. glabrata versus C. albicans; 13, S. castellii versus C. albicans.
DISCUSSION
Genome-wide profiling of nucleosome occupancy in the budding yeast S. cerevisiae revealed several global features, including the organization of nucleosomes along genes and the classification of promoters into those which contain NDR and those which do not. By comparing the genome-wide nucleosome occupancy of four yeast species separated by various evolutionary distances, we found that these global features are maintained. However, two notable differences were observed between the sensu stricto species and C. glabrata: a global shift of nucleosome positions in C. glabrata, and the lack of conservation in gene-specific nucleosome patterns.
Global shift of nucleosome positions and promoter elements.
Although a similar periodic pattern of nucleosomes was observed in all species, this pattern was shifted upstream in C. glabrata by ∼20 bp. Alignment of orthologous coding sequences does not reveal a similar shift, and thus the shift in nucleosome positions cannot be dictated by the coding sequence (data not shown). Promoter sequences have diverged extensively and therefore cannot be directly aligned, yet analysis of promoter features that may influence nucleosome positioning does reveal a similar shift of the TSS, TFBS, and DNA bendability. These results suggest that evolutionary changes in promoter architecture have directly influenced the positions of the NDR and surrounding nucleosomes (e.g., −1 and +1) and that these initial shifts further influence nucleosome positioning along the entire coding region, since the positions of adjacent nucleosomes are linked by statistical positioning (20, 25) and by an active packing mechanism (45). Notably, the shift of TSS positions only partially accounts for the shift in nucleosome positioning, indicating that the linkage between these features is not complete. The shifts of TFBSs and DNA bendability cannot be determined with high resolution, yet both changes are comparable in magnitude to the observed shifts of nucleosomes rather than TSS positions (35) (see Fig. S1 in the supplemental material). This may indicate that nucleosome positioning diverged primarily through the location of NDRs, which are partially encoded by TFBSs and low DNA bendability, and that these evolutionary changes facilitated similar, yet smaller, movements of TSS positions. The fact that multiple types of promoter elements have shifted in the same direction at thousands of promoters may suggest that different promoter organizations were selected for in these two evolutionary lineages.
Extensive divergence of gene-specific nucleosome patterns.
Gene-specific nucleosome patterns diverged extensively, even among genes with conserved expression patterns. This finding echoes previous reports showing that orthologous promoters diverged extensively in sequence while still driving similar expression patterns (7, 12, 24, 29–31, 38, 40). Furthermore, recent studies have shown that transcription factor binding diverged extensively among closely related species (4, 27). Thus, both promoter sequences and nucleosome occupancy can diverge to the point of no detectable similarity, while gene expression patterns are still conserved. This does not imply that promoter sequences and nucleosome patterns are not important for gene regulation. Rather, it suggests that the same functional outcome can be obtained by widely different combinations of promoter sequences and nucleosome occupancy patterns. These findings suggest a scenario in which specific changes in nucleosome positioning are purged by purifying selection, while many others that do not (or only weakly) affect gene expression are free to evolve, and with sufficient evolutionary time, extensive neutral evolutionary changes accumulate while the functional outcome—gene expression pattern—is maintained.
While individual orthologs between sensu stricto species and C. glabrata typically show extensive divergence in nucleosome patterns, there are also specific examples of conservation. In particular, genes with high expression variability and a TATA box preferentially maintain their OPN patterns, suggesting that these patterns are functionally important for dynamic regulation of gene expression. Increased conservation is also observed when nucleosome patterns are averaged over functionally related gene sets (see Fig. S8 in the supplemental material) (39). Analysis of functional gene sets is thus able to average out the extensive changes while highlighting conserved features that are often coherent among related genes.
Our results suggest that nucleosome divergence is often driven by neutral drift and is, overall, not a major driver of gene expression evolution. However, this does not exclude the possibility that, in many specific instances, changes in nucleosome occupancy have played a significant role in expression and phenotypic divergence. Indeed, two recent studies have demonstrated correlations between divergence of promoter nucleosome depletion and gene expression (10, 39). These studies highlighted the potentially adaptive role of nucleosome patterns but have focused primarily on specific sets of respiration-related genes that diverged in expression between fermentative (e.g., S. cerevisiae) and respirative (e.g., C. albicans) yeasts (16). When all genes are analyzed, the correlation between divergence of promoter nucleosome depletion and divergence of gene expression becomes very low (R ∼ 0.1), both in our comparison of S. cerevisiae to C. glabrata and in previous data sets comparing S. cerevisiae to C. albicans (10, 39) (see Fig. S7 and S9 in the supplemental material). Thus, nucleosome occupancy may play a role in gene expression and phenotypic evolution, but these cases appear to constitute an exception (albeit an important one) rather than the rule. We also note that even in the case of respiration genes, the observed correlation does not imply causality: nucleosome divergence at respiration genes is linked to the differential occurrence of an AT-rich binding site for the Stb3 transcription factor (16, 23). Therefore, expression divergence could have been driven by differential binding of Stb3 to its sequence-specific binding site, while changes in nucleosome positioning might reflect a by-product of the AT richness of these binding sites.
Clearly, a stronger case could have been made if we were able to distinguish neutral from adaptive nucleosome changes, yet tests for neutrality are based on specific features of coding sequences and thus cannot be applied to patterns of nucleosomes. Recent work suggested that given a sequence-dependent code for nucleosome positioning, one could define which mutations influence nucleosome positioning (analogous to nonsynonymous mutations) and which do not (analogous to synonymous mutations) and then inspect the dN/dS ratios as is done with tests of coding sequences (1). However, such an approach is not suitable for the work described here, as (i) it can be applied only to closely related species and cannot be used for comparisons with more distant species, like C. glabrata, (ii) it examines sequence evolution (and its predicted effects on nucleosomes) but cannot be used with actual data for nucleosome positioning, (iii) it assumes that sequence-dependent models of nucleosome positioning are completely accurate, and (iv) it assumes that natural selection acted only on the effects of mutations on nucleosome positioning, while clearly this is not the case, e.g., as suggested above in the case of Stb3 binding sites.
