Abstract
The Autographa californica multiple nucleopolyhedrovirus (AcMNPV) GP64 protein mediates membrane fusion during entry. Fusion results from a low-pH-triggered conformational change in GP64 and subsequent interactions with the membrane bilayers. The low-pH sensor and trigger of the conformational change are not known, but histidine residues are implicated because the pKa of histidine is near the threshold for triggering fusion by GP64. We used alanine substitutions to examine the roles of all individual and selected clusters of GP64 histidine residues in triggering and mediating fusion by GP64. Three histidine residues (H152, H155, and H156), located in fusion loop 2, were identified as important for membrane fusion. These three histidine residues were important for efficient pore expansion but were not required for the pH-triggered conformational change. In contrast, a cluster of three histidine residues (H245, H304, and H430) located near the base of the central coiled coil was identified as a putative sensor for low pH. Three alanine substitutions in cluster H245/H304/H430 resulted in dramatically reduced membrane fusion and the apparent loss of the prefusion conformation at neutral pH. Thus, the H245/H304/H430 cluster of histidines may function or participate as a pH sensor by stabilizing the prefusion structure of GP64.
INTRODUCTION
Membrane fusion is an essential step in the entry of enveloped viruses into host cells. Typically, the fusion process is mediated by one or more viral envelope glycoproteins, including one identified as a fusion protein. Although viral fusion proteins from different virus groups may differ substantially in structure, they appear to share some fundamental similarities in mechanism. The best-developed models of virus-mediated membrane fusion include several basic steps, as follows. An initial interaction with the viral receptor serves to attach the virus particle to the cell plasma membrane. This is followed by a triggering event that results in a series of conformational changes in the viral fusion protein. The viral fusion protein, which is anchored in the viral envelope, then extends and inserts a hydrophobic domain (fusion peptide or fusion loops) into the host cell membrane. With this insertion, the fusion protein forms a bridge between the virus envelope and the cellular membrane. In some models, such as the well-studied influenza hemagglutinin (HA) protein, the trimeric fusion protein folds back on itself to form a trimer of hairpins, pulling and forcing the viral and cellular membranes toward each other and into close proximity. This is thought to be followed by disruption or perturbation of the outer membrane leaflets and membrane merger, a step referred to as hemifusion. Next, the inner membrane leaflets merge by an unknown mechanism to form a fusion pore (15, 48), and the fusion pore subsequently expands. In many viruses, this precisely controlled and ordered series of events appears to be mediated by a single viral fusion protein and its interaction with the cellular receptor(s) and host membrane. The process is regulated by a critically important triggering event. Several different fusion-triggering mechanisms have been identified, and they involve various combinations of interactions of the fusion protein with a receptor(s), other viral proteins, and/or exposure to low pH. In the latter and perhaps the simplest scenario, a fusion protein that has previously bound a host cell receptor is triggered to initiate fusion by exposure to low pH, which typically occurs in the endosome during virus entry (48).
To date, three classes of viral membrane fusion proteins have been identified largely on the basis of key structural features of the proteins. Class I fusion proteins include envelope proteins from orthomyxoviruses, paramyxoviruses, retroviruses, filoviruses, myxoviruses, and coronaviruses and F proteins from some baculoviruses. Class II includes fusion proteins from alphaviruses and togaviruses. Class III is a recently described group that includes the herpesvirus gB protein, the rhabdovirus G protein, and the baculovirus GP64 protein (4, 48). Although not all fusion proteins are triggered by low pH, each of the three classes of fusion proteins has members that are triggered by low pH (48). While some of the low-pH-triggered fusion proteins have been studied in great detail, the specific mechanism of low-pH triggering in any of them is not clearly understood. Because the low-pH threshold for fusion protein triggering is typically around pH 5 to 6, histidine residues are obvious candidates as pH sensors. Histidine residues have a side-chain pKa of 6.04 in solution and thus may become positively charged below pH 6. Thus, histidine residues may represent the sensor or “switch” for initiating the conformational change in the fusion protein, which leads to membrane fusion (20, 30). At neutral pH, histidine is considered uncharged. At pH values below 6, histidine is doubly protonated and positively charged and thus may interact with negatively charged amino acid side chains. Thus, it is believed that the pH-modulated charge state of histidine residues may be critical for triggering or initiating a pre- to postfusion conformation change in some or many low-pH-triggered fusion proteins (20, 30). Although histidine protonation as a low-pH trigger has not been definitively demonstrated for any viral fusion protein, recent studies provide evidence that histidine residues play critical roles in low-pH-triggered conformation changes in a number of viral fusion proteins (7, 12, 21, 37, 39, 42, 47).
The Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the type species of the Baculoviridae, a family of large, enveloped, double-stranded DNA viruses that replicate only in arthropods and are described almost exclusively from insects (46). Budded virions of AcMNPV enter host cells by endocytosis (26), and virus entry requires a low-pH-triggered membrane fusion event that is mediated by the major envelope glycoprotein GP64 (6). GP64 is a type I membrane protein that is present on the viral envelope as a disulfide-associated homotrimer (23, 33). GP64 has receptor binding activity, although the nature of the receptor is not known (16, 50). In addition to its essential role in virus entry, GP64 is also important for efficient virion budding (32). Based on the crystal structure, GP64 is classified as a class III viral fusion protein (4, 19). The low-pH structure (presumably the postfusion structure) of GP64 is an elongated molecule that is composed mostly of β-sheets with a long central coiled coil, and the overall structure is highly similar to that of herpesvirus gB proteins. Although the class III fusion proteins share no recognizable amino acid sequence similarities and are identified from viruses in different virus families (4), structural similarities show a clear relationship and imply functional similarities.
Within the class III fusion proteins, baculovirus GP64 and herpesvirus gB proteins share close structural similarities, but triggering differs dramatically. Membrane fusion by HSV-1 gB requires its interaction with envelope proteins gD, gH, and gL and may or may not require a low-pH step, depending on the mode of virus entry and the cell type (11, 43). In contrast to herpesvirus gB, however, GP64 independently mediates membrane fusion (requiring no other viral protein interactions) and is triggered by low pH (6). GP64 is an attractive model for studies of class III membrane fusion because GP64-mediated membrane fusion results in very rapid formation of large fusion pores. In comparison, the well-studied influenza HA protein forms smaller fusion pores that flicker and open over a prolonged time period (34–36). Based on the postfusion conformation of AcMNPV GP64, it was previously suggested that the charge state of histidines located at the trimer interfaces of GP64 may function as the pH sensor (19). In the current study, we experimentally examined the roles of all histidine residues in the GP64 ectodomain as potential sensors of low pH and triggers of GP64-induced membrane fusion. We examined each of the 14 ectodomain histidine residues using single and multiple alanine substitutions. We found that histidine residues located within or adjacent to the second potential fusion loop region (H152, H155, and H156) were important for fusion pore expansion or enlargement. In addition, we found that histidine residues located at the base of the long central coiled coil (H245, H304, and H430) may play a role in stability of the neutral-pH conformation of GP64 and thus may represent either the sensor or a component of the low-pH sensor for triggering the conformational change of GP64.
MATERIALS AND METHODS
Cells, transfections, and infections.
Spodoptera frugiperda (Sf9) cells and the cell line Sf9Op1D, a line that constitutively expresses an Orgyia pseudotsugata MNPV (OpMNPV) GP64 protein (34), were cultured at 27°C in TNMFH medium (18) containing 10% fetal bovine serum (FBS). Cells were transfected using CaPO4 precipitation (5). For viral infections, budded virions from cell culture supernatants were incubated on cells at the indicated MOI for 1 h, and then cells were washed once and incubated in TNMFH medium. Times postinfection (p.i.) were calculated from the time the viral inoculum was added.
Site-directed mutagenesis.
Histidine-to-alanine (His-to-Ala) mutations in AcMNPV GP64 were generated using an overlap PCR method, with the plasmid pGEM3ZGP64 (24) as the template. Primer sequences are available upon request. Briefly, the PCR products were purified and digested with unique restriction enzymes (XbaI and NotI or NotI and EcoRI) and used for subcloning into plasmid pBieGP64 (24). Recombinant baculoviruses expressing the modified GP64 proteins were generated by first subcloning overlap PCR products into plasmid pGEM3ZGP64 (using KpnI and NotI or NotI and HindIII sites) and then excising the promoter and modified GP64 open reading frame (ORF) with KpnI and EcoRI and subcloning into the same sites of pFastBac1 (Invitrogen). The construct containing a modified gp64 gene was inserted into the polyhedrin locus of an AcMNPV gp64-null bacmid (vAc64−) by Tn7-mediated transposition (27). All constructs were confirmed by restriction enzyme analysis and DNA sequencing.
