Abstract
We assessed the relative susceptibilities to disease of the DBA.2 and C57BL/6 mouse models upon infection with a range of influenza A and B viruses. DBA.2 mice were more susceptible to disease upon inoculation with human H1N1 influenza A virus strains, several swine influenza viruses, and influenza B viruses but were not overtly susceptible to infection with human seasonal H3N2 strains. Hemagglutination inhibition and immunoglobulin isotype profiling indicated that DBA.2 and C57BL/6 mice generate comparable humoral responses upon equivalent 50% mouse lethal dose (MLD50) challenges with influenza virus. Our data demonstrate the utility of DBA.2 mice for the elucidation of influenza virus pathogenicity determinants and the testing of influenza vaccines.
TEXT
Seasonal influenza constitutes a significant public health problem, causing an estimated 200,000 hospitalizations (25) and an average of 25,000 to 35,000 excess deaths yearly in the United States (6, 7, 26). Additionally, infrequent influenza pandemics cause considerable disease, with associated mortality ranging from approximately 50 million deaths during the 1918 pandemic (11) to 1 to 4 million deaths in 1957 and approximately 1 million deaths in 1968 (5). In 2009, a novel H1N1 virus emerged and spread rapidly in humans (17, 21), causing severe disease in susceptible populations, as well as an unusually high number of hospitalizations of young people (8).
The most practical model for the study of influenza virus pathogenicity is the mouse; nevertheless, commonly utilized mouse strains such as C57BL/6 and BALB/c carry a significant drawback in that there is typically an absence or relative lack of disease generated by human seasonal and animal influenza viruses (4). For this reason, pathogenicity studies are conducted predominantly using highly pathogenic or mouse-adapted strains of influenza virus (14, 23, 27).
The low virulence of primary influenza virus isolates in mouse models further limits the number of challenge viruses that can be used in the development of influenza vaccines and therapeutics. Efforts to circumvent this problem by adapting influenza virus strains to mice can lead to mutations in the viral genome which are not reflective of human influenza viruses and which may alter the susceptibility of the virus to vaccines or therapeutics, as has been shown to be the case for egg-adaptive changes (15).
Recent reports indicate that DBA.2 mice are highly susceptible to H5N1 influenza viruses (2) as well as the mouse-adapted strain A/Puerto Rico/8/1934 (H1N1) (PR8) (23). Furthermore, a majority of avian viruses with low pathogenicity in natural hosts are pathogenic in this model and are more pathogenic than in C57BL/6 mice (3). For these reasons, use of the DBA.2 model in influenza virus research has increased recently (1, 9, 10, 12, 22). Despite the rising popularity of this model, the breadth of susceptibility of DBA.2 mice to influenza viruses isolated from mammalian hosts is as yet unclear. Herein we addressed this knowledge gap in the field. Specifically, we demonstrated that relative to C57BL/6 mice, DBA.2 mice are more susceptible to seasonal human H1N1 subtype influenza viruses, pandemic strains of both H1 and H3 subtypes, swine influenza viruses of H1 and H3 subtypes, and some influenza B viruses. In contrast, DBA.2 mice were found not to have increased susceptibility to seasonal human H3N2 subtype viruses. Further, we showed that the DBA.2 mouse mounts a humoral immune response to influenza virus that is qualitatively similar to that of C57BL/6 mice, suggesting that in principle, DBA.2 mice present a suitable model for influenza vaccination studies.
A disparate range of influenza virus isolates cause disease in DBA.2 mice.
To evaluate the utility of the DBA.2 mouse for the study of human influenza viruses, we assessed the relative sensitivities of DBA.2 and C57BL/6 mice to a panel of human isolates. The 50% mouse lethal dose (MLD50) was determined once each for pandemic, seasonal, and mouse-adapted human H1N1 and H3N2 subtype influenza viruses, as well as for influenza B virus strains (Table 1). Significance was ascertained by determination of 95 percent confidence intervals (CIs) (19, 24). Overall, the results obtained demonstrate that the DBA.2 mouse represents a more sensitive system in which to evaluate influenza virus pathogenicity than the C57BL/6 mouse, although several observations that limit this generalization were noted. In DBA.2 mice, all mouse-adapted, human pandemic, and human seasonal H1N1 virus strains were shown to be lethal, with MLD50 values of 106.1 PFU or lower. In each case, the MLD50 values were significantly lower for DBA.2 mice than for C57BL/6 mice. Seasonal H3N2 viruses, however, showed no pathogenicity, either in C57BL/6 mice or in DBA.2 mice (Table 1). Among the influenza B virus strains tested, B/Lee/1940 and B/Victoria/2/1987 were significantly more pathogenic in DBA.2 mice than in C57BL/6 mice. The strain B/Yamagata/16/1988, however, showed similar lethality in the two mouse models (Table 1).
Table 1.
