Abstract
VP23 is a key component of the triplex structure. The triplex, which is unique to herpesviruses, is a complex of three proteins, two molecules of VP23 which interact with a single molecule of VP19C. This structure is important for shell accretion and stability of the protein coat. Previous studies utilized a random transposition mutagenesis approach to identify functional domains of the triplex proteins. In this study, we expand on those findings to determine the key amino acids of VP23 that are required for triplex formation. Using alanine-scanning mutagenesis, we have made mutations in 79 of 318 residues of the VP23 polypeptide. These mutations were screened for function both in the yeast two-hybrid assay for interaction with VP19C and in a genetic complementation assay for the ability to support the replication of a VP23 null mutant virus. These assays identified a number of amino acids that, when altered, abolish VP23 function. Abrogation of virus assembly by a single-amino-acid change bodes well for future development of small-molecule inhibitors of this process. In addition, a number of mutations which localized to a C-terminal region of VP23 (amino acids 205 to 241) were still able to interact with VP19C but were lethal for virus replication when introduced into the herpes simplex virus 1 (HSV-1) KOS genome. The phenotype of many of these mutant viruses was the accumulation of large open capsid shells. This is the first demonstration of capsid shell accumulation in the presence of a lethal VP23 mutation. These data thus identify a new domain of VP23 that is required for or regulates capsid shell closure during virus assembly.
INTRODUCTION
Herpesviruses encode six proteins that come together in a highly coordinated and regulated fashion to form a protective coat surrounding the virus genome. The assembly pathway of herpes simplex virus 1 (HSV-1) produces three capsid structures detected in sucrose gradients of lysates from infected cells. They are called “A,” “B,” and “C” capsids, because of their sedimentation profiles in the gradient (10). C capsids contain the virus genome, B capsids contain the internal scaffold proteins, and A capsids are empty (reviewed in references 11, 26, and 31). A fourth capsid particle, which is sensitive to temperature and sedimentation but has been observed by ultrastructural analysis of infected cells, is the procapsid and is thought to be the earliest spherical structure formed in HSV-1-infected cells (16, 28, 34).
Seven proteins make up B capsids: VP5, VP19C, p21, p22a, VP23, VP24, and VP26. VP5, VP19C, VP23, and VP26 form the outer capsid shell, whereas the scaffold proteins (pre22a and pre21) occupy the internal space of the capsid (reviewed in references 11, 26, and 31). Protein-protein interactions drive the assembly process, and, as shown in a number of studies, the capsid proteins have an inherent ability to self-assemble into capsid shells (32, 33). Interactions have been detected between VP5 and the scaffold protein; this interaction is important for closure of the capsid shell into an icosahedral structure (7, 13, 19, 35), between the triplex proteins VP19C and VP23, which is important for stabilization of the shell structure (1, 7, 21, 27, 30), and a self-interaction between the scaffold proteins (7, 20, 22, 25), which is important for the production of the capsid. Interactions have also been inferred or detected between VP5 and VP26 (3, 27) and VP5 and VP19C (27) and a self-interaction between VP23 molecules (14, 30). These interactions have been determined using the yeast two-hybrid assay, cellular colocalization, sedimentation, coimmunoprecipitation, capsid binding, and cryoelectron microscopy (cryo-EM) analysis. The interaction between the two triplex proteins VP23 and VP19C is important for assembly of the herpesvirus icosahedron. This interaction was first discovered using the yeast two-hybrid assay (7), inferred by cryo-EM analysis (18, 37), and confirmed using biochemical approaches using purified proteins (17, 30). VP19C (50 kDa) and VP23 (33 kDa) bind together to form a hetero-trimer and a three-pronged structure termed the triplex. This structure, of which there are 320 copies per capsid, consists of one molecule of VP19C and two molecules of VP23 (34). It is important for stabilizing the capsid shell by interactions with the adjoining capsomeres made up of the major capsid protein, VP5.
Several studies have reported the identification of important functional domains of VP19C using N- and C-terminal truncation mutations and random transposition mutagenesis (1, 21, 30). We utilized a random mutagenesis approach with VP23 initially and discovered many sites of this protein, which were important (21). Although some of the mutations had been analyzed in a baculovirus self-assembly system, none had been studied in the context of HSV-1-infected cells. To this end, we undertook a more precise mutagenesis approach using our results from the transposition mutations and have made 79 single-amino-acid substitutions in this protein of 318 residues. All the mutations were analyzed for interaction with VP19C using the yeast two-hybrid assay and for genetic complementation of the VP23 null mutant. Several mutations were identified which did not affect VP23-VP19C interaction but failed to complement the growth of the VP23 null mutant virus. These mutations were transferred into the KOS genome using a marker rescue/marker transfer method. The outcome has been the identification of a new functional domain of VP23, which is required for capsid shell closure.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and transformed Vero cell lines were grown in minimum essential medium-alpha supplemented with 10% fetal calf serum (Gibco-Invitrogen) and passaged as described by Desai et al. (5). C32 cells complement the growth of mutants in VP5 (UL19) and VP23 (UL18) (24), whereas K20 cells complement the growth of only VP23 mutants. Virus stocks of KOS (HSV-1) and the mutant viruses were prepared as previously described (5). KΔ18/19CG is a mutant virus that contains a deletion in both UL18 (VP23) and UL19 (VP5) genes. This virus required the C32 cell line for propagation and served as the recipient genome for all marker rescue (of VP5) and marker transfer (of VP23 mutants) experiments. K23R was the marker-rescued virus of KΔ18/19CG.
Plasmids.