Detection of cell surface GP64 by cELISA.
Relative levels of cell surface-localized GP64 protein were analyzed by a cell surface enzyme-linked immunosorbent assay (cELISA) (22, 24). Briefly, at 36 h posttransfection (p.t.), Sf9 cells were fixed in 0.5% glutaraldehyde. Relative levels of cell surface-localized GP64 were measured by cELISA using monoclonal antibody (MAb) AcV5 as described previously (22, 24). To determine whether modified GP64 proteins are capable of undergoing low-pH-induced conformational change, cELISA was performed with MAb AcV1, which recognizes the neutral-pH conformation of GP64 but not the low-pH conformation (51). After 36 h p.t., the transfected Sf9 cells were incubated for 30 min in phosphate-buffered saline (PBS), adjusted to various pH values (4.5 to 7), and then fixed in 4% (wt/vol) paraformaldehyde. The following steps in cELISA were similar to those used for MAb AcV5 as described previously (22, 24).
Analysis of membrane fusion by syncytium formation assay.
Membrane fusion activity of modified GP64 proteins expressed on the surfaces of transfected cells was measured by syncytium formation assays as previously described (22, 25). In brief, Sf9 cells were seeded in 12-well plates and transfected with plasmids encoding wild-type (WT) or modified forms of GP64. After incubation at 27°C for 36 h, cells were exposed to PBS at pH 5.0 for 3 min and then incubated in TNMFH medium for 4 h. A syncytium was defined as a group or mass of fused cells containing at least 5 nuclei. Relative levels of fusion activity were determined by dividing the total number of nuclei found in syncytia by the total number of nuclei in the same field, normalized to parallel data from cells expressing WT GP64, and localized to the cell surface at equivalent levels.
Detection of hemifusion and pore formation.
To detect hemifusion and pore formation by WT GP64 and GP64 constructs containing histidine substitution mutations, a dual dye transfer assay was performed as described previously (25). Briefly, sheep red blood cells (RBCs; HemoStat Laboratories) were colabeled with the membrane-restricted lipid probe (octadecyl rhodamine B chloride or R18) and the cytosolic aqueous dye calcein-AM (Molecular Probes, Invitrogen). At 36 h p.t., transfected Sf9 cells (4 × 105 cells/well in 12-well plates) were washed once with PBS (pH 7.4) and then incubated with dually labeled RBCs for 20 min at room temperature (RT) for binding. Unbound RBCs were removed by washing three times with PBS (pH 7.4). Sf9 cells with bound RBCs were then incubated with PBS at pH 5.0 for 3 min at RT, washed in PBS at pH 7.4, and transferred into TNMFH medium. After incubation for 20 min at 27°C, the transfer of fluorescence was observed by epifluorescence microscopy. Five randomly selected fields were scored for dye transfer. The efficiency of hemifusion was calculated as the ratio of R18 dye-transferred Sf9 cells to RBC-bound Sf9 cells. Similarly, the efficiency of pore formation was estimated as the ratio of calcein dye-transferred Sf9 cells to RBC-bound Sf9 cells.
Transfection-infection assay.
Bacmid DNAs carrying modified gp64 gene constructs (and no WT gp64 gene) were used to transfect Sf9 cells using calcium phosphate precipitation. At 96 h p.t., the supernatant was removed, clarified by centrifugation (10 min at 2,200 × g), and then used to infect Sf9 cells, which were subsequently examined for β-glucuronidase (GUS) activity (expressed under the control of the p6.9 late promoter) at 96 h p.i., as described previously (22, 25). Both transfected and infected cells were stained for GUS activity. To confirm the viability of each bacmid genome and preparation, the same AcMNPV bacmids were used to transfect Sf9Op1D cells (which express a wild-type GP64 protein), followed by infection of Sf9 cells.
Virus growth curves.
To generate virus growth curves, Sf9 cells (1 × 106) were infected in triplicate with each virus in 6-well plates, at the indicated multiplicity of infection (MOI). After a 1-h incubation period, the inoculum was removed and exchanged with TNMFH medium. Supernatants were collected at the indicated times postinfection, and the titers of all supernatants were determined by 50% tissue culture infective dose (TCID50) measurements on Sf9 cells.
Western blot analysis.
Analysis of GP64 proteins by reducing and nonreducing SDS-PAGE (6% or 10% polyacrylamide gels) was described previously (24). GP64 was detected on Western blots using MAb AcV5 at a dilution of 1:1,000.
RESULTS
Conservation of GP64 histidine residues.
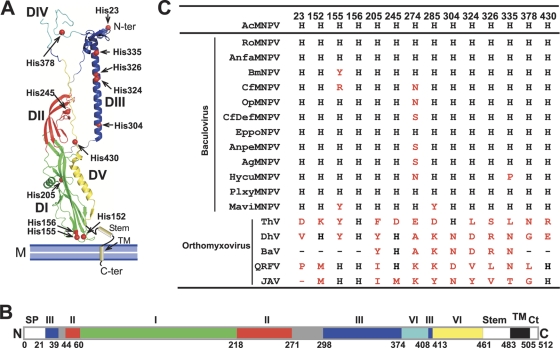
Histidine residues are dispersed among various structural domains of the AcMNPV GP64 protein (19) (Fig. 1A), and several potential histidine clusters are evident. One group of three histidines (H152, H155, and H156) is located in domain DI within and near a region known as fusion loop 2 (Fig. 1A). Residues in fusion loop 2 are critical for membrane fusion and appear to play a role in virus attachment (19, 50). A group of four histidines (H304, H324, H326, and H335) is located along the length of the long central core helix B, which forms a triple-stranded α-helical coiled coil at the heart of the low-pH (postfusion) trimer structure. An additional cluster of three histidine residues (H245, H304, and H430) is found at the base of the long central core helix and in the center of the molecule (Fig. 1A; see also Fig. S2 in the supplemental material). Two histidine residues, H274 and H285, are located in a region that is not visible in the low-pH crystal structure, the so-called variable region, which has the lowest level of conservation among baculovirus GP64 proteins. The variable region forms part of the conformational epitope of neutralizing antibody AcV1, an epitope found only in the prefusion form of GP64 (51).
Fig. 1.
Locations and conservation of histidine residues in the AcMNPV GP64 protein ectodomain. (A) Locations of histidine residues on the postfusion structure of the AcMNPV GP64 ectodomain monomer (Protein Data Bank [PDB] no. 3DUZ). Histidine residues are shown as red spheres on a monomer structure. Domains DI to DV are shown in green, red, blue, cyan, and yellow, respectively, and correspond to the linear domain map shown below in panel B. Two histidine residues (H274 and H285) are absent from the crystal structure. The illustrated structure was prepared with the PyMOL molecular graphics system (version 1.3; Schrödinger, LLC). (B) Linear domain map of GP64. The signal peptide (SP; residues 1 to 20), membrane proximal stem domain (Stem; residues 461 to 482), transmembrane (TM; residues 483 to 504), and cytoplasmic tail (Ct; residues 506 to 512) are absent in the postfusion structure of GP64. (C) Alignment of histidine residues of baculovirus GP64 proteins and their homologues. Conserved histidine residues are shown in black, and variable residues are shown in red. Virus abbreviations are as follows: baculoviruses, Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Rachiplusia ou MNPV (RoMNPV), Anagrapha falcifera MNPV (AnfaMNPV), Bombyx mori NPV (BmNPV), Choristoneura fumiferana MNPV (CfMNPV), Orgyia pseudotsugata MNPV (OpMNPV), Choristoneura fumiferana DEF NPV (CfDefNPV), Epiphyas postvittana NPV (EppoNPV), Antheraea pernyi NPV (AnpeNPV), Anticarsia gemmatalis MNPV (AgMNPV), Hyphantria cunea NPV (HycuNPV), Plutella xylostella MNPV (PlxyMNPV), and Maruca vitrata MNPV (MaviMNPV); orthomyxoviruses, Thogoto virus (ThV), Dhori virus (DhV), Batken virus (BaV), Quaranfil virus (QRFV), and Johnston Atoll virus (JAV).
