Abstract
Influenza A virus encodes M2, a proton channel that has been shown to be important during virus entry and assembly. In order to systematically investigate the role of the membrane-proximal residues in the M2 cytoplasmic tail in virus replication, we utilized scanning and directed alanine mutagenesis in combination with transcomplementation assays and recombinant viruses. The membrane-proximal residues 46 to 69 tolerated numerous mutations, with little, if any, effect on virus replication, suggesting that protein structure rather than individual amino acid identity in this region may be critical for M2 protein function.
INTRODUCTION
Influenza A virus is a member of the Orthomyxoviridae family whose genome consists of 8 negative-sense RNA segments which encode 10 or 11 proteins. The highly conserved integral membrane protein M2 is encoded by segment 7 via a spliced mRNA, consists of 97 amino acids, and forms disulfide-linked tetramers which have a pH-gated, proton-selective ion channel activity. M2 is a type III integral membrane protein with an extracellular amino terminus and an intracellular carboxy terminus. M2 is required for virus entry, during which it translocates protons into the virion interior, which allows for the dissociation of viral ribonucleoprotein complexes (vRNPs) from the site of virus-cell membrane fusion, thereby allowing vRNP transport to the nucleus (12, 18, 26). M2 is also required during virus assembly, as its cytoplasmic tail is required for proper incorporation of vRNPs into budding virions (9, 19, 20). The M2 protein and a peptide corresponding to an amphipathic helix in the membrane-proximal cytoplasmic tail of the protein have been shown to induce membrane curvature and scission in unilamellar vesicles and were suggested to be necessary for membrane scission and virion release (31). However, virus-like particles (VLPs) (3, 4, 8, 15, 17, 39) and virions (6, 9, 13) can be released in the absence of M2.
Various deletions and mutations in the M2 protein, particularly in the cytoplasmic tail, attenuate influenza A virus replication significantly (9, 14, 19, 20) and can be complemented with full-length M2 expressed in trans (3, 9, 19, 20, 35). Residues within the cytoplasmic tail form a canonical cholesterol binding motif (CRAC motif) and have been shown to mediate cholesterol binding in purified bacterially expressed protein and during virus infection but not during expression of the protein alone in mammalian cells (30, 33, 37). Mutation of residues within the CRAC motif caused no defect in virus replication in tissue culture but resulted in a slight attenuation of virus in vivo (35). Although structural studies have not been performed on the full-length M2 sequence, nuclear magnetic resonance (NMR) studies of peptides corresponding to the transmembrane domain and portions of the cytoplasmic tail revealed that residues adjacent to the transmembrane domain form an amphipathic helix (25, 32, 34).
To systematically investigate the role of the membrane-proximal residues (amino acids 46 to 69) in the M2 cytoplasmic tail on virus replication, we substituted alanine residues at a number of positions and assessed M2 protein function with a transcomplementation assay and reverse genetics. We show here that residues 46 to 69 tolerate numerous mutations, with little, if any, attenuation of virus replication, as shown by both complementation assays and growth curves of recombinant viruses. Therefore, despite being highly conserved and forming a stable structure, this region can tolerate a large number of amino acid substitutions without significantly affecting influenza A virus replication.
MATERIALS AND METHODS
Plasmids.
The plasmid pCAGGS (24) M2Ud, expressing the M2 cDNA from influenza A/Udorn/72, has been described previously (20). All M2 mutations were introduced into the expression plasmid by overlap extension PCR (35). The pHH21 M segment plasmid, which carries the entire M gene segment used to generate recombinant viruses (20), was mutated via QuikChange site-directed mutagenesis (Stratagene) (35). All inserts in mutant plasmids were confirmed by sequencing. Primer sequences are available upon request.
Cells.
Madin-Darby canine kidney (MDCK) cells were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals), 100 U/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), 1 mM sodium pyruvate (Sigma), and 2 mM Glutamax (Invitrogen). Cells were maintained at 37°C in a humidified 5% CO2 environment.
MDCK cells stably expressing wild-type M2Ud or M2WSN N31S have been described previously (9, 35). The N31S mutation conveys amantadine sensitivity to the M2 protein encoded by influenza A/WSN/33 virus (11, 36). The pBABE plasmid, which expresses puromycin N-acetyltransferase (21), was used to generate MDCK cells stably expressing mutant M2 proteins as described previously (35). Cells expressing M2 proteins were maintained with 5 μM amantadine hydrochloride (Sigma) and 7.5 μg/ml puromycin dihydrochloride (Sigma).
Viruses.
The wild-type viruses used in this study, rUd and rWSN (recombinant versions of A/Udorn/72 [H3N2] and A/WSN/33 [H1N1]), as well as viruses carrying functional deletions of the M2 open reading frame, have been described previously (20, 22, 36). Recombinant viruses were generated using a 12-plasmid rescue system as described previously (19, 20, 22, 36). Recombinant viruses expressing mutant M2 proteins were generated by replacing the pHH21 M segment plasmid with one carrying the indicated mutant M2 open reading frame, as described previously (9). Viruses expressing mutant M2 proteins were plaque purified and grown in MDCK cells stably expressing M2WSN N31S in order to alleviate any selective pressure on the virus to revert the M2 mutations. The entire coding region of the M segment was confirmed by sequencing for all rescued viruses.
Virus infections.
Multistep virus growth curve experiments were performed at a multiplicity of infection (MOI) of 0.001 50% tissue culture infectious dose (TCID50) per cell. For complementation assays, MDCK cells expressing mutant M2 proteins were infected with recombinant viruses that do not carry the full-length M2 protein (M2Stop viruses), as described previously (19). For recombinant viruses, mutant M2-expressing viruses were used to infect MDCK cells. Cells were infected by twice washing confluent 12-well plates with phosphate-buffered saline with calcium and magnesium (PBS+; Invitrogen) and then infecting the cells with the indicated viruses in 250 μl infection medium (DMEM supplemented with 0.5% bovine serum albumin [BSA; Sigma], 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 2 mM Glutamax, and 4 μg/ml N-acetyl trypsin [NAT; Sigma]) at room temperature (RT) with rocking for 1 h. Cells were then washed twice with PBS+ and incubated with 500 μl infection medium at 37°C. At the indicated times, the medium was removed, stored at −80°C, and replaced with fresh medium. Infectious virus titers were determined by TCID50 assay on MDCK cells expressing M2WSN N31S.