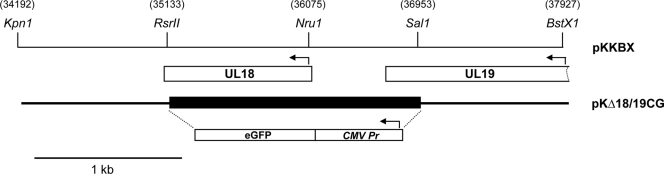
The template for the mutagenesis of the VP23 gene was a plasmid designated pUC-UL18 which carried the UL18 open reading frame (ORF) derived from pGBT9-VP23 (7) as an EcoRI-BamHI fragment cloned into the same sites of the pUC19 plasmid. pKKBX is a plasmid which carries the KpnI-BstX1 3.7-kb fragment of strain KOS cloned into the KpnI and HincII sites of pUC19 and encompasses the UL18 gene and flanking regions. The double deletion in UL18 and UL19 was made by digesting pKKBX with RsrII (in UL18) and SalI (in UL19), blunt ended, and a similarly treated Ase1-NotI cytomegalvirus (CMV)-enhanced green fluorescent protein (eGFP) fragment from pEGFP-N1 (Clontech) was ligated into the deletion site in both orientations (see Fig. 3). This plasmid was designated pKΔ18/19CG.
Fig. 3.
Marker rescue/marker transfer of the VP23 alanine substitution mutants into the virus. Plasmid pKKBX (KpnI-BstX1) was the backbone for generation of a double mutant in the UL18 and UL19 genes. The deletion (filled box) was replaced with a CMV-eGFP reporter cassette to facilitate the identification of recombinant plaques. The virus isolated and purified was designated KΔ18/19CG and was the recipient genome for all the alanine substitution mutants. Numbers in parentheses refer to HSV-1 genome nucleotide numbers, the arrows indicate direction of transcription, and a 1-kb size marker is shown.
Mutagenesis.
Site-directed mutations were made by the “QuikChange” mutagenesis protocol (Stratagene). The use of this method is described in detail by Walters et al. (35). Positive clones carrying the mutation were confirmed by restriction enzyme and sequence analysis. The mutant VP23 sequence was then cloned into the yeast two-hybrid vector pGBT9 and pcDNA 3.1 as an EcoRI-BamHI fragment and into the genomic plasmid pKKBX as an Nru1-RsrII fragment. The mutant genes were all sequenced to confirm the presence of the specified mutation as well as authentic amplification by the polymerase.
Yeast two-hybrid assays.
The protocol for yeast two-hybrid assays is described by Desai and Person (7). Sequences encoding VP23 were fused to the yeast Gal4 DNA-binding domain (pGBT9-VP23), and the gene encoding VP19C was fused to the Gal4 transactivation domain (pGAD424-VP19C) (7). Both plasmids were cotransformed into SFY526 cells (9), and the transformants were allowed to grow on nitrocellulose membranes in order to assay each mutation using the X-gal filter assay (2).
Genetic complementation assay.
Vero cells (5 × 105) were transfected with 1 μg of plasmid DNA and 6 μl of Lipofectamine reagent (Invitrogen) diluted in 200 μl of serum-free medium. The DNA-lipid complexes were allowed to form at room temperature for 30 min and incubated with the cells for 5 h. Twenty-four hours after the addition of the complexes to the culture, the cells were infected with K23Z (VP23 null mutant) (4) at a multiplicity of infection (MOI) of 5 PFU/cell. The following day (24 h later) the infected cells were harvested following freeze-thawing and sonicated and the virus titers were determined on C32 cell monolayers.
Construction of transformed Vero cell lines.
Transformed cell lines were made as documented by Desai et al. (6). Early-passage Vero cells (1 × 106 in 100-mm petri dishes) were cotransfected with pSV2-neo (1.0 μg) (30), and a molar 3- or 5-fold excess of the plasmid encoding the UL18 gene (pKKBX; see Fig. 3). A total of 54 G418-resistant colonies were harvested, passaged, and then tested for their ability to plaque K23Z, a null mutant of the UL18 gene (6). Six cell lines were able to complement K23Z, and one line designated K20 was chosen for the marker rescue/marker transfer experiments.
Marker rescue and market transfer.
Marker rescue/marker transfer assays were performed as described by Person and Desai (24). C32 cells (1 × 106) in 60-mm dishes were cotransfected with KΔ18/19CG-infected cell DNA and linearized pKKBX mutant plasmids (0.1 to 1 μg). Titers of the transfection progeny harvested 3 days later were determined on Vero, C32, and K20 cell monolayers to determine the growth properties of each mutant virus. Mutant viruses were isolated following 2 to 3 rounds of single-plaque isolation on K20 or Vero cells. The mutant virus genomes were used to amplify the UL18 gene and sequenced to confirm the presence of the correct mutation.
Burst-size growth assays.
Vero cells (5 × 105) in 12-well trays were infected with K23R and the mutant viruses at an MOI of 10 PFU/cell. Twenty-four hours postinfection, the cells were harvested and sonicated and virus progeny numbers were determined by plaquing on C32 monolayers.
Radiolabeling and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Procedures for radiolabeling of infected cells and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and Western blots are described by Person and Desai (24).
Sedimentation analysis of capsids.
Sedimentation analysis of capsids from infected cells was performed as described by Walters et al. (35).
Electron microscopy.
For transmission electron microscopy and negative-stain analysis of sucrose gradient fractions, the procedures described by Huang et al. (12) and Perkins et al. (23) were used, except samples were stained with 1% phosphotungstic acid.
Data and figure preparation.
Scanned autoradiographs and digital electron micrographs were imported into Adobe Photoshop for figure compilation.
RESULTS
Site-directed mutagenesis of VP23 amino acids using alanine-scanning method.