Histidine residues are highly conserved in baculovirus GP64 proteins. Of the 14 histidine residues of AcMNPV GP64, 10 positions (H23, H152, H156, H205, H245, H304, H324, H326, H378, and H430) appear to be absolutely conserved among the baculovirus GP64 proteins (Fig. 1C). Four additional histidine positions show high (H155, H285, H335) to moderate (H274) levels of conservation. Alignments of baculovirus GP64 and the more distantly related orthomyxovirus GP75 proteins show that His residues are not as highly conserved between baculovirus and orthomyxovirus GP75 proteins. Various levels of conservation are observed between the baculovirus GP64 and orthomyxovirus GP75 proteins, with a high level of conservation at position 156 but various levels of conservation at positions H152, H155, H245, H304, and H430.
GP64 proteins with histidine mutations.
In prior studies, it was reported that His residues in the so-called second fusion loop region (amino acids [aa] 148 to 157) play roles in membrane fusion and receptor binding (19, 50). His residues at the central trimer interfaces were also proposed to possibly function as pH-sensitive triggers of GP64 conformational change. The latter potentially include positions H245, H304, H324 and H326, H335, and H430 (19). To experimentally examine the roles of His residues in the low-pH-triggered conformational change, we generated three sets of histidine mutations. First, we generated a series of GP64 constructs that each contained a single His-to-Ala replacement at each ectodomain histidine position. Next, we generated a number of double or triple His-to-Ala mutations within the trimer interface regions (H245A/H304A, H245A/H335A, H304A/H335A, H324A/H326A, H335A/H430A, H245A/H304A/H335A, and H245A/H304A/H430A), the second fusion loop (H152A/H155A), and in positions where histidine residues are in close proximity (H23A/H378A, H205A/H245A, and H274A/H285A) (Fig. 1A). Finally, we introduced conservative (aromatic and positively charged) amino acid substitutions for histidines in the second fusion loop (H156K, H156Y, H152K/H155K, and H152Y/H155Y).
Expression and cell surface levels.
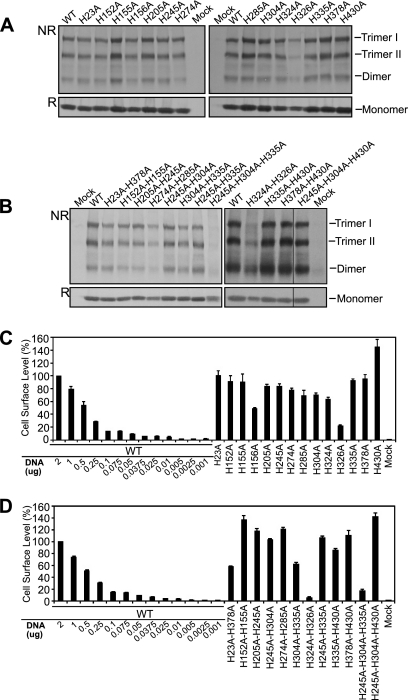
To determine whether substitution mutations in GP64 had unanticipated effects on protein production or surface display, we first examined the effects of His-to-Ala replacements on GP64 protein expression and trimerization. Each GP64 construct was transiently expressed in Sf9 cells and then examined by Western blot analysis under reducing and nonreducing SDS-PAGE (Fig. 2A and B, bottom and top panels, respectively). With the exception of construct H245A/H304A/H335A (which appeared to be expressed at substantially reduced levels), all the single or multiple His-to-Ala replacement constructs were expressed at levels similar to, or slightly lower than, that of wild-type (WT) GP64 (Fig. 2A and B, bottom panels). Analysis by nonreducing SDS-PAGE showed that trimerized GP64 was detected for each construct except for H245A/H304A/H335A (Fig. 2A and B, top panels). On nonreducing gels, the typical distribution of oligomers (trimers I and II and dimer) (33) was observed in all cases except for construct H245A/H304A/H335A. These data suggest that expression and oligomerization were normal for most constructs, but the combination of substitutions in construct H245A/H304A/H335A affected folding or stability of the GP64 trimer prior to or during formation of the intermolecular disulfide.
Fig. 2.
Transient expression of GP64 constructs containing His-to-Ala mutations. (A, B) Analysis of transient expression and trimerization of GP64 constructs. Sf9 cells were transfected with 2 μg of plasmid DNA encoding wild-type (WT) GP64 or a GP64 construct containing a His-to-Ala mutation (indicated above each lane). Cell lysates were prepared 36 h after transfection and electrophoresed under nonreducing (NR) or reducing (R) conditions. GP64 proteins were detected by Western blot analysis using monoclonal antibody AcV5. The positions of oligomeric forms of GP64 (trimers I and II and dimer) are indicated on the right. (C, D) Analysis of cell surface levels of GP64 constructs. The relative cell surface level of each GP64 construct was measured by cELISA, using monoclonal antibody AcV5. Each GP64 construct was expressed by transfecting Sf9 cells with 2 μg of the appropriate plasmid DNA. Values represent the means obtained from triplicate transfections and are normalized to those of cells transfected with 2 μg of the plasmid DNA expressing WT GP64 (representing 100%). A standard curve generated by transfecting decreasing quantities of the plasmid encoding WT GP64 is shown on the left. Plasmid DNA quantities for the plasmid expressing WT GP64 are indicated by the numbers below the graph (in μg). Error bars represent the standard deviations from the means.
To determine if modified GP64 constructs were transported to and displayed at the cell surface, transfected Sf9 cells displaying GP64 constructs were fixed at 36 h p.t. and analyzed by cELISA. Most of the modified GP64 constructs were expressed at the cell surface at a level similar to, or sometimes higher than, that of wild-type GP64 (Fig. 2C and D; Table 1). Moderate reductions of surface levels (levels of approximately 22 to 63% relative to WT levels) were observed for constructs H156A, H326A, H23A/H378A, and H304A/H335A. More dramatic reductions in cell surface levels for constructs, H324A/H326A, and H245A/H304A/H335A were observed. However, in all cases, the cell surface levels that were detected were sufficient for detection and analysis of fusion activity by WT GP64.
Table 1.