High-MOI infections (MOI of 0.5 or 5) were performed by using the protocol for multistep growth curves, with the exception that both NAT and BSA were omitted during all steps of infection. For protein expression studies, the medium was removed at 16 h postinfection (hpi), and cells were processed for Western blot analysis. For determination of virion composition, the cell lysates and supernatants were collected at 12 hpi. Cell debris was removed from supernatants by centrifugation at 1,300 × g for 10 min at 4°C. Virus particles were then concentrated through a 35% sucrose cushion with centrifugation at 182,000 × g in a Sorvall TH-641 rotor for 1 h at 4°C. Virus pellets were resuspended in 200 μl PBS, mixed 3:1 in 4× SDS-PAGE sample buffer, and analyzed along with the cell lysates by Western blot analysis.
Plaque assay.
Plaque assays were carried out by infecting confluent 6-well plates of MDCK cells with the indicated viruses serially diluted in infection medium. Cells were washed twice with PBS+ and infected with 250 μl of virus dilutions for 1 h at RT with rocking, the inoculums were aspirated, and the cells were overlaid with DMEM (Invitrogen) containing 1% agarose (Invitrogen), 0.5% BSA, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM Glutamax, and 4 μg/ml NAT. After overlays had solidified, the cells were incubated at 37°C for 3 days. Cells were then fixed with 4% formaldehyde (Fisher Scientific) in PBS for 1 h at RT and stained with a Naphthol Blue Black solution overnight. Individual plaque diameters were measured from scanned images by using ImageJ (NIH).
Microscopy.
MDCK cells were grown to confluence on tissue culture-treated glass coverslips (Fisher Scientific) in 12-well plates, and the medium was changed every 2 days. Four days after confluence, cells were infected with 500,000 TCID50 per well (MOI of ∼0.9) in 500 μl of infection medium as in the high-MOI infections described above and then incubated at 37°C. At 15 hpi, the cells were incubated on ice for 15 min, washed twice with cold PBS+, and blocked for 30 min on ice in PBS with 0.5% BSA and 3% normal goat serum (Sigma). Surface staining was performed for 1 h on ice in blocking solution containing goat anti-H3 serum raised against A/Aichi/2/68 (1:500 dilution; National Institute of Allergy and Infectious Diseases). Cells were then washed 3 times with cold PBS+, fixed for 10 min with 2% paraformaldehyde (Sigma) in PBS at RT, and incubated for 1 h at RT with blocking solution containing donkey anti-goat IgG conjugated to Alexa Fluor 555 (4 μg/ml; Invitrogen). Cells were then washed twice with PBS+ and mounted on slides by using ProLong Gold antifade solution (Invitrogen).
Samples were imaged on a Nikon Eclipse 90i epifluorescence microscope. Twenty nonoverlapping images were taken of each sample, using a 20× objective. For each image, the total number of infected cells and the number of cells expressing filaments were counted and used to determine the percentage of infected cells expressing filaments.
TCID50 assay.
MDCK cells expressing M2WSN N31S were plated in 96-well plates. When the monolayer was confluent, cells were washed twice with PBS+, infected with 100 μl of virus serially diluted in infection medium, and incubated for 4 days at 37°C. Cells were then fixed by adding 50 μl of 4% formaldehyde in PBS, stained with a Naphthol Blue Black solution, and scored for cytopathic effect. The TCID50 was calculated by the method of Reed and Muench (27).
SDS-PAGE and Western blotting.
Cells were lysed in 1% SDS (Fisher Scientific) in PBS and mixed 1:1 in 2× SDS-PAGE sample buffer. Purified virus particles were mixed 3:1 with 4× SDS-PAGE sample buffer. Sample buffer for samples analyzed for total protein expression contained the reducing agent dithiothreitol (DTT), while samples analyzed for oligomerization did not. Proteins were resolved in 17.5% polyacrylamide gels with 4 M urea and transferred to polyvinylidene difluoride membranes (PVDF-FL; Millipore). Wash buffer contained PBS with 0.3% Tween 20 (Sigma), and the blocking buffer was wash buffer with 5% dry milk added. Membranes were blocked for 30 min at RT, incubated for 2 h at RT with primary antibody, washed three times each for 5 min, incubated for 1 h at RT with secondary antibody, and washed four times each for 5 min. Primary and secondary antibodies were diluted in blocking buffer. The primary antibodies used were mouse anti-M2 14C2 monoclonal antibody (MAb) (1:500 dilution) (40), goat anti-A/Udorn/72 (1:500 dilution) (41), and mouse anti-β-actin AC-15 MAb (1:10,000 dilution; Abcam). The Alexa Fluor 647-conjugated secondary antibodies used were a goat anti-mouse immunoglobulin G (IgG), a donkey anti-goat IgG, and a donkey anti-mouse IgG (all at a 1:500 dilution; Invitrogen). For visualization, membranes were scanned using an FLA-5000 phosphorimager (FujiFilm).
Sequence alignments.
All M2 protein sequences from H1N1 and H3N2 influenza A virus strains (excluding 2009 pandemic H1N1 and laboratory strains) were obtained from the NCBI Influenza Virus Sequence Database (1). Sequences were aligned using ClustalW 2.0.10 (16). The percentage of sequences encoding the most conserved residue at each amino acid position was determined using WebLogo 3 (7).
Statistical analysis.