Lethal doses of influenza A and B viruses in DBA.2 and C57BL/6 mice
| Virusa | MLD50b (PFU) for indicated mouse model |
95% CId for MLD50 for indicated mouse model |
||
|---|---|---|---|---|
| DBA.2 | C57BL/6 | DBA.2 | C57BL/6 | |
| Human pandemic viruses | ||||
| A/Hong Kong/1/68 (H3N2) | 105.5 | >106 | 104.9–106.1 | N/Ae |
| A/Netherlands/602/09 (H1N1) | 100.5 | 104.3 | 10−0.1–101.1 | 103.4–105.3 |
| Human seasonal viruses | ||||
| A/Brisbane/10/07 (H3N2) | >106 | >106 | N/A | N/A |
| A/Wisconsin/67/05 (H3N2) | >106 | >106 | N/A | N/A |
| A/Panama/2007/99 (H3N2) | >106 | >106 | N/A | N/A |
| A/Brisbane/59/07 (H1N1) | 105 | >106 | 104.1–105.9 | N/A |
| A/New Caledonia/20/99 (H1N1)* | 103.8, 104.7 | 105.5, 106.3 | 102.8–104.7 | 105.2–106.4f |
| A/Solomon Islands/03/06 (H1N1) | 103.9 | >105.3c | 103–104.9 | N/A |
| B/Victoria/2/87 | 102.5 | 104.7 | 101.8–103.2 | 103.7–105.6 |
| B/Yamagata/16/88* | 104.5, 105.4 | 105 | 104.3–105.5 | 104.1–105.9 |
| Swine influenza viruses | ||||
| A/swine/Spain/53207/04 (H1N1) | 102.2 | 105.7 | 101.6–102.8 | 104.7–106.6f |
| A/swine/Kansas/77778/07 (H1N1) | 101 | 102.5 | 100.13–101.87 | 101.88–103.1 |
| A/swine/Spain/40564/04 (H1N2) | 101.3 | 104.7 | 100.4–102.3 | 103.7–105.6 |
| A/swine/Spain/54008/04 (H3N2) | 105.5 | >106 | 104.9–106.1 | N/Ae |
| A/swine/Texas/4199-2/98 (H3N2) | 105.7 | 106.2 | 104.7–106.6 | 105.6–106.8 |
| Avian influenza viruses | ||||
| A/duck/Alberta/76 (H1N1) | >106 | >106 | N/A | N/A |
| A/duck/Ukraine/63 (H3N8) | >106 | >106 | N/A | N/A |
| Recombinant viruses | ||||
| A/duck/Alberta/76 (H1N1) (6:2 virus with PR8 internal genes) | 101.2 | 103.5 | 100.4–101.9 | 102.9–104.1 |
| A/duck/Ukraine/63 (H3N8) (6:2 virus with PR8 internal genes) | 105.5 | >106 | 104.9–106.1 | N/Ae |
| Mouse adapted viruses | ||||
| A/Puerto Rico/8/34 (H1N1)* | 100.3, 100.5 | 101.3, 101.5 | 100.01–100.6 | 100.9–101.65 |
| X-31 (H3N2) | 100.7 | 105.3 | 10−0.4–101.8 | 104.4–106.3 |
| B/Lee/40* | 103.5, 103.5 | 104.3, 104.5 | 103.2–103.8 | 104–104.6 |
MLD50 values were determined twice for a subset of viruses, marked with an asterisk, and CI values were calculated based on both data sets where appropriate. Abbreviations: HK, Hong Kong; NL, Netherlands.
For determination of MLD50, groups of four mice were inoculated intranasally with symmetrically spaced serial dilutions of the indicated strains. Mice were scored as dead if >25% weight loss was observed.
The stock titer of the A/Solomon Islands/03/06 virus did not allow inoculation at doses greater that 105.3 PFU.
N/A, not applicable.
For the A/Hong Kong/1/68, A/swine/Spain/54008/04, and A/duck/Ukraine/63 (6:2) viruses, marginal or no weight loss was observed in C57BL/6 mice inoculated at 106 PFU. For this reason, the MLD50 was assumed to be greater than 106.1 PFU for the purpose of assessing the significance of the differences between the DBA.2 and C57BL/6 groups.
For determination of the 95% confidence interval for A/New Caledonia/20/99 and A/swine/Spain/53207/04 in C57BL/6 mice, 0% survival was assumed for a 107-PFU inoculum.