Phenotypes of wild-type and histidine-mutated AcMNPV GP64 proteinsa
| Construct | Trimer | Surface (%) | Fusion (syncytium) (%) | Dye transfer (%) |
pH of SF | Infectivity |
||
|---|---|---|---|---|---|---|---|---|
| R18 | Calcein-AM | Rescue | Virus titer (TCID50/ml) | |||||
| WT | + | 100 | 100 | 56.6 ± 0.7 | 56.6 ± 0.7 | 0 (pH 5.7) | + | (2.2 ± 0.5) × 108 |
| H23A | + | 101.7 ± 6.5 | 100.0 ± 0.2 | ND | −0.1 | + | (2.1 ± 0.5) × 108 | |
| H152A | + | 91.2 ± 8.6 | 29.1 ± 2.8 | 64.7 ± 12.4 | 56.6 ± 9.2 | ND | − | NA |
| H155A | + | 90.8 ± 11.9 | 90.7 ± 1.3 | ND | −0.2 | + | (0.4 ± 0.1) × 108 | |
| H156A | + | 48.8 ± 2.0 | 5.1 ± 1.4 | 59.3 ± 7.2 | 50.0 ± 8.2 | ND | − | NA |
| H156K | + | 66.3 ± 3.0 | 1.7 ± 0.8 | 47.4 ± 5.9 | 45.1 ± 6.0 | ND | − | NA |
| H156Y | + | 85.6 ± 5.4 | 5.5 ± 1.6 | 56.1 ± 3.2 | 52.2 ± 3.0 | ND | − | NA |
| H205A | + | 83.9 ± 3.2 | 98.9 ± 0.9 | ND | 0 | + | (1.9 ± 0.2) × 108 | |
| H245A | + | 83.4 ± 4.2 | 100.0 ± 0.2 | ND | −0.1 | + | (2.0 ± 0.5) × 108 | |
| H274A | + | 78.1 ± 2.8 | 99.7 ± 0.3 | ND | 0 | + | (4.2 ± 0.4) × 108 | |
| H285A | + | 69.6 ± 7.6 | 99.9 ± 0.2 | ND | 0 | + | (2.3 ± 0.2) × 108 | |
| H304A | + | 70.7 ± 2.6 | 98.9 ± 0.9 | ND | 0 | + | (1.6 ± 0.2) × 108 | |
| H324A | + | 63.3 ± 3.1 | 94.7 ± 1.9 | ND | −0.1 | + | (3.4 ± 0.2) × 108 | |
| H326A | + | 22.4 ± 1.3 | 70.2 ± 3.2 | ND | −0.1 | + | (2.6 ± 0.3) × 108 | |
| H335A | + | 93.1 ± 1.7 | 100.0 ± 0.4 | ND | 0 | + | (2.4 ± 0.1) × 108 | |
| H378A | + | 95.9 ± 5.8 | 99.2 ± 0.6 | ND | 0 | + | (2.7 ± 0.4) × 108 | |
| H403A | + | 145.2 ± 11.5 | 99.9 ± 0.3 | ND | 0 | + | (2.2 ± 0.3) × 108 | |
| H23A/H378A | + | 58.0 ± 1.1 | 99.9 ± 0.5 | ND | −0.2 | + | (2.9 ± 0.2) × 108 | |
| H152A/H155A | + | 137.1 ± 7.3 | 0 | 28.3 ± 4.1 | 23.6 ± 1.8 | ND | − | NA |
| H152K/H155K | + | 78.0 ± 2.6 | 0 | 51.8 ± 2.2 | 48.1 ± 2.1 | ND | − | NA |
| H152Y/H155Y | + | 124.7 ± 2.0 | 0 | 39.1 ± 3.3 | 37.3 ± 3.3 | ND | − | NA |
| H205A/H245A | + | 118.7 ± 2.8 | 98.8 ± 1.0 | ND | −0.2 | + | (1.9 ± 0.3) × 108 | |
| H245A/H304A | + | 103.3 ± 1.6 | 88.2 ± 3.2 | ND | −0.7 | + | (5.3 ± 0.6) × 105b | |
| H274A/H285A | + | 121.8 ± 2.4 | 99.8 ± 0.2 | ND | −0.2 | + | (4.4 ± 0.4) × 108 | |
| H245A/H335A | + | 106.5 ± 2.4 | 93.4 ± 2.5 | ND | −0.3 | + | (0.3 ± 0.0) × 108 | |
| H304A/H335A | + | 62.6 ± 3.1 | 81.0 ± 2.6 | ND | −0.2 | + | (2.0 ± 0.2) × 108 | |
| H324A/H326A | + | 5.9 ± 1.7 | 0 | 0 | NA | − | NA | |
| H335A/H430A | + | 86.1 ± 2.2 | 99.9 ± 0.2 | ND | −0.1 | + | (1.1 ± 0.1) × 108 | |
| H378A/H430A | + | 111.2 ± 7.4 | 99.8 ± 0.2 | ND | −0.1 | + | (4.2 ± 0.1) × 108 | |
| H245A/H304A/H335A | − | 17.7 ± 2.2 | 0 | 0 | NA | − | NA | |
| H245A/H304A/H430A | + | 142.6 ± 5.6 | 5.6 ± 2.0 | 6.8 ± 1.3 | 4.3 ± 1.1 | −0.7 | + | (2.8 ± 0.4) × 103b |
ND, not done; NA, not avaliable; pH of SF, pH threshold of syncytium formation.
Infection at an MOI of 1.
Fusion activity of GP64 constructs with histidine mutations. (i) Syncytium formation assays.
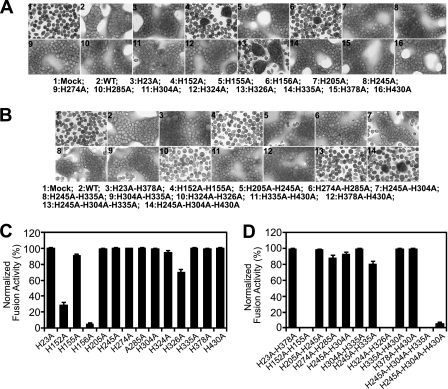
Membrane fusion activity by GP64 constructs containing His-to-Ala replacements was first examined and measured by syncytium formation assays. For most GP64 constructs that contained a single His-to-Ala replacement, the replacement had no substantial effect on syncytium formation (Fig. 3A and C). However, substitutions at three positions showed significant effects. GP64 construct H326A showed a slight reduction in normalized fusion activity (resulting in 75% of WT GP64 activity). Constructs H152A and H156A resulted in more dramatic reductions in fusion activity, with only 25% and 5% of WT GP64 fusion activity remaining, respectively (Fig. 3A and C; Table 1).
Fig. 3.
Analysis of fusion activity by GP64 proteins with His-to-Ala mutations. (A, B) Syncytium formation assays. Sf9 cells were transfected with 2 μg of a plasmid expressing WT GP64 or expressing GP64 proteins containing histidine mutations (indicated below each panel). At 36 h p.t., cells were incubated in PBS at pH 5.0 for 3 min, returned to TNMFH medium (pH 6.2), incubated for 4 h, and then photographed under phase-contrast microscopy (×200). (C, D) Analysis of fusion activity in syncytium formation assays. The relative fusion activity, as determined by syncytium formation assays, was determined for each GP64 construct containing a His-to-Ala mutation. Fusion efficiency was determined for each construct by measuring the percentages of cells found in syncytia. For each field of cells, the number of nuclei in syncytia was divided by the total number of nuclei in the field. Percentages were normalized to parallel syncytium formation data from WT GP64 (100%) that was localized to the cell surface at equivalent levels. For each replicate transfection, 5 fields were analyzed. The means and standard deviations of triplicate transfections are shown.
Like the single His-to-Ala replacements, many of the double and triple alanine substitutions had no substantial effect on membrane fusion activity in syncytium formation assays (Fig. 3D; Table 1). However, dramatic reductions in fusion activity were detected in four constructs. No fusion activity was observed for constructs H152A/H155A, H324A/H326A, and H245A/H304A/H335A, and only 5% of WT activity was observed for construct H245A/H304A/H430A (Fig. 3D; Table 1). In the case of construct H245A/H304A/H335A, the defect in membrane fusion may result from a folding or conformation defect since the trimerized form of GP64 was not detected in the prior analysis (Fig. 2B, lane 10).
(ii) Hemifusion assays.
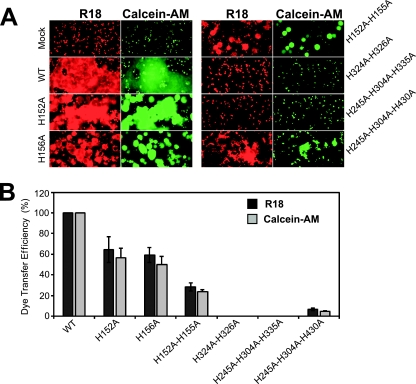
The process of membrane fusion can be subdivided into steps that include the following: (i) the initial mixing of the outer leaflets of the opposing membranes, also known as hemifusion; (ii) the merger of the inner leaflets of the two membranes to form a fusion pore; and (iii) the expansion of the fusion pore. To identify the stage in fusion that was affected by GP64 constructs that were deficient in membrane fusion, we used a previously described dual fluorescent dye labeling assay (24) to distinguish between membrane merger, pore formation, and pore enlargement. Red blood cells (RBCs) were dually labeled with membrane-restricted dye (R18) and a cytosolic dye (calcein-AM) and then bound to Sf9 cells displaying GP64 constructs that were previously found to be defective for fusion in syncytium formation assays (H152A, H156A, H152A/H155A, H324A/H326A, H245A/H304A/H335A, and H245A/H304A/H430A) or a control WT GP64. After exposure to low pH (5.0), transfer of membrane and cytosolic dyes was monitored, and efficiencies were calculated (Fig. 4B; Table 1). Surprisingly, GP64 constructs H152A and H156A, which showed severely reduced fusion in syncytium formation assays, resulted in transfer of both membrane and cytosolic dyes in the hemifusion assay. We noted, however, that cytosolic dye transfer was observed with mostly single Sf9 cells (Fig. 4A, H152A and H156A with calcein-AM), and the cytosolic dye was not transferred into a large syncytial mass, as observed with WT GP64. This suggests that the defect in syncytium formation occurs after the formation of the fusion pore and may be related to fusion pore expansion. A similar result that was even more pronounced was observed when GP64 containing the double mutation (H152A/H155A) was examined. In these three cases, dye transfer efficiencies for both dyes were moderate (24 to 65%) in comparison with WT GP64-mediated membrane fusion (Fig. 4B and Table 1, see H152A, H156A, and H152A/H155A).