Plaque diameters and percentages of infected cells expressing filaments were compared using the Student t test. Growth curves were analyzed using mixed analyses of variance (ANOVAs) and Bonferroni posttests, with time and virus as independent variables for transcomplementation assays and growth assays of recombinant viruses. Statistically significant differences are indicated in the figures (*, P < 0.05; **, P < 0.01; ***, P < 0.001). All analyses were done with Prism 4.0 (GraphPad Software Inc.).
RESULTS
Oligomerization and expression of mutant M2 proteins in stable cell lines.
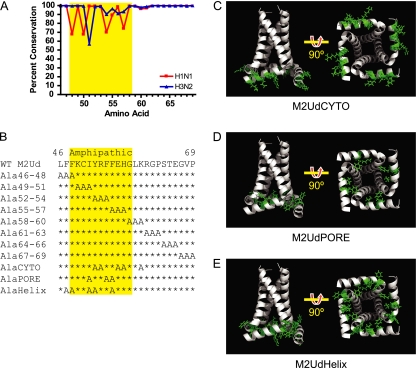
The membrane-proximal region of the M2 protein cytoplasmic tail consists of a region that displays some sequence variability (amino acids 47 to 57) and a region that shows very high conservation (amino acids 58 to 69) among seasonal influenza A virus strains (Fig. 1A). In order to determine if residues 46 to 69 of the M2 protein cytoplasmic tail are required for protein function, two types of alanine substitution mutations were generated (Fig. 1B). First, triple-scanning alanine mutations were generated across the region. Second, an NMR structure (34) of the M2 protein was utilized to select amino acids corresponding to two separate faces (creating the M2UdCYTO and M2UdPORE mutants) of the amphipathic helix within the membrane-proximal region of the cytoplasmic tail. Figure 1C shows residues 52, 53, 56, 57, and 60 of the helix, which face the cytoplasm and were mutated to alanines in the M2UdAlaCYTO mutant. Figure 1D shows residues 51, 54, and 55 of the helix, which face inward toward the amphipathic helixes of the other M2 peptides and were mutated to alanines in the M2UdAlaPORE mutant. Alanine substitutions were utilized in order to minimize structural perturbations in the amphipathic alpha helix while still assessing the role of amino acid side chains in interactions with viral or cellular factors.
Fig. 1.
Sequence and structural location of M2 cytoplasmic tail amino acids. (A) Conservation of residues 46 to 69 of the M2 protein among H1N1 (virus sequences prior to 2009) and H3N2 influenza A virus strains. (B) Sequences of residues 46 to 69 in wild-type M2 protein and alanine substitution mutants. The amphipathic helix is highlighted in yellow (34). (C to E) Structures of M2 mutant proteins (residues 22 to 62), with mutated amino acids shown in green. Residue 22 appears at the top of the structures on the left. The structures on the right are rotated 90°, as indicated, in order to show the cytosolic face of the M2 tetramer. (C) M2UdCYTO mutant (mutations in Y52, R53, E56, H57, and K60). (D) M2UdPORE mutant (mutations in I51, F54, and F55). (E) M2UdHelix mutant (mutations in F47, F48, I51, Y52, and F55). Structures were generated based on the structure under Protein Data Bank accession number 2LOJ by using PyMol.
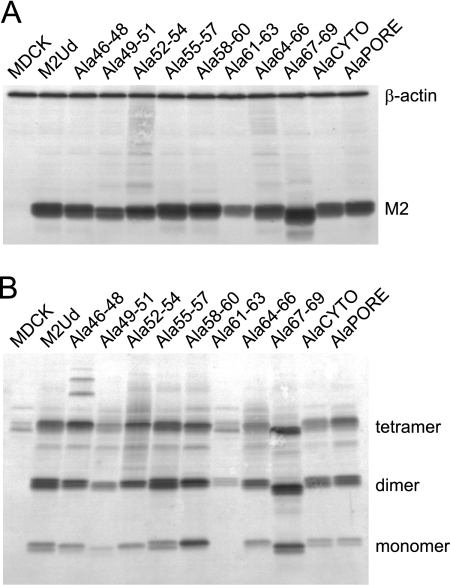
Stably transfected MDCK cells were generated to express the mutant M2 proteins. All cell lines expressed M2 above the level required to complement M2-deficient viruses (Fig. 2A) (20). Additionally, the mutations did not alter the oligomerization potential of M2, as determined by SDS-PAGE and Western blot analysis under nonreducing conditions (Fig. 2B).
Fig. 2.
Expression and oligomerization of mutant M2 proteins. MDCK cells stably expressing the indicated M2 proteins were analyzed by Western blotting in order to determine the overall expression under reducing conditions (A) and the presence of disulfide-linked oligomers under nonreducing conditions (B). Monomeric M2 and the cell protein loading control, β-actin, are indicated in panel A, and monomers, dimers, and tetramers of M2 are indicated in panel B.
M2 proteins are able to complement M2-null viruses.
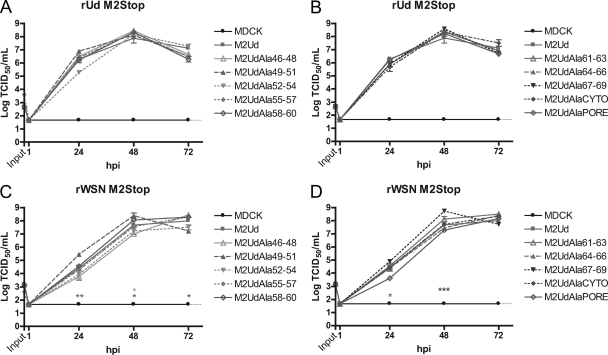
To study mutations which may be deleterious to M2 function, a complementation assay was utilized (20). Stably transfected MDCK cells were infected with two strains of M2-null virus, rUd M2Stop (Fig. 3A and B) and rWSN M2Stop (Fig. 3C and D) (19). Neither rUd M2Stop nor rWSN M2Stop was able to produce infectious virus on MDCK cells that did not express M2 (Fig. 3). The replication of rUd M2Stop in all mutant M2-expressing cell lines was not different from what was observed in wild-type M2-expressing cells (Fig. 3A and B), indicating that all of the mutated M2 proteins were capable of supporting rUd virus replication.