Previous studies using random transposition mutagenesis of the gene encoding VP23 (UL18) identified several domains of this protein important for interaction with VP19C and for capsid assembly in insect cells using recombinant baculoviruses (Fig. 1) (21). As an extension of those studies, an extensive mutagenesis was performed to identify the key interactive residues of VP23. An alanine-scanning mutagenesis approach was carried out; the amino acids targeted for this mutagenesis reside in the functional domains previously identified as well as additional regions that were uncovered during this scanning mutagenesis (blue residues in Fig. 1). Site-directed mutations were made by the QuikChange mutagenesis protocol (Stratagene). Most mutations also introduced a “silent” restriction enzyme site for detection of the mutant. Seventy-nine alanine substitution mutations were made in this study. In almost all cases the residues targeted were hydrophobic in nature. The mutant UL18 genes were transferred into both a yeast two-hybrid vector (pBGT9) and a eukaryotic expression plasmid (pcDNA3.1−).
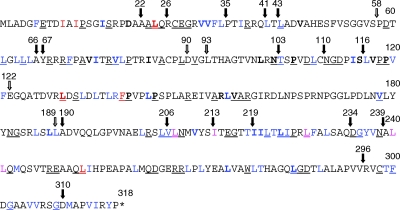
Fig. 1.
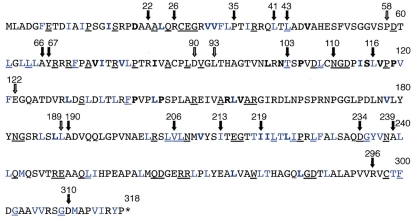
VP23 molecule. Shown is the VP23 polypeptide sequence (318 amino acids) and key mutational landmarks. The filled arrows show sites of TN mutants that abolished interaction with VP19C and genetic complementation of the VP23 null mutant. The open arrows show sites of TN insertions, which did not affect these two activities of VP23. The TN insertion (5 amino acids) was after the residue numbered above the arrow (21). The alanine substitution mutations made in this study are shown in blue. Underlined amino acids are conserved in the alphaherpesvirus family, and the residues in bold are conserved in all three herpesvirus families.
Yeast two-hybrid analysis of the VP23 mutants.
Most of the mutations were analyzed using the yeast two-hybrid assay (9) to determine the effect of these changes on the interaction with VP19C. The yeast two-hybrid assay originally permitted the first confirmation of the interaction between these two proteins. The Bluo-gal filter assay was used to assay the interaction between the mutant VP23 and wild-type VP19C (Table 1). As shown in Table 1, the mutations were classified into three classes: those that interact with VP19C with wild-type kinetics of blue color development of the yeast transformants (+++), those that interact with VP19C with decreased or slowed kinetics of blue color development (++/+), and those that fail to interact with VP19C as judged by lack of blue color of the colonies (−). This analysis identified several single residues which when changed to alanine completely abolished the triplex interaction. These residues include I10, I12, L24, L130, F139, and L253.
Table 1.
Yeast two-hybrid assay of the VP23 alanine substitution mutantsa
| Group 1 |
Group 2 |
Group 3 |
Group 4 |
Group 5 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP23 (UL18) protein | Y2H assay result | VP23 (UL18) protein | Y2H assay result | VP23 (UL18) protein | Y2H assay result | VP23 (UL18) protein | Y2H assay result | VP23 (UL18) protein | Y2H assay result | ||||
| Wild type | +++ | Wild type | +++ | Wild type | +++ | Wild type | +++ | Wild type | +++ | ||||
| F6A | +++ | T103A | +++ | L202A | +++ | G235A | ++ | F300A | +++ | ||||
| I10A | − | L108A | +++ | L205A | ++ | Y236A | + | G302A | +++ | ||||
| I12A | − | I114A | ++ | V206A | +++ | V237A | +++ | V305A | +++ | ||||
| I16A | +++ | L116A | ++ | L207A | ++ | L240A | + | V306A | +++ | ||||
| L24A | − | V120A | +++ | V210A | +++ | L241A | + | G309A | +++ | ||||
| V31A | +++ | F121A | ND | I213A | + | M243A | ++ | M311A | +++ | ||||
| V32A | +++ | L130A | − | T218A | +++ | L253A | − | V314A | +++ | ||||
| F33A | ND | L133A | ND | I219A | +++ | I254A | + | I315A | +++ | ||||
| L34A | +++ | L135A | ND | I220A | ++ | L261A | +++ | Y317A | +++ | ||||
| I37A | +++ | L137A | ND | L221A | ++ | L269A | +++ | ||||||
| L41A | +++ | F139A | − | T222A | +++ | L271A | ND | ||||||
| L43A | ND | L143A | +++ | L223A | +++ | L275A | + | ||||||
| L61A | ++ | L155A | +++ | I224A | +++ | L279A | +++ | ||||||
| L63A | +++ | L178A | ++ | L227A | + | T280A | ND | ||||||
| L64A | ND | L186A | ND | F228A | + | L285A | +++ | ||||||
| L65A | +++ | L188A | ND | L230A | ++ | L289A | + | ||||||
| F71A | + | L189A | +++ | T299A | +++ | ||||||||
| I75A | ND | ||||||||||||
| V78A | +++ | ||||||||||||
| L79A | +++ | ||||||||||||
| Vector | − | Vector | − | Vector | − | Vector | − | Vector | − | ||||
Group 1, amino acids 6 to 79; group 2, amino acids 103 to 189; group 3, amino acids 202 to 230; group 4, amino acids 235 to 299; group 5, amino acids 300 to 317. Plus signs are used to indicate the kinetics of the bimolecular interaction between VP23 and VP19C. +++, wild-type interaction (the colonies turned blue within 45 min of the start of the assay); ++, the colonies took between 1-2 h to turn blue; +, the colonies took longer than 2 h to turn blue; −, the colonies remained white. The data in the table were derived from >5 independent transformations. ND, not determined.