Fig. 4.
Analysis of hemifusion and fusion-pore formation by fusion-deficient GP64 proteins. (A) Red blood cells (RBCs) that were dually labeled with R18 and calcein-AM dyes were bound to Sf9 cells transiently expressing GP64 protein constructs. The cells were exposed to acidic PBS (pH 5.0) for 3 min to induce membrane fusion and then examined after a 20-min incubation period. Membrane dye transfer (R18; left panels) or cytosolic dye transfer (calcein-AM; right panels) were monitored by fluorescence microscopy. Each GP64 construct (or control WT GP64) is indicated on the left or right of the panels (Mock, no GP64 expressed; WT, wild-type GP64 expressed). (B) Analysis of dye transfer efficiency. The efficiency of lipid or cytosolic dye transfer was estimated from the ratio of the number of R18-transferred or calcein-AM-transferred Sf9 cells to the number of Sf9 cells with RBCs bound. Five fields were examined for each GP64 construct. The data are the means from three independent experiments, and error bars represent the standard errors of the means.
In the case of the triple His-to-Ala replacement construct H245A/H304A/H430A, a severely reduced level of dye transfer was observed for both the membrane and cytosolic dyes (approximately 6% compared with that of WT GP64) (Fig. 4A and B, H245A/H304A/H430A). This is consistent with the low level of syncytium formation observed previously (Fig. 3D) and suggests a general inefficiency in membrane fusion, perhaps indicating loss of function at or prior to the initial step of membrane merger and consistent with a defect in pH sensing or triggering. For two constructs that were negative in the syncytium formation assay, H324A/H326A and H245A/H304A/H335A, no dye transfer was detected in the hemifusion assay (Fig. 4A). Lack of fusion in these latter cases may have resulted from failure of the protein to trimerize (Fig. 2B, lane 10, H245A/H304A/H335A) or low levels of surface expression (Fig. 2B and D, H324A/H326A).
(iii) pH threshold measurements.
Mutations in residues involved in pH sensing or triggering may affect the pH required for triggering fusion. To examine possible effects of histidine mutations on the pH threshold required for triggering GP64-mediated membrane fusion, Sf9 cells expressing GP64 constructs with His-to-Ala replacements were exposed to a range of pH values, and fusion was analyzed by syncytium formation assays. For each construct, the pH threshold of fusion was defined as the highest pH value at which syncytium formation was observed. Because cell surface expression levels differed for various GP64 constructs (and cell surface levels may affect the measured threshold of low-pH-triggered fusion), Sf9 cells were transfected with various quantities of plasmid DNA expressing WT GP64 to titrate cell surface levels of GP64 and to measure the corresponding pH thresholds. pH thresholds for membrane fusion for each GP64 construct containing a His-to-Ala mutation were determined and compared to the threshold determined for WT GP64 expressed at a similar cell surface level. For all expression levels of WT GP64, the pH threshold for fusion in this assay was in a very narrow range, from pH 5.6 to 5.7. For alanine substitution constructs, a shift in the pH threshold for initiating syncytium formation of >0.2 pH units was defined as a substantial change. For 19 of the 26 GP64 His-to-Ala constructs, we detected no substantial change in the pH threshold for initiation of membrane fusion (Table 1). In the case of substitutions in the fusion loop (H152A, H156A, H152A/H155A), our prior analysis showed that syncytia were inefficiently formed or failed to form even though membrane merger and pore formation occurred (Fig. 3 and 4, H152A, H156A, and H152A/H155A). Therefore, constructs H152A, H156A, and H152A/H155A were excluded from this analysis. Only three His-to-Ala replacement constructs with substantial reduction in the pH threshold (≥0.3 units) for fusion were observed, and these included constructs with mutations at or near the base of the long central coiled coil (Table 1, H245A/H304A, H245A/H335A, and H245A/H304/H430A). These results show that histidine residues that are associated with the central coiled-coil domain influence the pH threshold required for GP64-mediated membrane fusion activity and therefore may be directly involved in pH triggering of the conformational change.
Low-pH-induced conformational change in GP64 constructs.
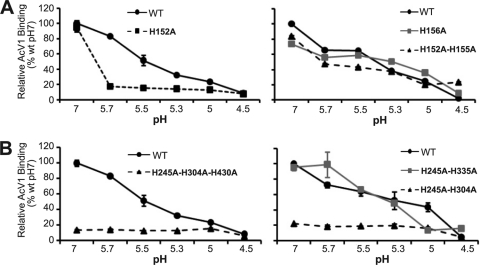
To further examine the GP64 His-to-Ala constructs that resulted in a significant shift in the pH threshold for membrane fusion, we asked whether the pH threshold shift reflected a change in the threshold for initiating a conformational change in GP64. To identify changes in GP64 conformation, we used the conformation-specific monoclonal antibody AcV1, which binds to the neutral (prefusion) conformation of AcMNPV GP64 but not to the low-pH (postfusion) conformation (50, 51). The AcV1 epitope has been mapped to the central variable domain of GP64 (aa 271 to 294) (51). The region containing the AcV1 epitope is not visible in the postfusion structure of GP64 (19). AcV1 is the only conformation-specific MAb that is available for GP64. To examine the pH-triggered conformation change in selected GP64 His-to-Ala constructs, each construct was transiently expressed in Sf9 cells and then incubated at selected pH values for 30 min before adding the conformation-specific AcV1 antibody and assessing binding (see Materials and Methods). As a control for cell surface localization, MAb AcV5 (a MAb that binds to denatured GP64) was used to detect GP64 at the cell surface (after denaturation and fixation with 0.5% glutaraldehyde) in parallel transfections (not shown). Successively lowering the pH of the medium from 7.0 to 4.5 resulted in an incremental decrease in AcV1 binding to WT GP64 but with a relatively dramatic decrease of between pH 5.7 and 5.3 (Fig. 5, WT). For GP64 constructs that contain His replacements in the fusion loop region (H152A, H156A, and H152A/H155A), AcV1 binding was generally similar to that of WT GP64 (Fig. 5A). For GP64 constructs H156A and H152A/H155A, the loss of AcV1 binding at various pH values was similar to, and paralleled, that of WT GP64. However, in the case of construct H152A, AcV1 binding was dramatically lower at pH 5.7, suggesting that the H152A construct may be somewhat more sensitive to the pH-induced conformation change. Importantly though, near-WT levels of AcV1 binding to constructs containing fusion loop His-to-Ala mutations at pH 7 suggest that these mutations had no detectable effect on the prefusion conformation of GP64 (Fig. 5A).
Fig. 5.
Analysis of low-pH-induced conformational changes in GP64 constructs containing His-to-Ala replacements. Each graph shows binding of the (prefusion) conformation-specific MAb AcV1 to transiently expressed GP64 constructs or WT GP64 over the indicated range of pH values. GP64 constructs were transiently expressed in Sf9 cells, and AcV1 binding was determined by cELISA at 36 h posttransfection (see Materials and Methods). Values represent the means from triplicate transfections and are normalized to that of Sf9 cells transfected with 2 μg of plasmid DNA expressing WT GP64 at neutral pH (pH 7). For all constructs, expression was confirmed by parallel cELISA using MAb AcV5, which is not conformation specific.
Next, we examined GP64 constructs with substitutions along the central coiled-coil core of the trimer. We observed two different effects of the substitutions. AcV1 binding to construct H245A/H335A closely mirrored that of WT GP64 (Fig. 5B, right), suggesting that the conformational response to pH was similar to that of WT GP64. In contrast, AcV1 binding to constructs H245A/H304A and H245A/H304A/H430A was exceptionally low throughout the pH range (pH 7 to 4.5), even though binding by a control antibody (AcV5, which is not conformation specific) was similar to that for WT GP64 (data not shown). Thus, although GP64 was detected on the cell surface (as indicated by AcV5 binding), the pH 7 conformation of constructs H245A/H304A and H245A/H304A/H430A was altered, resulting in severely reduced AcV1 binding and no substantial reduction in AcV1 binding at lower pH values (Fig. 5B). These data indicate that little of the H245A/H304A or H245A/H304A/H430A protein at the cell surface was in the prefusion conformation and suggest that these mutations may destabilize the neutral-pH conformation of GP64.