Fig. 3.
Complementation of M2-deficient viruses by expression of mutant M2 proteins. MDCK cells stably expressing the indicated M2 proteins were infected at an MOI of 0.001 with a recombinant influenza A virus that does not encode the full-length M2 protein. The amount of infectious virus at each time point was determined by TCID50 assay on cells expressing wild-type M2. Complementation of rUd M2Stop virus (A and B) or rWSN M2Stop virus (C and D) infectivity was determined with cells expressing the indicated M2 proteins. The means and standard errors of the means for triplicate samples from a representative experiment are shown. The limit of detection is marked by a horizontal dotted line.
rWSN M2Stop showed a minor but statistically significant increase in virus replication on cell lines expressing M2UdAla49-51 and M2UdAla67-69, while slightly reduced replication was observed on cell lines expressing M2UdAla46-48 and M2UdAlaPore (Fig. 3C and D). However, no titers at 24 hpi or 48 hpi were >1 log different from the titer of rWSN M2Stop on cells expressing wild-type M2 protein. Despite these minor differences in replication, all cell lines expressing mutant M2 proteins supported high levels of replication of rWSN M2Stop, consistent with the data obtained utilizing the rUd M2Stop virus (Fig. 3A and B). Taken together, the data suggest that the region of M2 from residues 46 to 69 is highly amenable to mutation, despite being highly conserved.
Replication of recombinant influenza A viruses expressing M2 alanine mutations.
In order to further investigate the importance of M2 residues 46 to 69 in virus replication, several recombinant viruses were generated in both the rUd and rWSN backgrounds. Because expression of M2UdAla49-51 supported slightly greater replication of rWSN M2Stop at two time points, a recombinant virus expressing this mutant was generated. A virus expressing M2Ala67-69 was generated because its expression supported higher levels of rWSN M2Stop at 48 hpi (P < 0.001). Lastly, because M2 has been shown to interact with M1 (3, 19) and the face of the M2 amphipathic helix exposed to the cytosol could mediate interactions with M1 or other proteins, viruses were also generated which expressed M2AlaCYTO.
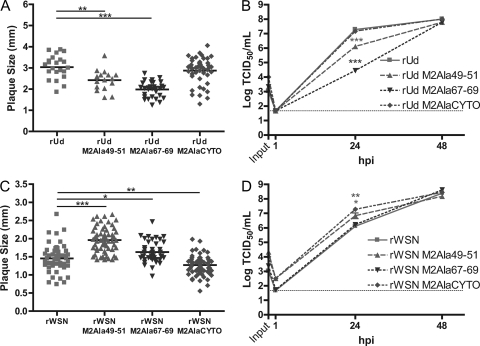
Plaque assays were performed on MDCK cells to determine the plaque diameter of each recombinant virus. Recombinant viruses expressing M2Ala49-51 or M2Ala67-69 formed smaller plaques in the rUd background but larger plaques in the rWSN background than the respective wild-type viruses (Fig. 4A and C). Expression of M2AlaCYTO resulted in smaller plaques of rWSN but caused no difference in the plaque diameter of rUd compared to wild-type virus.
Fig. 4.
Plaque sizes and replication kinetics of recombinant viruses expressing mutated M2 proteins. (A and C) Plaque diameters of MDCK cells infected with the indicated recombinant viruses at 3 days postinfection (dpi). The cells were fixed and stained with a Naphthol Blue Black solution, and individual plaque diameters were measured from scanned images by using ImageJ. Representative data from triplicate experiments are shown. The Student t test was performed to determine significant differences compared to wild-type virus. (B and D) MDCK cells were infected with the indicated recombinant viruses at an MOI of 0.001. At the indicated hpi, cell supernatants were collected and the numbers of infectious virus particles were determined by TCID50 assay on M2-expressing cells. The means and standard errors of the means for triplicate samples from a representative experiment are shown. The limit of detection is marked by a horizontal dotted line.
The decreased plaque diameters of mutant M2-expressing rUd viruses correlated with the production of infectious virus at 24 hpi in a low-MOI growth curve experiment (Fig. 4B), but by 48 hpi, there were no differences in infectious virus production between any of the viruses. The ability of mutant M2-expressing rWSN viruses to form large or small plaques did not fully correlate with the ability to replicate in a low-MOI growth curve experiment (Fig. 4D). rWSN expressing M2Ala49-51 formed larger plaques and had higher infectious virus titers at 24 hpi in the multistep growth curve experiment than those of wild-type virus. However, rWSN expressing M2Ala67-69 formed slightly larger plaques than wild-type virus but resulted in similar levels of infectious virus at both 24 hpi and 48 hpi. Additionally, expression of M2AlaCYTO in rWSN resulted in smaller plaques but more infectious virus at 24 hpi. Interestingly, despite some statistically significant changes at 24 hpi, all mutant M2-expressing rUd and rWSN viruses formed similar amounts of infectious virus at 48 hpi compared to their respective wild-type recombinant viruses, suggesting that the membrane-proximal region (residues 46 to 69) of M2 can tolerate significant mutations without eliminating the ability of influenza A virus to replicate.
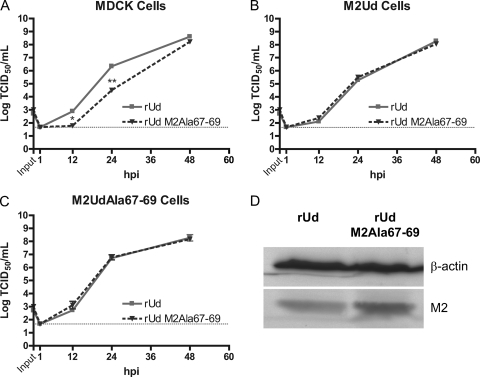
Expression of M2UdAla67-69 is able to complement a recombinant virus expressing M2Ala67-69.