Genetic complementation of the VP23 null mutant virus.
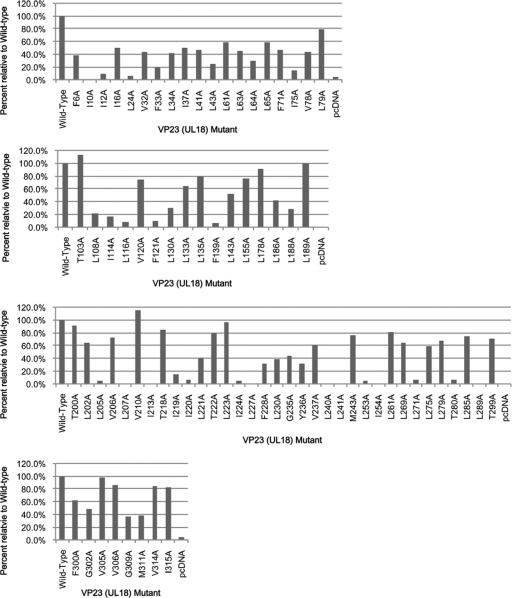
In order to correlate the in vitro results from the yeast two-hybrid assay (which measures a bimolecular interaction) with the ability of the VP23 mutants to support virus growth (which measures capsid assembly), most of the mutants were assayed for their ability to complement the growth of the VP23 null mutant virus K23Z (4). The complementation assay (Fig. 2) was able to distinguish between mutants that support the replication of the null mutant (complementation of 20 to 100% relative to wild-type protein) and those that do not (complementation of 2 to 5% relative to wild-type protein; this level was similar to the number seen with empty vector). In general, there was very good correlation between the ability of the mutants to participate in a molecular interaction (Y2H assay) and to complement the growth of the null mutant virus. Thus, the complementation levels derived for mutants I10A and L24A all were lower than 10% of the wild-type level, and those for V305A and I315A were similar to that of the wild-type protein. Even the mutants that gave a weak interaction in the yeast assay were able to support the replication of the null mutant, for example, F71A, F228A, Y236A and L275A. There were some exceptions (which interact with VP19C but fail to complement the VP23 null mutant virus), and these were alanine substitutions at I114, L116, and a cluster of residues that span amino acids 205 to 241.
Fig. 2.
Genetic complementation of K23Z by the VP23 alanine substitution mutants. Transient transfections were performed using cloned genes in the pcDNA vector. Twenty-four hours after transfection, the cells were infected with the VP23 null mutant, K23Z, and the virus yield was determined after another 24 h postinfection. Progeny yields were quantitated by titration on the complementing cell line, C32. Data are averages of results of two to three independent transfections. The level of complementation derived with the expressed wild-type gene product was set at 100% and the levels achieved with the empty vector were generally 1 to 10% of that.
Introduction of specific VP23 mutations into the virus genome.
Our initial goal was to characterize the VP23 mutants using just the yeast two-hybrid and the genetic complementation assays because these two methods have been shown to be highly indicative of the outcome of a VP23 mutation during the replication cycle. However, because we discovered a number of mutants that interacted with VP19C but failed to complement the growth of K23Z, we hypothesized that there maybe another key interaction in which VP23 participates. Thus, our goal changed to introduce these specific mutations into the virus genome.
We used the genetic marker rescue/marker transfer method (6) to place VP23 mutations into the virus genome (Fig. 3). Using existing restriction enzymes a deletion was made that includes all the N-terminal 307 residues of VP23 and deletes the C-terminal 182 residues of VP5 (Fig. 3). In place of the deleted sequence a CMV-eGFP reporter cassette was inserted which would facilitate isolation of this virus following homologous recombination (Fig. 3). This plasmid, designated pKΔ18/19CG, was cotransfected into C32 (UL18- and UL19-transformed cell line) cells together with KOS DNA, and single green fluorescing plaques were picked and purified. This virus was designated KΔ18/19CG. This virus plaqued only on C32 cells but not on cells transformed with just UL19 or just UL18 (data not shown). To derive a cell line that is transformed with just UL18, we cotransformed Vero cells with pSV2Neo and a plasmid that encodes just the gene encoding VP23, pKKBX. Cell line K20 was chosen for further studies because it displayed the best plaquing and complementation of K23Z.
To test the system, we used the wild-type plasmid pKKBX to rescue the UL19 mutation and restore wild-type gene function at the UL18 locus. We were able to readily isolate virus on K20 cells, and one such isolate was purified and designated K23R. This virus plaqued and replicated on Vero cells in a manner comparable to that of the wild-type parental KOS virus. We then proceeded to use this genetic method to introduce several mutations into the virus genome. The mutants introduced into the virus genome include L24A, F33A, L43A, L64A, I75A, L108A, L116A, F121A, L205A, L207A, I213A, I219A, I224A, L227A, L240A, and L241A. Initially the viruses were checked for their ability to plaque on Vero cells. Mutant viruses L24A, L207A, I213A, L227A, L240A, and L241A failed to plaque on Vero cells, whereas L205A and I219A gave small plaques on Vero cells and I224A gave rise to very small plaques that were discernible only after extended incubation. The other mutants all plaqued normally on Vero cells. In order to quantitate this, a burst size determination was made for all the mutants (Table 2). Almost all the progeny yields for the mutants correlated with the genetic complementation data, except for L108A and I219A, which gave higher yields in the single-step growth assay than the complementation assay. Even though the I224A mutant gave only a minimal burst size yield (1% of wild-type level), this virus can still form plaques on Vero cells after prolonged growth; thus the assembly process appears to be slowed significantly in this mutant. The genomes of the viruses were also used as templates for PCR of the UL18 gene, and the PCR product was sequenced to confirm that the correct mutation was present in each virus. In this analysis we discovered that the L24A mutant gene had incorporated a stop codon in the sequence. The mutant viruses were also examined for their ability to accumulate stable amounts of VP23 protein. Infected cells were examined by Western blot methods using polyclonal antisera to VP23. The results of the blotting experiments show that all the mutants are able to synthesize and accumulate wild-type levels of VP23 (Fig. 4).