Conserved substitutions of His residues in a fusion loop domain.
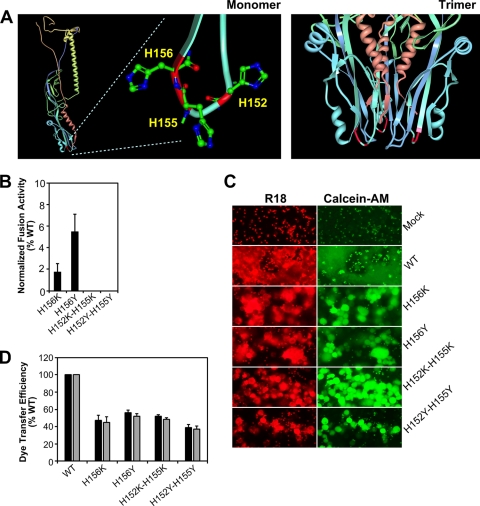
Although alanine substitutions for histidines in the fusion loop region dramatically reduced fusion activity, as observed by loss of syncytium formation (Fig. 3), they did not inhibit membrane or cytosolic dye transfer (Fig. 4), nor did they abrogate the pH-triggered conformational change (Fig. 5A). This suggests that fusion loop histidine residues are unlikely to function as the trigger of the conformational change, although H152A appears to influence the pH threshold for the conformational change. To determine whether effects on membrane fusion were related to the aromatic or charged nature of the histidine side chains in the fusion loop region, we generated conservative substitutions in these positions and examined the resulting GP64 constructs in membrane fusion and dye transfer assays. We initially generated GP64 constructs H156K and H152K/H155K, which contain positively charged lysine substitutions for histidine residues at H156 or H152/H155. We also generated constructs H156Y and H152Y/H155Y, which substitute tyrosine, an amino acid with an aromatic side chain. GP64 constructs containing the above-described substitutions were transiently expressed in Sf9 cells. Expression, trimerization, and cell surface display were confirmed to be at or near WT GP64 levels, using the previously described methods. For the four constructs containing conservative substitutions in the fusion loop histidines (Fig. 6A), syncytium formation was either dramatically reduced (approximately 2 to 6% of WT GP64) or undetectable (Fig. 6B; Table 1), indicating that neither the charged nor the aromatic amino acid could substitute for histidine in these positions. We also examined each construct for membrane merger and pore formation using dual dye-labeled RBCs, as described earlier. Neither charged nor aromatic substitutions in fusion loop histidine residues resulted in normal membrane fusion (Fig. 6C), and results were similar to those observed for alanine substitutions in these positions (Fig. 4A). Although syncytium formation was severely reduced or absent (Fig. 6B), GP64 constructs containing charged or aromatic substitutions in H152, H155, and H156 mediated transfer of both membrane and cytosolic dyes at moderate efficiencies (Fig. 6C and D; Table 1). Thus, GP64 proteins containing both neutral (alanine) and conservative (lysine and tyrosine) substitutions in fusion loop histidines were fusion defective, and in all those substitution constructs, the defect appears to represent a defect in fusion pore expansion.
Fig. 6.
Analysis of conserved substitutions for histidine residues at the second fusion loop of GP64. (A) Location of three histidine residues on the second fusion loop of the postfusion structure of the AcMNPV GP64 monomer and trimer. The positions of histidine residues are shown in red on the ribbon diagrams of the monomer (left) and trimer (right), and histidine side chains are shown as sticks on the monomer (left). (B) Analysis of relative fusion activity of GP64 constructs containing conserved fusion loop histidine mutations. Relative fusion activity was evaluated by measuring the efficiency of syncytium formation from representative random fields and normalized to parallel syncytium formation data from wild-type GP64 that was localized to the cell surface at equivalent levels, as described for Fig. 3C and D. The means and standard deviations from triplicate transfections are shown. (C, D) Analysis of hemifusion and fusion-pore formation by GP64 constructs containing conserved fusion loop histidine mutations. Dually labeled red blood cells (RBCs) were bound to Sf9 cells that were transiently expressing GP64 protein constructs (as labeled on the right). Exposure to low pH and the analysis of dye transfer efficiency were carried out as described for Fig. 4. (C) Fluorescence microscopy and dye transfer to Sf9 cells. (D) The means and standard deviations of dye transfer efficiency obtained from triplicate transfections are shown.
Virus infectivity.
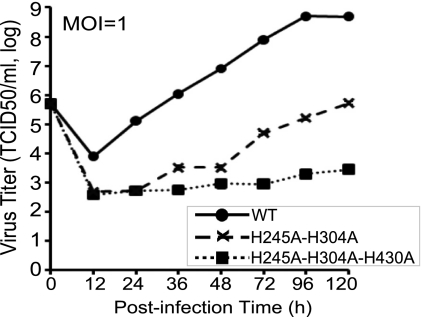
GP64 is an essential structural protein (29). In addition to its function in membrane fusion, GP64 is also important for virus attachment (receptor binding) and virion egress (budding). Because individual histidine residues may be required for critical functions not related to membrane fusion, we asked whether GP64 constructs containing histidine replacements were capable of rescuing virus infectivity of a GP64-null virus. We first used a transfection-infection assay (23) to determine whether GP64 constructs containing histidine mutations were capable of rescuing virus infectivity. For these studies, each GP64 construct (plus a GUS reporter gene under a p6.9 late promoter) was inserted into the polyhedrin locus of a gp64-null bacmid. Each bacmid was used to transfect permissive Sf9 cells in a transfection-infection assay. Initiation of productive infection in the transfected cells was confirmed by analysis of GUS activity at 96 h p.i. (data not shown). After 96 h p.i., supernatants from the transfected cells were collected and used to infect Sf9 cells, which were then stained for GUS activity at 5 days p.i. to identify cells infected by progeny viruses generated from the initial transfection. Results of the transfection-infection assay are summarized in Table 1 (infectivity rescue). We found that, in general, constructs that were positive for membrane fusion in syncytium formation assays (Fig. 3C and D) also rescued virus infectivity. GP64 constructs H152A, H156A, H156K, H156Y, H152A/H155A, H152K/H155K, H152Y/H155Y, H324A/H326A, and H245A/H304A/H335A showed no detectable rescue of viral replication, consistent with the defect in membrane fusion observed for those constructs (Table 1). To further examine rescue of infectivity, bacmids containing GP64 constructs that rescued infectivity were amplified in Sf9Op1D cells and then used to infect Sf9 cells at an MOI of 5. Supernatants were collected at 120 h p.i., and infectious virus titers were determined. The virus titers for most of those viruses were comparable to that of wild-type AcMNPV (Table 1). Two viruses, containing GP64 constructs H245A/H304A and H245A/H304A/H430A, replicated to much lower titers. Because it was not possible to produce sufficient titers of those viruses for the above-described experiment, Sf9 cells were infected at an MOI of 1 with viruses expressing constructs H245A/H304A and H245A/H304A/H430A, and growth curves for those viruses were compared with that of a control virus expressing WT GP64 and also infected at an MOI of 1 (Fig. 7). Maximal viral titers of constructs H245A/H304A and H245A/H304A/H430A reached only 5.3 × 105 and 2.8 × 103 at 120 h p.i., respectively, compared with 2.2 × 108 for the control virus expressing WT GP64. Thus, single His-to-Ala mutations in fusion loop histidines H152 and H156 had severe effects on viral replication, while the substitution at H155 alone had little effect. Failure of constructs H324A/H326A, and H245A/H304A/H335A to support viral replication (Table 1) appears to result from a combination of low expression at the cell surface, failure to trimerize, and lack of membrane fusion activity. Constructs H245/H304 and H245A/H304A/H430A rescued the gp64-null virus, but virus titers were very low. Parallel infections at an MOI of 1 and viral growth curves suggest that these low titers may result from lower rates of virion production, low virion infectivity, or both.
Fig. 7.
Virus one-step growth curves. Budded virus (BV) preparations generated from bacmids encoding the WT GP64 or GP64 construct with the histidine replacements (H245A-H304A and H245A-H304A-H430A) were used to infect Sf9 cells (MOI = 1). Infectious BV yields were determined at the indicated time points by the TCID50 assays. Data points indicate that averages obtained from infections were performed in triplicate, and error bars represent standard deviations.