Because rUd M2Ala67-69 produced fewer infectious virus particles at 24 hpi but rUd M2Stop replicated to similar levels on cells expressing M2UdAla67-69, the growth of rUd M2Ala67-69 was compared to that of wild-type rUd in MDCK, M2Ud-expressing, and M2UdAla67-69-expressing cells (Fig. 5). In MDCK cells, rUd M2Ala67-69 replicated to lower titers at 12 hpi and 24 hpi but had formed similar amounts of infectious virus at 48 hpi (Fig. 5A). In M2Ud- or M2UdAla67-69-expressing cells, both viruses produced similar amounts of infectious virus particles at all times measured (Fig. 5B and C). This indicates that expression of wild-type M2 or M2UdAla67-69 is able to rescue any potential defect in production of infectious virus at early times postinfection. In order to determine if the level of M2 expressed by rUd M2Ala67-69 was equivalent to that of wild-type virus, high-MOI infections were performed on MDCK cells. Expression of M2 was similar in cells infected by both viruses (Fig. 5D), indicating that the small defect in the growth of rUd M2Ala67-69 at 12 hpi and 24 hpi was not due to decreased expression of M2 and suggesting that perhaps a larger amount of M2 (either wild type or mutated) is needed for this virus to replicate efficiently.
Fig. 5.
Complementation of a recombinant virus expressing M2Ala67-79. MDCK (A), M2Ud-expressing (B), and M2UdAla67-69-expressing (C) cells were infected with the indicated recombinant viruses at an MOI of 0.001. At the indicated hpi, cell supernatants were collected and the numbers of infectious virus particles were determined by TCID50 assay on cells expressing wild-type M2. The means and standard errors of the means for triplicate samples from a representative experiment are shown. The limit of detection is marked by a horizontal dotted line. (D) MDCK cells were infected with the indicated recombinant viruses at an MOI of 0.5. At 16 hpi, expression in cell lysates of M2 and the cell protein loading control, β-actin, was compared by Western blotting.
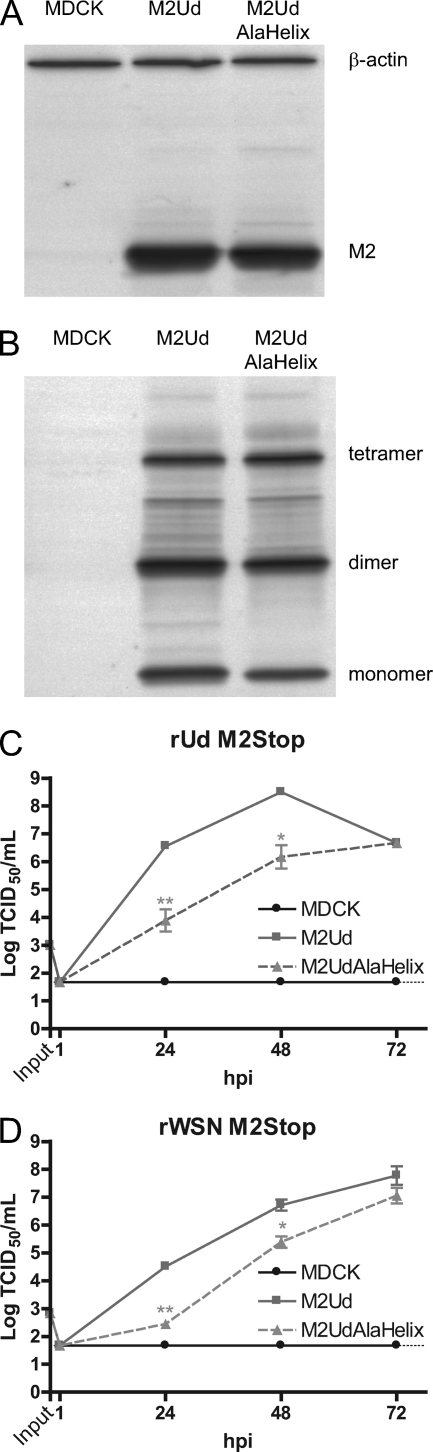
An M2 protein with alanine mutations at F47, F48, I51, Y52, and F55 is partially able to complement M2-deficient viruses.
Rossman et al. determined that alanine substitutions of residues F47, F48, I51, Y52, and F55 in the rUd M2 protein (M2AlaHelix) (Fig. 1B and E) resulted in a recombinant virus that was severely debilitated in growth (30). Given the limited effect of the mutations we introduced into the M2 protein on virus replication, we sought to compare this mutant to our M2 mutants. Stably transfected MDCK cells were generated which expressed the M2UdAlaHelix protein. The MDCK cell line expressing M2UdAlaHelix expressed a similar level of M2 to that in a cell line expressing wild-type M2Ud (Fig. 6A). Additionally, the M2UdAlaHelix mutant formed oligomers, as determined by SDS-PAGE and Western blot analysis under nonreducing conditions (Fig. 6B).
Fig. 6.
Expression and function of M2UdAlaHelix protein. Western blot analysis was performed with MDCK cells stably expressing either wild-type M2Ud or M2UdAlaHelix. (A) Total expression of M2 protein under reducing conditions. (B) Presence of disulfide-linked oligomers under nonreducing conditions. Monomeric M2 and the cell protein loading control, β-actin, are indicated in panel A, and monomers, dimers, and tetramers of M2 are indicated in panel B. (C and D) MDCK cells stably expressing the indicated M2 protein were infected with the indicated recombinant virus that does not encode the full-length M2 protein at an MOI of 0.001. The amount of infectious virus at each time point was determined by TCID50 assay on cells expressing wild-type M2. The means and standard errors of the means for triplicate samples from a representative experiment are shown. The limit of detection is marked by a horizontal dotted line.