Table 2.
Single-step growth of the VP23 mutant virusesa
| VP23 virus | Burst size (% of wild-type) | Y2H assay resultb | % Complementation |
|---|---|---|---|
| Wild type | 100 | +++ | 100 |
| L24A | 0.25 | − | 6.3 |
| F33A | 28.26 | ND | 19.3 |
| L43A | 28.26 | ND | 23.8 |
| L64A | 53.62 | ND | 30.2 |
| I75A | 13.76 | ND | 14.6 |
| L108A | 76.81 | +++ | 21.4 |
| L116A | 9.42 | ++ | 8.4 |
| F121A | 17.39 | ND | 9.6 |
| L205A | 11.59 | ++ | 4.2 |
| L207A | 0.38 | ++ | 1.6 |
| I213A | 0.22 | + | 1.8 |
| I219A | 37.68 | +++ | 14.7 |
| I224A | 1.00 | +++ | 5.1 |
| L227A | 0.31 | + | 2.9 |
| L240A | 0.28 | + | 1.5 |
| L241A | 0.41 | + | 1.3 |
Data were derived from two or three independent experiments. ND, not determined.
See Table 1 for an explanation of Y2H scoring.
Fig. 4.
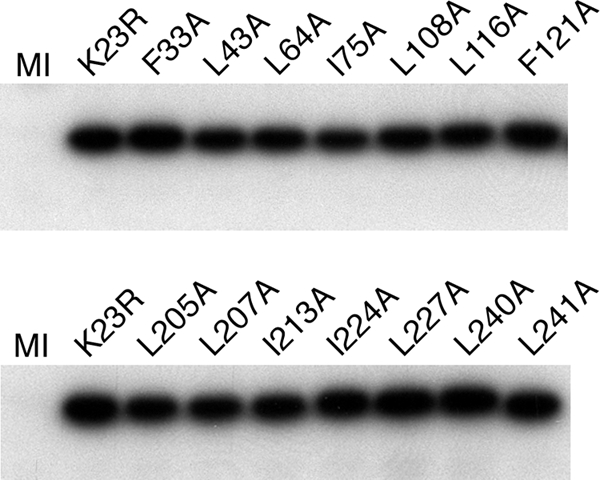
Stable accumulation of VP23 in the mutant infected cells. Vero cells infected with each lethal VP23 mutant were examined by Western blot methods using a rabbit antibody to VP23. VP23 polypeptide accumulation was detected for the marker-rescued wild-type virus (K23R) as well as for each mutant. Mock-infected cells are shown in lane MI.
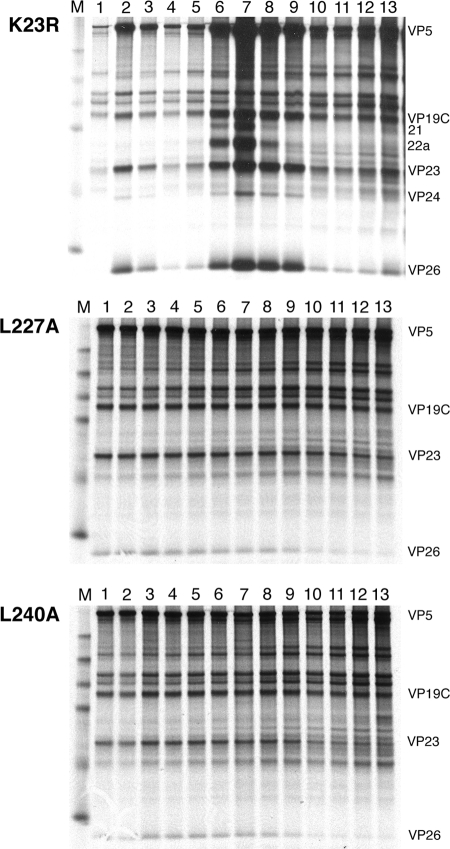
We next examined the mutants for their ability to assemble capsids following infection of nonpermissive cells. Previously we had shown that the VP23 null mutant virus K23Z cannot assemble capsids or capsid precursors using EM and sedimentation methods (4). Vero cells infected with different mutants were metabolically labeled with 35S-methionine, and lysates prepared were subjected to rate-velocity sedimentation (Fig. 5). For the wild-type rescuant K23R, we observed three peaks of radioactivity corresponding to A (fraction 9), B (fraction 7), and C (fraction 2) capsids. The scaffold protein (protein 22a) present in B capsids was for the most part present in the fractions (fractions 6 and 7) containing these capsids. For mutant viruses L227A and L240A, capsid proteins were observed to cosediment in several of the gradient fractions. The proteins detected were the capsid shell proteins VP5, VP19C, VP23, and VP26. The scaffold protein and the protease (VP24) were not detected in the fractions where the shell proteins were seen to sediment, indicating that the scaffold was not part of this assembled structure. This was also seen in gradients for mutants L207A, I213A, and L241A (data not shown). Furthermore, similar to scaffold null mutants (8), several fractions contained these cosedimenting proteins; that is, there was not a specific peak of radioactivity that is seen with A, B, and C capsids. This indicated that the structures detected in the gradients may be of varied size and shape.