DISCUSSION
In the current study, we used single and multiple amino acid substitutions to examine the roles of GP64 histidine residues in triggering and mediating membrane fusion. The only single histidine mutations that substantially affected membrane fusion were substitutions of fusion loop histidines H152 and H156 (Fig. 3C and 6A). While single amino acid substitutions in those positions dramatically inhibited syncytium formation, further examination showed that GP64 constructs with those substitutions underwent a pH-triggered conformation change (Fig. 5A), followed by membrane merger and pore formation (Fig. 4). Dye-labeling experiments suggest that these histidine residues play a role in fusion pore expansion. One substitution (H152A) did, however, result in a notable change in the GP64 conformational response to low pH (Fig. 5A), suggesting that the charge state of H152 may also have a role in sensing and responding to low pH. The protonated form of H152 forms a salt bridge with D119 within the same protomer, stabilizing the loop in the postfusion structure (see Fig. S1B in the supplemental material). No similar perturbation of the structure (as detected by AcV1 binding) was observed for mutation constructs H156A and H152A/H155A (Fig. 5A), which also inhibited syncytium formation but appeared to closely mirror WT GP64 in structural response to low pH (Fig. 5). Thus, fusion loop histidines (H152 and H156) appear to be important for fusion pore expansion, and in addition, the charge state of H152 also may play a role in triggering, propagating, or stabilizing the conformational change. The functional importance of H152 and H156 is reflected in the conservation of these residues in the GP64 family proteins from baculoviruses and orthomyxoviruses (Fig. 1).
In contrast to the effects from substitutions of the fusion loop histidines, a cluster of histidines at the base of the central coiled coil was identified as playing a more direct role in the GP64 conformational change. Histidines 245, 304, and 430 flank and ring the outside of the base of the long central helix in the GP64 trimer (Fig. 1; see also Fig. S2 in the supplemental material). Substitutions for these three histidines (H245A/H304A/H430A) did not interfere with protein trimerization, cellular transport, or display of GP64 at the cell surface, but this construct was defective for membrane fusion at a step prior to membrane merger. Combined with a loss of recognition by the conformation-specific MAb AcV1 (which recognizes the prefusion conformation), these data suggest that histidines 245, 304, and 430 may be more directly involved as a pH sensor and/or trigger of the conformational change. While the prefusion conformation has not yet been solved, ionic interactions between these three histidines and nearby charged amino acid side chains stabilize the postfusion conformation (see Fig. S2D in the supplemental material). Histidine 245 forms salt bridges with aspartate 314 and glutamate 307 from an adjacent protomer in the trimer. In addition, histidine 430 forms a salt bridge and a hydrogen bond with an aspartate residue (D301) from an adjacent promoter. Histidine 304 also forms a hydrogen bond with isoleucine 252 from the adjacent protomer. Thus, while it is not known if protonation of these three histidines destabilize the prefusion conformation of GP64 to initiate a conformational change, the salt bridges formed as a result of the protonated side chains of H245 and H430 stabilize the protomer-protomer interactions in the postfusion conformation of the trimer. However, we cannot rule out important roles for other histidine residues in triggering fusion. In the current study, we found that in some cases (constructs H324A/H326A and H245A/H304A/H335A) multiple histidine mutations resulted in low production or transport of GP64 to the cell surface, and we were not able to analyze those constructs in detail. Those histidines residues are all involved in ionic interactions that appear to stabilize the postfusion structure (Fig. S2B), and some or all could be involved in destabilizing the prefusion conformation.
While GP64 constructs H245/H304 and H245A/H304A/H430A had severely reduced membrane fusion activity, some remaining activity was detected, and viruses expressing those constructs were able to replicate, although very inefficiently (Fig. 7). This further confirms that fusion was not abrogated in the presence of these substitutions but only severely reduced.
Accumulating experimental evidence from a number of viral fusion proteins provides support for the so-called histidine switch model (7, 8, 10, 12, 14, 20, 21, 30, 37, 39, 42, 44, 47). However, no single mechanistic paradigm has emerged to describe the triggering of conformational changes in all low-pH-triggered fusion proteins. In some cases, a single critical or dominant histidine residue has been proposed to serve as a functional pH sensor and trigger (12, 14). In other studies, data suggest that multiple histidines (and other residues) play key roles in low-pH sensing and initiation of the conformational changes associated with triggering membrane fusion (31, 39, 45, 49). Detailed studies of a variety of models will be required to develop a clear understanding of the variation and mechanistic complexity of low-pH sensing and the forces that initiate and propagate the conformational changes required for viral membrane fusion. In the case of the fusion protein of tick-borne encephalitis virus (TBEV; a flavivirus), a single histidine residue was identified as a critical and perhaps dominant component of the pH sensor for conformational change (12, 14). The model developed from those studies suggests that protonation of a critical histidine residue results in disruption or destabilization of domain-domain interactions and exposure of the fusion peptide. In the case of the extensively studied influenza virus HA protein, mutations that affect the pH of fusion have been identified in residues involved in interactions between domains within monomers, between monomers in the trimer, and near the cavity associated with the fusion peptide in the prefusion structure (9, 38, 47, 49). The prefusion structure of HA shows an extensive network of salt bridges that stabilize the prefusion structure (38). Adding further to the complexity of understanding pH sensing, a recent study that examined an early structural intermediate of HA suggested that amino acid residues and ionic interactions involved in stabilizing and precisely determining the pH of triggering may involve numerous balanced interactions that vary in conservation between different clades of HA proteins (41, 49). Because single histidine replacements in GP64 did not identify a single residue associated with triggering, it is likely that the trigger for GP64 does not involve a single dominant histidine residue as a trigger but rather a balance of several or numerous ionic interactions. This type of variation and balancing in ionic interactions associated with triggering membrane fusion may explain the lack of conservation of H245, H304, and H430 between baculovirus GP64 and orthomyxovirus GP75 proteins. Ectodomains of the baculovirus GP64 and orthomyxovirus GP75 proteins show only approximately 20% similarity between the groups, and the two groups have evolved under distinctly different host selective pressures.
The baculovirus GP64 protein is a class III viral fusion protein (4, 19). Two members of this group, GP64 and vesicular stomatitis virus glycoprotein (VSV-G), appear to undergo a reversible conformational change (28, 40, 51) in response to low-pH treatment. The other class III fusion protein, herpesvirus gB, is structurally very similar to GP64 (4) but is triggered in a complex manner. gB fusion is initiated at either neutral or low pH and requires interactions with other viral envelope proteins (gD, gH, and gL) that appear to regulate the fusion activity of gB (1–4). Thus, triggers for conformational change and the degree of conformational changes between gB and GP64 may differ substantially. Recently it was demonstrated that the low-pH conformational change in HSV-1 gB was comprised primarily of an outward shift in fusion loop 2 and no global conformational change (43). In other pH-triggered fusion proteins, expansive and global conformational changes occur. The shift in fusion loop 2 of gB was likely caused by protonation of H263, a histidine at the periphery of fusion loop 2 that was previously shown to affect membrane fusion (13). Based on structural analysis of neutral- and low-pH forms of gB, it was proposed that H263 may serve as a histidine switch in gB (43). Of the three histidines found in fusion loop 2 of GP64, protonated H152 forms a salt bridge with an aspartate residue (D119), while the other two histidines (H155, H156) form hydrogen bonds with N78 and G122, respectively (see Fig. S1B in the supplemental material). Because the prefusion structure is not available, it is not yet known whether a global conformational change occurs in the GP64 protein and/or whether the fusion loop may hinge as described for herpesvirus gB. Although we found that H152 and H156 were clearly important for membrane fusion (dramatically affecting fusion pore expansion), the potential role of fusion loop histidines in low-pH triggering remains unresolved. We identified an important cluster of histidines flanking the base of the long coiled coils that comprise the core of the trimer. Although single substitutions had no substantial effect on membrane fusion activity, when substitutions for histidines 245, 304, and 430 were combined, constructs were trimerized and displayed at the cell surface, but fusion activity was severely reduced. Our studies suggest that the prefusion conformation of that construct was destabilized. The loss of recognition by the conformational MAb AcV1 suggests that histidines H245/H304/H430 (in their neutral-pH charge state) are necessary for a functional prefusion GP64 structure and that substitutions in these positions may result in subtle structural changes. Thus, changes in the charge state in these histidine residues (in response to low pH) may similarly disrupt the functional prefusion GP64 structure, perhaps triggering the further conformational changes leading to membrane fusion. Whether these specific histidines are necessary for interactions in subsequent intermediate steps is not known, but analysis of the crystal structure shows direct interactions of H245 with D314 and E307, H304 with I252, and H430 with D301 in the postfusion structure (Fig. S2), suggesting that the charged state of these residues may be important for stabilizing the postfusion structure. Thus, these data provide experimental evidence that histidines 245, 304, and 430 may comprise a component of a histidine switch required for triggering membrane fusion. Because the herpesvirus gB and baculovirus GP64 proteins are closely similar in structure yet differ substantially in their requirements for protein-protein interactions and pH for initiating fusion activity, detailed studies of fusion triggering in these two systems will provide a unique opportunity for understanding both the variety of activating mechanisms and the mechanistic requirements of fusion proteins in regulating this critical function.