In order to determine if constitutive expression of M2UdAlaHelix was able to complement M2-null viruses, MDCK or M2-expressing MDCK cells were infected at a low MOI, and the amounts of infectious virus were determined at the indicated times postinfection. There was a reduction in the ability of M2UdAlaHelix to complement either rUd M2Stop (Fig. 6C) or rWSN M2Stop (Fig. 6D). Both M2-null viruses grew to infectious virus titers that were approximately 1 to 2 log lower on M2UdAlaHelix-expressing cells than on wild-type M2-expressing cells at both 24 hpi and 48 hpi, which was a greater attenuation than that seen on any other M2-expressing cell lines analyzed.
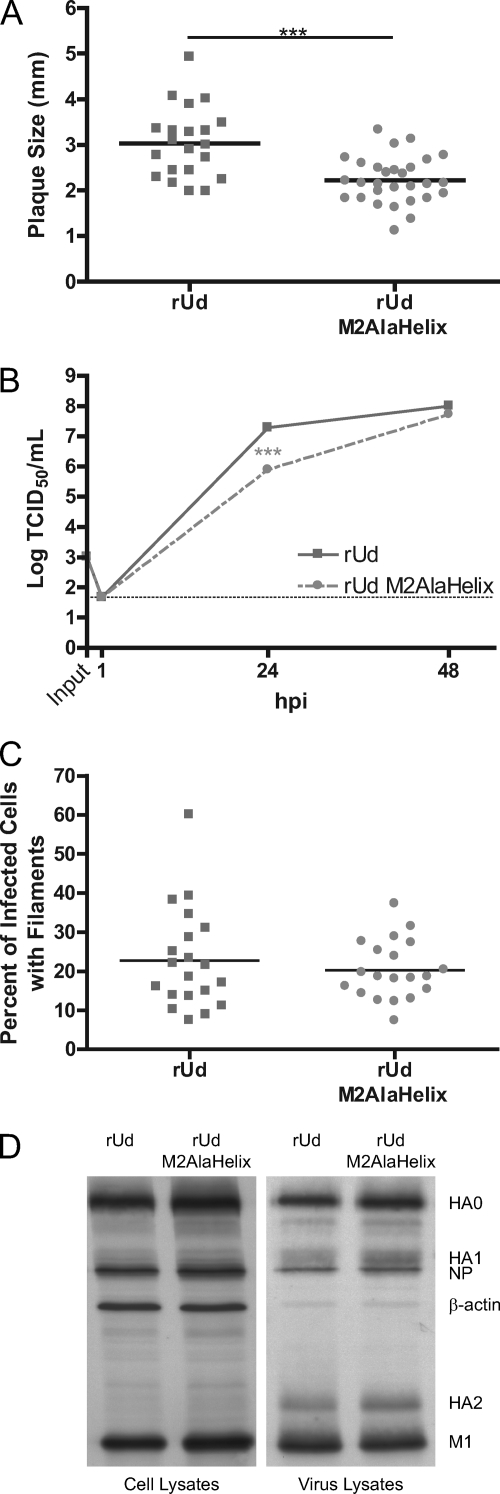
Recombinant influenza A virus expressing the M2UdAlaHelix protein is attenuated at early time points but grows to similar peak titers as the wild type.
Because the attenuation of rUd M2Stop in cells expressing M2UdAlaHelix was less than what was expected based on data for a recombinant virus expressing this mutant M2 protein (30), we generated an rUd virus expressing the M2UdAlaHelix protein to further characterize this mutation. Plaque assays of this recombinant, rUd M2AlaHelix, on MDCK cells showed significantly smaller plaques than those of wild-type rUd (Fig. 7A). Low-MOI growth curves revealed a decrease in production of infectious virus particles at 24 hpi, but by 48 hpi, rUd M2AlaHelix had produced as much infectious virus as recombinant wild-type rUd. Sequence analysis of the rUd M2AlaHelix virus produced at this time point demonstrated that no changes in the M segment sequence had accumulated, indicating that a revertant virus had not emerged (data not shown). The ability of rUd M2AlaHelix to form filaments in virus-infected cells was not significantly different from that of rUd (Fig. 7C). The expression levels of the hemagglutinin (HA), NP, and M1 proteins were similar in cells infected with either wild-type rUd or rUd M2AlaHelix (Fig. 7D). Western blot analysis of pelleted rUd M2AlaHelix virus particles showed no major defect in the secretion of virus particles as well as no defect in the incorporation of HA, NP, or M1 (Fig. 7D). The modest level of attenuation and lack of significant changes in structural protein packaging seen with rUd M2AlaHelix were not consistent with what was observed in previous reports (30). However, these data are consistent with the data on other M2 mutations generated in this region of the protein (Fig. 3 and 4), indicating a limited role for membrane-proximal amino acids 46 to 69 of M2 in supporting influenza A virus infectious virus production.
Fig. 7.
Characterization of a recombinant virus expressing M2AlaHelix. (A) Plaque diameters of MDCK cells infected with recombinant viruses expressing either wild-type M2 or M2AlaHelix at 3 dpi. The individual and average diameters of plaques were determined. Representative data from triplicate experiments are shown. (B) MDCK cells were infected with the indicated viruses at an MOI of 0.001. At the indicated hpi, cell supernatants were collected and the numbers of infectious virus particles were determined by TCID50 assay on M2-expressing cells. The means and standard errors of the means for triplicate samples from a representative experiment are shown. The limit of detection is marked by a horizontal dotted line. (C) MDCK cells infected with 500,000 TCID50 of the indicated virus were stained for immunofluorescence at 15 hpi and visualized by microscopy. The percentage and average percentage of infected cells showing filaments were determined from 20 nonoverlapping images taken with an epifluorescence microscope. Data for one representative experiment are shown. (D) MDCK cells were infected with the indicated virus at an MOI of 5. At 12 hpi, supernatants were collected, virus particles were concentrated by ultracentrifugation through 35% sucrose in PBS and resuspended in PBS, and equal volumes were analyzed along with the cell lysates by Western blotting under reducing conditions.