We next assessed the relative sensitivities of the DBA.2 mouse model to infection with swine and avian influenza viruses of the H1 and H3 subtypes. The MLD50 values of five swine and two low-pathogenicity avian viruses were determined once in DBA.2 mice and once in C57BL/6 mice (Table 1). H1 subtype swine viruses were lethal in both strains of mice, but the MLD50 values were significantly lower for DBA.2 mice than for C57BL/6 mice. As was observed with the human strains, the H1 subtype swine viruses were in general more virulent than the H3 subtype swine viruses (Table 1). The two avian viruses tested were not lethal in DBA.2 or C57BL/6 mice, although both corresponding 6:2 reassortant viruses carrying the avian HA and NA genes in the PR8 background were significantly more lethal in the DBA.2 model (Table 1). For virus strains that caused mortality in both mouse models, the mean times to death were not found to be significantly different (data not shown).
Pathogenicity in DBA.2 mice correlates with viral lung titers.
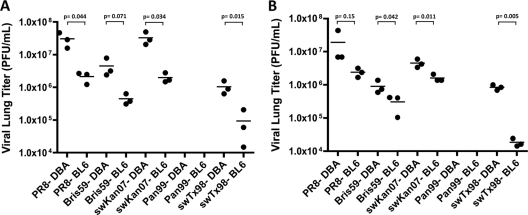
To address whether the range of pathogenicity phenotypes observed was related to the ability of the viruses to replicate in mice or was due rather to specific virulence factors, we determined titers in the lungs of mice infected with five selected viruses. DBA.2 mice were inoculated with one of the following strains at a dose of 103.7 PFU: A/Panama/2007/1999 (H3N2), A/Brisbane/59/2007 (H1N1), PR8, A/swine/77778/Kansas/2007 (H1N1), and A/swine/Texas/4199-2/1998 (H3N2). Consistent with the hypothesis that virulence in the DBA.2 model would correlate with replicative capacity, lung tissue collected from mice given A/Panama/2007/1999 virus did not carry detectable viral loads. In contrast, all other virus strains tested grew efficiently (Fig. 1). The highly pathogenic PR8 and A/swine/Kansas/77778/2007 (H1N1) viruses produced the highest titers, with each reaching approximately 107.5 PFU/lung on day 2 postinfection. A/Brisbane/59/2007 (H1N1) virus yielded somewhat lower titers, with a peak of 106.7 PFU/lung on day 2 postinfection, while the H3N2 strain A/swine/Texas/4199-2/1998 grew to about 106 PFU/lung. The differences between the day 2 lung titers reached by the highly pathogenic viruses and those obtained by the A/Brisbane/59/2007 and A/swine/Texas/4199-2/1998 viruses in the DBA.2 mice were significant (P < 0.05); furthermore, these differences correlated with the virulence observed.
Fig. 1.
Human H1N1 and swine influenza viruses grow to high titers in DBA.2 and C57BL/6 mouse lungs. DBA.2 and C57BL/6 mice were infected intranasally with 103.7 PFU of the indicated human or swine influenza viruses in 50-μl inoculum volumes. Viral titers obtained in the homogenates of lungs harvested on day 2 (A) and day 4 (B) postinfection are shown. The experiment was performed once with three mice per group, and Student's t test was applied, with Welch's correction when appropriate, to ascertain the significance of the data. Abbreviated strain names are as follows: PR8, A/Puerto Rico/8/1934 (H1N1); Bris59, A/Brisbane/59/2007 (H1N1); swTx98, A/swine/Texas/4199-2/1998 (H1N1); Pan99, A/Panama/2007/1999 (H3N2); swKan07, A/swine/Kansas/77778/2007 (H1N1).
When the same experiment was carried out in C57BL/6 mice, very similar results were obtained, except that viral titers in C57BL/6 mice were up to 10-fold lower than those seen in DBA.2 mice. On day 2, the PR8, A/swine/Kansas/77778/2007, and A/swine/Texas/4199-2/1998 viruses grew to higher titers (P < 0.05) in DBA.2 mice than in C57BL/6 mice. The difference following infection with the A/Brisbane/59/2007 virus approached significance (P = 0.071). A similar trend was seen on day 4, when all viruses tested (with the exception of A/Panama/2007/1999) grew to higher titers in DBA.2 mice than in C57BL/6 mice (P < 0.05).
In the case of the C57BL/6 mice, two viruses that were not lethal did give moderately high lung titers: A/Brisbane/59/2007 (H1N1) and A/swine/Texas/4199-2/1998 (H3N2). These titers were, however, lower (P < 0.05) than lung titers of mice infected with the lethal virus PR8 or A/swine/Kansas/77778/2007, suggesting that a threshold level of viral growth must be reached to result in severe disease.
Viral-receptor-binding preference does not correlate with virulence.
We hypothesized that replication efficiency and, in turn, pathogenicity in the DBA.2 model might correlate with the receptor binding preference of the virus. Using a cell-based assay employing α-2,3-linked or α-2,6-linked N-acetyl neuraminic acid substrates obtained from the Consortium of Functional Glycomics (20), we determined the receptor binding preferences of a number of viruses (data not shown). We did not, however, observe any clear correlation between sialic acid binding specificity and virulence in DBA.2 mice.