Fig. 5.
Cosedimentation of the capsid shell proteins in the VP23 mutant-infected cells. Vero cells were infected with KOS and VP23 mutants L227A and L240A and metabolically labeled with 35S-methionine from 8 to 24 h postinfection. These lysates, prepared, were sedimented on 20 to 50% sucrose gradients, and the fractions collected were examined by gel electrophoresis (4 to 12% Bis-Tris NuPage gels; Invitrogen). Fraction 1 corresponds to the bottom of the centrifuge tube. The position of capsid proteins in the gel is indicated on the right of the gel. Protein standards are shown in lane M, and the visible markers are 97.4, 66, 46, 30, and 14.3 kDa.
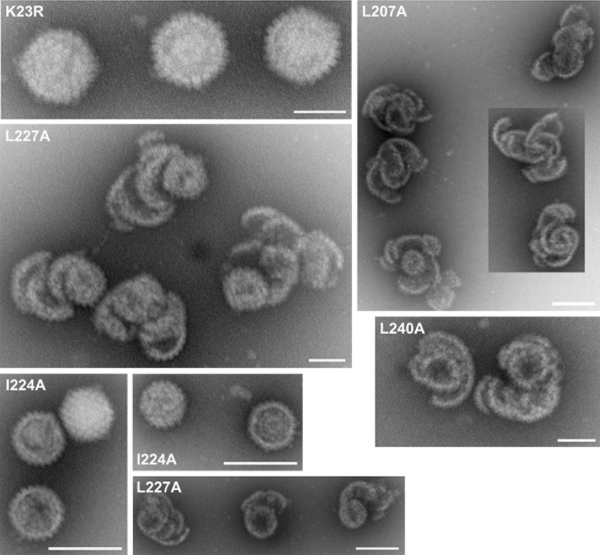
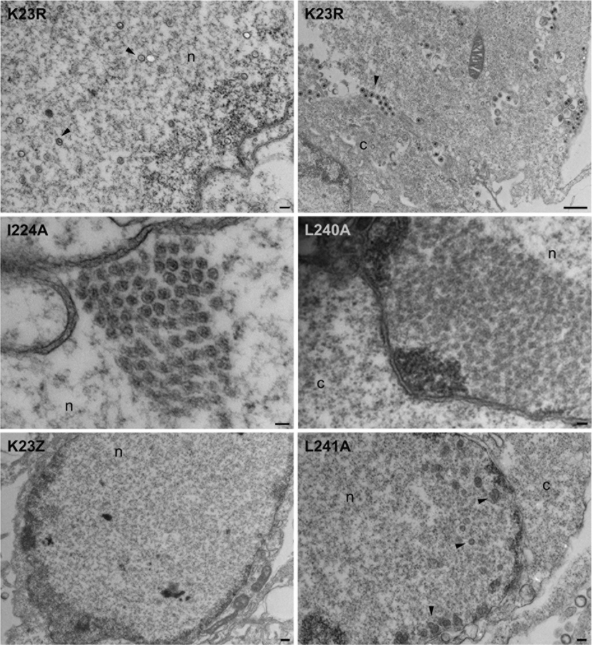
Because the biochemical data showed that the VP23 mutants may be accumulating shell structures, material from similar gradients was examined by EM following negative stain (Fig. 6). Icosahedral capsids were evident when material from K23R was examined; however, when the material from the mutant-infected cells was examined, large open shells were observed in which the capsomeres composed of VP5 were clearly visible. There was no uniformity to these shells; these structures were of varied size and shape, and this correlates with their sedimentation profile in sucrose gradients. These open shell structures visibly were similar in appearance to the shells that accumulate in the absence of the major capsid-scaffold protein interaction (12, 35). Icosahedral closed capsids were observed in sucrose gradients for mutant I224A albeit at reduced levels compared to wild-type capsids. These data were also confirmed by transmission EM (TEM) analysis of infected cells (Fig. 7). Assembled capsids in the nucleus and enveloped virions in the cytoplasm were evident in KOS-infected cells, whereas no structures were evident in the VP23 null mutant virus (K23Z)-infected cells. Almost complete shells were observed in the nucleus of I224A-infected cells, and capsid shells in the nuclei of L240A- and L241A-infected cells.
Fig. 6.
Ultrastructural examination of VP23 mutant assembled structures. Lysates derived from VP23 mutant and K23R-infected cells were sedimented in sucrose gradients and the fractions collected examined by TEM following negative staining. Scale bars: 100 nm (for K23R and middle panel of L227A) and 200 nm (for all other panels).
Fig. 7.
Ultrastructural analysis of VP23 mutant infected cells. Vero cells were infected with K23R and the mutant viruses at an MOI of 5 PFU/cell and the infected cells processed for thin sectioning followed by TEM at 20 h postinfection. Intranuclear capsids are evident in the nucleus of K23R-infected cells (left panel, arrowheads) and enveloped virions in the cytoplasm of K23R-infected cells (right panel, arrowhead). Shell structures were present in the nuclei of L240A- and L241A-infected cells, whereas near-complete capsids were evident in I224A-infected cells. n, nucleus; c, cytoplasm. Scale bars: 100 nm (for L240A and I224A), 200 nm (for K23R [left panel] and L241A), 400 nm (for K23Z), and 1,000 nm (for K23R [right panel]).