DISCUSSION
The influenza A virus M2 protein plays a crucial role during several steps of the viral life cycle. During virus entry, the ion channel activity of M2 mediates the release of vRNPs into the cytoplasm after HA-induced fusion of the viral and cellular membranes. Additionally, the M2 cytoplasmic tail is critical for proper incorporation of vRNPs into budding particles. The exact mechanism of incorporation might occur through binding of NP and/or M1. Indeed, there is support for the latter in that M1 and M2 have been shown to interact with each other (3, 19). Two separate M1 binding sites have been identified within M2, one within amino acids 70 to 77 (3, 19) and another within residues 45 to 69 (19).
The membrane-proximal region consisting of amino acids 46 to 69 is believed to mediate the association of M2 with sites of virus budding. This region consists of a palmitoylation site, a cholesterol binding motif, and an amphipathic helix that can insert into the lipid bilayer (23, 25, 32, 34, 38). Although palmitoylation was expendable for growth in vitro, mutation of the palmitoylated cysteine at residue 50 led to a reduction in pathogenesis in mice (10), and this cysteine was required for the partial raft partitioning of full-length M2 in giant plasma membrane vesicles (37). Mutation of the critical residues within the cholesterol binding motif caused no effect on in vitro growth but a small reduction of morbidity and mortality associated with infection in mice (35). This study sought to investigate which residues in the membrane-proximal region (residues 46 to 69) of the M2 protein cytoplasmic tail are required for the function of M2 during virus replication.
Recombinant forms of two distinct strains of influenza A virus, A/Udorn/72 and A/WSN/33, were utilized. rUd is an H3N2 strain that forms both spherical and filamentous virus particles, while rWSN is an H1N1 mouse-adapted strain that forms spherical virus particles almost exclusively (2, 28, 29). While some M2 mutations have been shown to be deleterious to virus replication in both virus strains, others have been shown to more deleterious in rUd (9, 19). During infection with rUd, less M2 protein is expressed in cells, and therefore less M2 protein is incorporated into virus particles than that for infection with rWSN (10, 35).
Triple-scanning alanine mutations were made across residues 46 to 69 to systematically test the necessity of these residues for the formation of infectious virus particles. Furthermore, two separate faces of the amphipathic helix found in the M2 protein cytoplasmic tail were also separately mutated to alanines (giving the M2UdAlaCYTO and M2UdAlaPORE mutants). All of the M2 mutants were able to complement two strains of influenza A viruses lacking functional M2 (Fig. 3). Moreover, recombinant viruses expressing several of these M2 mutants were generated and grew to similar peak titers at 48 hpi (Fig. 4). Taken together, these data show that membrane-proximal residues 46 to 69 of the M2 protein can tolerate significant mutations without eliminating the ability of the virus to replicate. While none of the mutated M2 proteins had any major effect on the replication of rUd viruses, it is interesting that two mutations, Ala49-51 and Ala67-69, caused slight but significant increases in replication in rWSN. This demonstrates the importance of assessing M2 mutations in different influenza A virus strains. The variable results for these mutations may map to other viral proteins, as demonstrated previously for virus budding (5) and infectious virus production (9, 19).
Analysis of a recent NMR structure revealed that some of the nonpolar residues within the amphipathic helix from one monomer interact with residues in the loop between the transmembrane domain and the amphipathic helix of another monomer (34). L46 was shown to interact with F54, and F48 was shown to interact with both F55 and L59. Interestingly, one of the large hydrophobic residues in each of those pairs (F54 and F55) was mutated in the M2AlaPORE mutant. Despite disruption of these interactions, these mutations did not abrogate the functions of the M2 protein during virus replication (Fig. 3B and D). However, it is conceivable that mutating these residues from phenylalanines to alanines was insufficient to fully disrupt the hydrophobic interactions. Additionally, the NMR structure revealed that the charged residues K49, R53, H57, K60, and R61 project outward and could interact with the negatively charged phospholipids. The M2AlaCYTO mutant had several of these amino acids altered from charged residues to alanines (R53A, H57A, and K60A), yet this mutant retained the ability to complement M2-null viruses (Fig. 3B and D), and recombinant viruses expressing it showed no defect in replication (Fig. 4B and D).
These data contrast somewhat with a study by Rossman et al., who demonstrated a critical role for the membrane-proximal cytoplasmic region of the M2 protein in virus budding and membrane scission (30). In that study, while single point mutations at positions 47, 48, 51, 52, and 55 did not affect virus assembly and budding, combining all those mutations led to a >5-log reduction in infectious virus production and to a decrease in the amount of NP incorporated relative to the other viral proteins (30). These mutations abrogated the ability of a peptide corresponding to this region of the M2 protein to induce membrane scission in unilamellar vesicles and also reduced the ability of full-length M2 to induce VLPs (31). The significance of this is not completely clear, since VLPs are formed in the absence of M2 expression (3, 4, 8, 15, 17, 39) and viruses carrying a mutation in the M2 5′ splice site or encoding a nonfunctional M2 protein express undetectable levels of M2 protein yet produce virus particles with decreased infectivity (6, 9, 13), thus demonstrating that M2 is not absolutely necessary for membrane scission of influenza A virus particles. We also generated an M2 protein with alanine substitutions at positions 47, 48, 51, 52, and 55 and demonstrated a 1- to 2-log decrease in infectious virus production from cells expressing M2UdAlaHelix compared to cells expressing wild-type M2 in a transcomplementation assay. The defect in infectious virus production was more pronounced in infections with rUd M2Stop than in those with rWSN M2Stop. An rUd virus encoding the M2AlaHelix mutant was able to form filaments and had no defect in NP incorporation (Fig. 7C and D), whereas the same virus reported by Rossman et al. was unable to form filaments and had a defect in NP incorporation (30). The rUd M2AlaHelix virus that we generated also showed reduced replication kinetics which were not changed as much (1- to 2-log reduction at 24 hpi and identical titers at 48 hpi) (Fig. 7B) as those reported by Rossman et al. (∼3-log reduction at 24 hpi and ∼5-log reduction at 48 hpi) compared to wild-type rUd (30). Although both growth curve experiments were performed at the same MOI (0.001), the amount of infectious virus was measured differently (plaque assay versus TCID50 assay). However, titration of stocks of rUd M2AlaHelix by the two assays showed comparable titers (data not shown), indicating that the method of detecting infectious virus was not responsible for the discrepancy in attenuation.