In summary, we examined the roles of each histidine residue, and several histidine clusters in baculovirus GP64 mediated membrane fusion. Histidines were analyzed for their effects on GP64-mediated membrane fusion and virus infection. We conclude that no single histidine residue plays a dominant critical role as a histidine switch. Fusion loop histidines 152 and 156 were identified as important for fusion pore expansion, and a cluster of histidines (H245/H304/H430) at the base of the central coiled coil was identified as a putative component of a histidine switch. The current data should yield a more definitive picture of low-pH triggering when the prefusion conformation of GP64 becomes available. It will then be possible to further examine the histidines identified in these functional studies in terms of their specific contributions toward structural stability of the prefusion conformation of GP64. In ongoing studies, we are also using purified soluble GP64 ectodomain constructs to examine GP64 interactions with model membranes before and after the pH-triggered conformational change. These studies should shed light on both triggering and interaction of GP64 with the target membrane during GP64-mediated membrane fusion.
Supplementary Material
ACKNOWLEDGMENTS
We thank Abigail McSweeney for technical assistance.
This work was supported by NIH grant AI33657 and BTI project R06-1255.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Atanasiu D., Saw W. T., Cohen G. H., Eisenberg R. J. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atanasiu D., et al. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718–18723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atanasiu D., et al. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Backovic M., Jardetzky T. S. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blissard G. W., Rohrmann G. F. 1991. Baculovirus gp64 gene expression: analysis of sequences modulating early transcription and transactivation by IE1. J. Virol. 65:5820–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blissard G. W., Wenz J. R. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH dependent membrane fusion. J. Virol. 66:6829–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carneiro F. A., et al. 2003. Membrane fusion induced by vesicular stomatitis virus depends on histidine protonation. J. Biol. Chem. 278:13789–13794 [DOI] [PubMed] [Google Scholar]
- 8. Chanel-Vos C., Kielian M. 2004. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J. Virol. 78:13543–13552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daniels R. S., et al. 1985. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40:431–439 [DOI] [PubMed] [Google Scholar]
- 10. Da Poian A. T., Carneiro F. A., Stauffer F. 2009. Viral inactivation based on inhibition of membrane fusion: understanding the role of histidine protonation to develop new viral vaccines. Protein Pept. Lett. 16:779–785 [DOI] [PubMed] [Google Scholar]
- 11. Dollery S. J., Delboy M. G., Nicola A. V. 2010. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J. Virol. 3759–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fritz R., Stiasny K., Heinz F. X. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hannah B. P., et al. 2009. Glycoprotein B of herpes simplex virus associates with target membranes via its fusion loops. J. Virol. 83:6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison S. C. 2008. The pH sensor for flavivirus membrane fusion. J. Cell Biol. 183:177–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harrison S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hefferon K., Oomens A., Monsma S., Finnerty C., Blissard G. W. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455–468 [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Hink W. F. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226:466–467 [DOI] [PubMed] [Google Scholar]
- 19. Kadlec J., Loureiro S., Abrescia N. G., Stuart D. I., Jones I. M. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024–1030 [DOI] [PubMed] [Google Scholar]
- 20. Kampmann T., Mueller D. S., Mark A. E., Young P. R., Kobe B. 2006. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure 14:1481–1487 [DOI] [PubMed] [Google Scholar]
- 21. Krishnan A., et al. 2009. A histidine switch in hemagglutinin-neuraminidase triggers paramyxovirus-cell membrane fusion. J. Virol. 83:1727–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z., Blissard G. W. 2009. The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) GP64 protein: analysis of transmembrane (TM) domain length and sequence requirements. J. Virol. 83:4447–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Z., Blissard G. W. 2010. Baculovirus GP64 disulfide bonds: the intermolecular disulfide bond of AcMNPV GP64 is not essential for membrane fusion and virion budding. J. Virol. 84:8584–8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z., Blissard G. W. 2008. Functional analysis of the transmembrane (TM) domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: substitution of heterologous TM domains. J. Virol. 82:3329–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z., Blissard G. W. 2009. The pre-transmembrane domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein is critical for membrane fusion and virus infectivity. J. Virol. 83:10993–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Long G., Pan X., Kormelink R., Vlak J. M. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luckow V. A., Lee S. C., Barry G. F., Olins P. O. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Markovic I., Pulyaeva H., Sokoloff A., Chernomordik L. V. 1998. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J. Cell Biol. 143:1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monsma S. A., Oomens A. G. P., Blissard G. W. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell to cell transmission of infection. J. Virol. 70:4607–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mueller D. S., et al. 2008. Histidine protonation and the activation of viral fusion proteins. Biochem. Soc. Trans. 36:43–45 [DOI] [PubMed] [Google Scholar]
- 31. Nelson S., Poddar S., Lin T. Y., Pierson T. C. 2009. Protonation of individual histidine residues is not required for the pH-dependent entry of West Nile virus: evaluation of the “histidine switch” hypothesis. J. Virol. 83:12631–12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oomens A. G. P., Blissard G. W. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297–314 [DOI] [PubMed] [Google Scholar]
- 33. Oomens A. G. P., Monsma S. A., Blissard G. W. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209:592–603 [DOI] [PubMed] [Google Scholar]
- 34. Plonsky I., Cho M. S., Oomens A. G. P., Blissard G. W., Zimmerberg J. 1999. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology 253:65–76 [DOI] [PubMed] [Google Scholar]
- 35. Plonsky I., Kingsley D. H., Rashtian A., Blank P. S., Zimmerberg J. 2008. Initial size and dynamics of viral fusion pores are a function of the fusion protein mediating membrane fusion. Biol. Cell 100:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plonsky I., Zimmerberg J. 1996. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J. Cell Biol. 135:1831–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin Z. L., Zheng Y., Kielian M. 2009. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J. Virol. 83:4670–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rachakonda P. S., et al. 2007. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J. 21:995–1002 [DOI] [PubMed] [Google Scholar]
- 39. Reed M. L., et al. 2009. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J. Virol. 83:3568–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roche S., Gaudin Y. 2002. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology 297:128–135 [DOI] [PubMed] [Google Scholar]
- 41. Russell C. J., Jardetzky T. S., Lamb R. A. 2004. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 78:13727–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schowalter R. M., Chang A., Robach J. G., Buchholz U. J., Dutch R. E. 2009. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 83:1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stampfer S. D., Lou H., Cohen G. H., Eisenberg R. J., Heldwein E. E. 2010. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J. Virol. 84:12924–12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stauffer F., et al. 2007. Inactivation of vesicular stomatitis virus through inhibition of membrane fusion by chemical modification of the viral glycoprotein. Antiviral Res. 73:31–39 [DOI] [PubMed] [Google Scholar]
- 45. Stiasny K., Fritz R., Pangerl K., Heinz F. 2009. Molecular mechanisms of flavivirus membrane fusion. Amino Acids doi:10.1007/s00726-009-0370-4 [DOI] [PubMed] [Google Scholar]
- 46. Theilmann D. A., et al. 2005. Baculoviridae, p. 177–185 In Van Regenmortel H. V., Bishop D. H. L., Van Regenmortel M. H., Fauquet C. M. (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, New York, NY [Google Scholar]
- 47. Thoennes S., et al. 2008. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology 370:403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. White J. M., Delos S. E., Brecher M., Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu R., Wilson I. A. 2011. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 85:5172–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou J., Blissard G. W. 2008. Identification of a GP64 subdomain involved in receptor binding by budded virions of the baculovirus AcMNPV. J. Virol. 82:4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou J., Blissard G. W. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.