DISCUSSION
We have carried out an extensive mutagenesis experiment on the essential VP23 polypeptide. Previously we had used random five-amino-acid insertions to identify key functional domains of VP23 for interaction with VP19C and capsid assembly (21). This time, using that information, we have made 79 amino acid substitutions in a polypeptide composed of 318 residues. The mutations substituted alanine for hydrophobic residues, and they were screened for interaction with VP19C using the Y2H assay and for genetic complementation of the VP23 null mutant; finally some were introduced into the virus genome to examine their effect on virus replication and capsid assembly. In all, there were six residues that, when changed, abolished completely the ability of the mutant VP23 to interact with VP19C and also rescue the VP23 null mutation. It is likely, as shown by Adamson et al. (1), that the mutant proteins that do not interact with VP19C will remain cytoplasmic in localization. The identification of a single change that can have a significant effect on virus assembly bodes well for future investigation of VP23 inhibition using a small molecule inhibitor. There were also additional mutations that displayed a range of phenotypes in their abilities to bind VP19C or complement the VP23 null mutant virus. A subset of such mutants that are concentrated in the C-terminal portion of VP23 gave a phenotype that we investigated further. These mutations were able to interact with VP19C, some weakly, but failed to complement the VP23 null mutant. When the same mutations were introduced into the virus genome and infected cells were examined, the defect in this set of mutants became apparent: all accumulated large open shells that could not attain closure. The mutants that displayed this phenotype were L207A, I213A, L227A, L240A, and L241A (Fig. 8), and because they cluster in a small region of VP23, we have thus identified a new domain of VP23 that is important for closed capsid assembly (Fig. 8).
Fig. 8.
Summary of the VP23 mutational analysis. Shown in the amino acid sequence of VP23 are all the residues that were changed to alanine (blue), the residues which when substituted for alanine abolished interaction with VP19C and complementation of K23Z (red), and the residues which are important for capsid shell closure (fuchsia). See Fig. 1 legend for other annotations of the amino acid sequence.
The triplex proteins of herpesviruses are essential structural elements of the capsid shells. Previous studies have shown that null mutations in either gene resulted in a complete absence of capsid shells or subcapsid assemblies (4, 24). Thus, it was recognized for many years that the triplex acts at an early step in the assembly pathway and a block in this step did not lead to the accumulation of higher-order assemblies. This study now shows that one of the triplex proteins is also required for a step subsequent to capsid shell accretion and that mutations in this key VP23 domain result in the accumulation of open shells. An alternative explanation could be that the mutant VP23 proteins have reduced affinities for the different components of capsid shell and what we are observing is the inability to complete capsid growth. This could then result in the accumulation of capsid shells in infected cells. While this possibility is clearly valid, previous results with VP5 mutants that have defects in capsid shell growth (because of decreased capsomere interactions) demonstrated that such mutants can be differentiated based on their sedimentation profiles (35).
Capsid assembly for herpesviruses begins in the cytoplasm. Here interactions between VP5 and the scaffold proteins and those between VP19C and VP23 occur, and these small complexes are transported into the nucleus using the cellular nuclear import machinery and a nuclear localization signal (NLS) present in one of the partners of each complex (1, 16, 17, 19, 27). VP26, by virtue of its association with VP5, “piggybacks” onto the VP5-scaffold complex and becomes concentrated in the assembly sites within the nucleus (3). Adamson et al. (1) identified a nonconsensus NLS in the N terminus of VP19C which is required for transporting the VP19C-VP23 complex into the nucleus. Thus, one function of the interaction between the triplex proteins is to ensure concentration of this complex at the site of assembly. Once in the nucleus, preformed triplex complexes bind to MCP-scaffold complexes and shell accretion begins, eventually resulting in the formation of a closed spherical structure (16). Shell accretion is an essential step for the synthesis of large capsid assemblies, and the function of the triplex is to facilitate the joining of the capsomeres.
What regulates and controls closure of an open shell into a closed stable structure? This is still not completely understood for herpesvirus assembly. Previous studies have demonstrated that the absence of the scaffold protein or mutations that abolish the interaction between the MCP and scaffold protein result in the accumulation of large open shells of undefined size and shape (8, 35). From these data it was proposed that the scaffold protein, as shown first in the large double-stranded DNA (dsDNA) phage P22, catalyzes the synthesis of a closed icosahedral structure. From the work presented here we have now have identified another component of this pathway. Thus, VP23 also plays an essential role in the transition from an open to closed shell. The exact role of VP23 in shell closure is purely speculative in the absence of other functional analysis. What is clear is that the triplex complex is functional in these mutants because we see the accumulation of significant quantities of open shells, which are composed of VP5, VP19C, VP23, and VP26. However, the triplex or VP23 in these mutants has a defect that prevents capsid closure. Interestingly, the SDS-PAGE analysis of these shells reveals very little or no scaffold protein associated with these structures. It is possible, then, that these mutant VP23 proteins may be affecting the interaction between VP5 and p22a and, in this way, abolishing closed capsid formation. In the baculovirus self-assembly system, small close structures (88-nm size) were formed when VP5 was coexpressed with VP19C (27, 29). However, upon the addition of VP23, the closed structures did not form, and it was suggested that VP23 was important for reconfiguration of the shell and modulating capsid shell assembly by interaction with both VP19C and VP5 (29).