This study sought to investigate whether the membrane-proximal residues 46 to 69 of the M2 protein cytoplasmic tail are required for the function of M2 during virus replication. Given the facts that numerous mutations were made in this region of the M2 protein and that in both complementation assays and recombinant viruses these mutations caused little, if any, attenuation in virus replication, we concluded that the region can tolerate numerous amino acid substitutions before protein function is compromised. Protein structure rather than individual amino acid identity may be critical for the function of this region of the M2 protein. It will be important to utilize a structural biology approach in order to gain further knowledge about the role of this region in M2 function.
ACKNOWLEDGMENTS
We thank the members of the Pekosz laboratory for helpful comments and discussions.
This study was supported by the Eliasberg and Marjorie Gilbert Foundations and by NIH grant R01 AI 053629.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Bao Y., et al. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bourmakina S. V., Garcia-Sastre A. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517–527 [DOI] [PubMed] [Google Scholar]
- 3. Chen B. J., Leser G. P., Jackson D., Lamb R. A. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen B. J., Leser G. P., Morita E., Lamb R. A. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen B. J., Takeda M., Lamb R. A. 2005. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79:13673–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheung T. K. W., et al. 2005. Generation of recombinant influenza A virus without M2 ion-channel protein by introduction of a point mutation at the 5′ end of the viral intron. J. Gen. Virol. 86:1447–1454 [DOI] [PubMed] [Google Scholar]
- 7. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez-Puertas P., Albo C., Perez-Pastrana E., Vivo A., Portela A. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538–11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grantham M. L., Stewart S. M., Lalime E. N., Pekosz A. 2010. Tyrosines in the influenza A virus M2 protein cytoplasmic tail are critical for production of infectious virus particles. J. Virol. 84:8765–8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grantham M. L., et al. 2009. Palmitoylation of the influenza A virus M2 protein is not required for virus replication in vitro but contributes to virus virulence. 83:8655–8661 [Google Scholar]
- 11. Hay A. J., Wolstenholme A. J., Skehel J. J., Smith M. H. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helenius A. 1992. Unpacking the incoming influenza virus. Cell 69:577–578 [DOI] [PubMed] [Google Scholar]
- 13. Hutchinson E. C., Curran M. D., Read E. K., Gog J. R., Digard P. 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82:11869–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwatsuki-Horimoto K., et al. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 80:5233–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai J. C. C., et al. 2010. Formation of virus-like particles from human cell lines exclusively expressing influenza neuraminidase. J. Gen. Virol. 91:2322–2330 [DOI] [PubMed] [Google Scholar]
- 16. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 17. Latham T., Galarza J. M. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin K., Helenius A. 1991. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 65:232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCown M. F., Pekosz A. 2006. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80:8178–8189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCown M. F., Pekosz A. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79:3595–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgenstern J. P., Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neumann G., et al. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen P. A., et al. 2008. pH-induced conformational change of the influenza M2 protein C-terminal domain. Biochemistry 47:9934–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niwa H., Yamamura K., Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 25. Pielak R. M., Chou J. J. 2010. Solution NMR structure of the V27A drug resistant mutant of influenza A M2 channel. Biochem. Biophys. Res. Commun. 401:58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pinto L. H., Holsinger L. J., Lamb R. A. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528 [DOI] [PubMed] [Google Scholar]
- 27. Reed L. J., Muench H. 1938. A simple method of estimating 50 percent endpoints. Am. J. Hyg. 27:493–499 [Google Scholar]
- 28. Roberts P. C., Compans R. W. 1998. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. U. S. A. 95:5746–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts P. C., Lamb R. A., Compans R. W. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127–137 [DOI] [PubMed] [Google Scholar]
- 30. Rossman J. S., et al. 2010. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84:5078–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rossman J. S., Jing X., Leser G. P., Lamb R. A. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schnell J. R., Chou J. J. 2008. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schroeder C., Heider H., Moncke-Buchner E., Lin T. I. 2005. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur. Biophys. J. 34:52–66 [DOI] [PubMed] [Google Scholar]
- 34. Sharma M., et al. 2010. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 330:509–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stewart S. M., Wu W.-H., Lalime E. N., Pekosz A. 2010. The cholesterol recognition/interaction amino acid consensus motif of the influenza A virus M2 protein is not required for virus replication but contributes to virulence. Virology 405:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takeda M., Pekosz A., Shuck K., Pinto L. H., Lamb R. A. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 76:1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thaa B., Levental I., Herrmann A., Veit M. 2011. Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem. J. 437:389–397 [DOI] [PubMed] [Google Scholar]
- 38. Tian C., Gao P. F., Pinto L. H., Lamb R. A., Cross T. A. 2003. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 12:2597–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tscherne D. M., Manicassamy B., García-Sastre A. 2010. An enzymatic virus-like particle assay for sensitive detection of virus entry. J. Virol. Methods 163:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zebedee S. L., Lamb R. A. 1988. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 62:2762–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J., Pekosz A., Lamb R. A. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]