Finally, our genome-wide data sets of nucleosome occupancy in four yeast species with high sequencing coverage will be useful for further analysis of chromatin structure and gene expression in these yeast species, and this data set can be accessed and viewed at http://barkai-serv.weizmann.ac.il/nuc_species/.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tanja Durbic and Larry Heisler of the Donnelly Sequencing Centre for help with library preparation and mapping sequencing reads and Mark Johnston for providing strains.
This work was supported by the Canadian Institutes for Health Research (MOP 86705 to C.N.), the Bi-national Science Foundation and the European Research Council (Ideas) (to N.B.), the Clore Center at the Weizmann Institute of Science (to I.T.), and the National Science Foundation (MCB0641776 to R.H.M.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Babbitt G. A., Kim Y. 2008. Inferring natural selection on fine-scale chromatin organization in yeast. Mol. Biol. Evol. 25:1714–1727 [DOI] [PubMed] [Google Scholar]
- 2. Badis G., et al. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32:878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basehoar A. D., Zanton S. J., Pugh B. F. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699–709 [DOI] [PubMed] [Google Scholar]
- 4. Borneman A. R., et al. 2007. Divergence of transcription factor binding sites across related yeast species. Science 317:815–819 [DOI] [PubMed] [Google Scholar]
- 5. Cairns B. R. 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461:193–198 [DOI] [PubMed] [Google Scholar]
- 6. Cantor C., Jukes T. 1969. Mammalian protein metabolism, p. 21–132 In Munro H. N. (ed.), Evolution of protein molecules. Academic Press, New York, NY [Google Scholar]
- 7. Chan E. T., et al. 2009. Conservation of core gene expression in vertebrate tissues. J. Biol. 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cliften P., et al. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71–76 [DOI] [PubMed] [Google Scholar]
- 9. Dujon B., et al. 2004. Genome evolution in yeasts. Nature 430:35–44 [DOI] [PubMed] [Google Scholar]
- 10. Field Y., et al. 2009. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat. Genet. 41:438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Field Y., et al. 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fisher S., Grice E. A., Vinton R. M., Bessling S. L., McCallion A. S. 2006. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312:276–279 [DOI] [PubMed] [Google Scholar]
- 13. Ganapathi M., et al. 2011. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 39:2032–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gasch A. P., et al. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartley P. D., Madhani H. D. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ihmels J., et al. 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309:938–940 [DOI] [PubMed] [Google Scholar]
- 17. Ihmels J., et al. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31:370–377 [DOI] [PubMed] [Google Scholar]
- 18. Jiang C., Pugh B. F. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaplan N., et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kornberg R. D., Stryer L. 1988. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 16:6677–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee W., et al. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235–1244 [DOI] [PubMed] [Google Scholar]
- 22. Li B., Carey M., Workman J. L. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 23. Liko D., Slattery M. G., Heideman W. 2007. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 282:26623–26628 [DOI] [PubMed] [Google Scholar]
- 24. Ludwig M. Z., Patel N. H., Kreitman M. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125:949–958 [DOI] [PubMed] [Google Scholar]
- 25. Mavrich T. N., et al. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mavrich T. N., et al. 2008. Nucleosome organization in the Drosophila genome. Nature 453:358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Odom D. T., et al. 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 39:730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roetzer A., et al. 2008. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol. Microbiol. 69:603–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Romano L. A., Wray G. A. 2003. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development 130:4187–4199 [DOI] [PubMed] [Google Scholar]
- 30. Ruvinsky I., Ruvkun G. 2003. Functional tests of enhancer conservation between distantly related species. Development 130:5133–5142 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi H., Mitani Y., Satoh G., Satoh N. 1999. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126:3725–3734 [DOI] [PubMed] [Google Scholar]
- 32. Tillo D., Hughes T. R. 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tirosh I., Barkai N. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 18:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tirosh I., Barkai N., Verstrepen K. J. 2009. Promoter architecture and the evolvability of gene expression. J. Biol. 8:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tirosh I., Berman J., Barkai N. 2007. The pattern and evolution of yeast promoter bendability. Trends Genet. 23:318–321 [DOI] [PubMed] [Google Scholar]
- 36. Tirosh I., Bilu Y., Barkai N. 2007. Comparative biology: beyond sequence analysis. Curr. Opin. Biotechnol. 18:371–377 [DOI] [PubMed] [Google Scholar]
- 37. Tirosh I., Sigal N., Barkai N. 2010. Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol. Syst. Biol. 6:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tirosh I., Weinberger A., Bezalel D., Kaganovich M., Barkai N. 2008. On the relation between promoter divergence and gene expression evolution. Mol. Syst. Biol. 4:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsankov A. M., Thompson D. A., Socha A., Regev A., Rando O. J. 2010. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 8:e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Q. F., et al. 2007. Detection of weakly conserved ancestral mammalian regulatory sequences by primate comparisons. Genome Biol. 8:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wapinski I., Pfeffer A., Friedman N., Regev A. 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 449:54–61 [DOI] [PubMed] [Google Scholar]
- 42. Weiner A., Hughes A., Yassour M., Rando O. J., Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20:90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitehouse I., Rando O. J., Delrow J., Tsukiyama T. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031–1035 [DOI] [PubMed] [Google Scholar]
- 44. Yuan G. C., et al. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626–630 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z., et al. 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332:977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.