High-resolution structural, biochemical, and genetic data show that VP23 is likely a dimer in the triplex complex. A dimer interface between the two VP23 polypeptides and an extensive interaction of each of the two VP23 molecules with VP19C was seen by cryo-EM analysis of the B capsid (37). Self-interaction of the CMV homolog of VP23 (36) has been detected using the yeast two-hybrid assay, and for HSV-1 a dimer has been observed using sedimentation assays of Sf9 infected cell lysates (30). VP23 purified from Escherichia coli was shown to be a monomer in solution, but at high protein concentrations it formed a multimer (14). Purified VP23 behaves as a molten globule: its tertiary structure is determined by its local environment and interactions, for example, when it is incorporated into the triplex (14). Thus, the functional state of VP23 is probably also a dimer, and we propose that this new essential domain in VP23 is potentially a region for protein dimerization. Using the Coils prediction program (15), a potential coiled-coil domain was discovered spanning amino acids 238 to 255. Hence this domain may be important for facilitating VP23 dimerization and thus capsid shell closure. If dimerization is defective in these mutant viruses, the fact that we still detect open shells as well as interaction with VP19C in the Y2H indicates that this protein transformation may not be required for triplex formation. It is required subsequent to capsid shell accretion at a step that has yet to be discovered.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI033077 and AI061382 and a summer student ARRA supplement, AI061382-05S1.
We thank Aaron Davis, Shawn Johnson, Grant Chen, and Kwun Wah Wen for help with parts of the experiments.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Adamson W. E., McNab D., Preston V. G., Rixon F. J. 2006. Mutational analysis of the herpes simplex virus triplex protein VP19C. J. Virol. 80:1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breeden L., Nasmyth K. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant Biol. 50:643–650 [DOI] [PubMed] [Google Scholar]
- 3. Desai P., Akpa J.-C., Person S. 2003. Residues of VP26 of herpes simplex virus type 1 that are required for its interaction with capsids. J. Virol. 77:391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desai P., DeLuca N. A., Glorioso J. C., Person S. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Desai P., DeLuca N. A., Person S. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115–124 [DOI] [PubMed] [Google Scholar]
- 6. Desai P., Homa F. L., Person S., Glorioso J. C. 1994. A genetic selection method for the transfer of HSV-1 glycoprotein B mutations from plasmid to the viral genome: preliminary characterization of transdominance and entry kinetics of mutant viruses. Virology 204:312–322 [DOI] [PubMed] [Google Scholar]
- 7. Desai P., Person S. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516–521 [DOI] [PubMed] [Google Scholar]
- 8. Desai P., Watkins S. C., Person S. 1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol. 68:5365–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fields S., Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246 [DOI] [PubMed] [Google Scholar]
- 10. Gibson W., Roizman B. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Homa F. L., Brown J. C. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107–122 [DOI] [PubMed] [Google Scholar]
- 12. Huang E., Perkins E. M., Desai P. 2007. Structural features of the scaffold interaction domain at the N terminus of the major capsid protein (VP5) of herpes simplex virus type 1. J. Virol. 81:9396–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kennard J., Rixon F. J., McDougall I. M., Tatman J. D., Preston V. G. 1995. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J. Gen. Virol. 76(Pt 7):1611–1621 [DOI] [PubMed] [Google Scholar]
- 14. Kirkitadze M. D., et al. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 72:10066–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lupas A. 1997. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 7:388–393 [DOI] [PubMed] [Google Scholar]
- 16. Newcomb W. W., et al. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432–446 [DOI] [PubMed] [Google Scholar]
- 17. Newcomb W. W., et al. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Newcomb W. W., et al. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499–511 [DOI] [PubMed] [Google Scholar]
- 19. Nicholson P., et al. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol. 75(Pt 5):1091–1099 [DOI] [PubMed] [Google Scholar]
- 20. Oien N. L., et al. 1997. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J. Virol. 71:1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okoye M. E., Sexton G. L., Huang E., McCaffery J. M., Desai P. 2006. Functional analysis of the triplex proteins (VP19C and VP23) of herpes simplex virus type 1. J. Virol. 80:929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pelletier A., Do F., Brisebois J. J., Lagace L., Cordingley M. G. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J. Virol. 71:5197–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perkins E. M., et al. 2008. Small capsid protein pORF65 is essential for assembly of Kaposi's sarcoma-associated herpesvirus capsids. J. Virol. 82:7201–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Person S., Desai P. 1998. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C). Virology 242:193–203 [DOI] [PubMed] [Google Scholar]
- 25. Preston V. G., McDougall I. M. 2002. Regions of the herpes simplex virus scaffolding protein that are important for intermolecular self-interaction. J. Virol. 76:673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rixon F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135–144 [Google Scholar]
- 27. Rixon F. J., et al. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77(Pt 9):2251–2260 [DOI] [PubMed] [Google Scholar]
- 28. Rixon F. J., McNab D. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714–5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saad A., Zhou Z. H., Jakana J., Chiu W., Rixon F. J. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 73:6821–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spencer J. V., Newcomb W. W., Thomsen D. R., Homa F. L., Brown J. C. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 72:3944–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steven A. C., Spear P. G. 1996. Herpesvirus capsid assembly and envelopment. Oxford University Press, New York, NY [Google Scholar]
- 32. Tatman J. D., Preston V. G., Nicholson P., Elliott R. M., Rixon F. J. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75(Pt 5):1101–1113 [DOI] [PubMed] [Google Scholar]
- 33. Thomsen D. R., Roof L. L., Homa F. L. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trus B. L., et al. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447–462 [DOI] [PubMed] [Google Scholar]
- 35. Walters J. N., Sexton G. L., McCaffery J. M., Desai P. 2003. Mutation of single hydrophobic residue I27, L35, F39, L58, L65, L67, or L71 in the N terminus of VP5 abolishes interaction with the scaffold protein and prevents closure of herpes simplex virus type 1 capsid shells. J. Virol. 77:4043–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wood L. J., Baxter M. K., Plafker S. M., Gibson W. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Z. H., et al. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877–880 [DOI] [PubMed] [Google Scholar]