C57BL/6 and DBA.2 mice mount similar humoral immune responses to infection with PR8 virus.
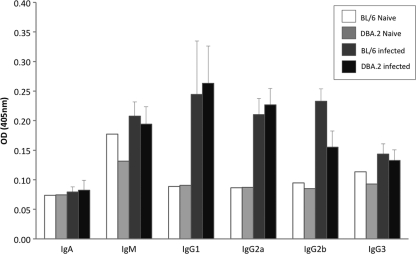
One aim of our study was to assess the utility of the DBA.2 mouse model for the testing of influenza vaccines. The neutralizing antibody response elicited by an influenza vaccine, as measured in the hemagglutination inhibition (HI) assay, is frequently used as a correlate of protection. Since one might argue that the increased susceptibility of DBA.2 mice to influenza disease is due to a defect in their immune response, it was important to test whether these animals would mount a humoral immune response following influenza virus infection. To this end, we infected groups of 10 DBA.2 and C57BL/6 mice with approximately 1 MLD50 of PR8 virus and monitored the infected animals for 2 weeks by taking daily weights. At 3 weeks postinfection, convalescent-phase sera were obtained and HI and serum antibody titers specific for influenza virus were measured. All surviving mice in each group were found to seroconvert (defined as a ≥4-fold increase in serum HI titer over that of a naïve mouse). Titers obtained in the HI assay ranged from 80 to 640 HI units for the C57BL/6 group (n = 4) and from 40 to 1,280 HI units for the DBA.2 group (n = 6). Geometric mean titers calculated for each group were not significantly different. Four surviving mice from each group that showed weight losses of between 10 and 25% were included in an analysis of influenza-specific antibody titers, as measured by enzyme-linked immunosorbent assay (ELISA). Titers were also found to be similar between strains, with elevated levels of influenza-specific IgM, IgG1, IgG2a, IgG2b, and IgG3 relative to those of naïve mice (Fig. 2). Thus, while a more comprehensive analysis of the immune response would further increase confidence in the utility of the DBA.2 model, both quantitatively and qualitatively, the humoral immune response to influenza virus infection was found to be similar in DBA.2 and C57BL/6 mice.
Fig. 2.
DBA.2 and C57BL/6 mice display similar influenza virus-specific antibody isotype profiles following sublethal infection. Sera were collected from mice 3 weeks following sublethal infection with PR8 virus, and ELISA was performed on plates coated with purified PR8 virus. Sera from infected mice (n = 4) were compared to that from a single naïve animal representing each mouse strain. Values shown are means, and error bars represent standard deviations. OD, optical density.
Conclusions.
Our study demonstrates that DBA.2 mice are susceptible to infection with a wide range of influenza A and B viruses. Our results are in agreement with those of Boon et al. and Srivastava et al., who previously reported the susceptibility of DBA.2 mice to infection with avian influenza A viruses (subtypes H2, H4 to H7, H9 to H10) and the mouse-adapted virus PR8, respectively (3, 23). Our data further extend these reports to show that influenza A viruses from human and porcine hosts, as well as influenza B viruses, are pathogenic in DBA.2 mice and that these mice are more susceptible to such infections than C57BL/6 mice. One limitation of the DBA.2 model was highlighted: in the case of the H3 subtype, mouse-adapted influenza virus variants are still needed in vaccine challenge tests. Nevertheless, the DBA.2 model presents a significant advantage over traditional mouse models in that it enables testing for heterologous protection against human H1 strains. This capability is highly desirable at a time when interest in cross-protective vaccines is increasing (13, 16, 18).
Acknowledgments
We thank the Consortium of Functional Glycomics (CFG) for providing reagents. A portion of the data will be published on the CFG website. We also thank the Flow Cytometry Shared Facility in the Mount Sinai School of Medicine.
John Steel was supported by a Career Development Fellowship from the Northeast Biodefense Center (U54-AI057158-Lipkin). N.M.B. was supported by an NIAID Mentored Clinical Scientist Research Career Development Award (5 K08 AI089940-02). This work was funded by the National Institutes of Health (NIH) (grant RC1 AI086061-01 to P.P.) and the NIH Center of Excellence for Influenza Research and Surveillance (CEIRS) (grant HHSN266200700010C to A.G.-S., P.P., and A.F.-S.).
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Alberts R., et al. 2010. Gene expression changes in the host response between resistant and susceptible inbred mouse strains after influenza A infection. Microbes Infect. 12:309–318 [DOI] [PubMed] [Google Scholar]
- 2. Boon A. C., et al. 2009. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J. Virol. 83:10417–10426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boon A. C., et al. 2010. Cross-reactive neutralizing antibodies directed against pandemic H1N1 2009 virus are protective in a highly sensitive DBA/2 mouse influenza model. J. Virol. 84:7662–7667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouvier N. M., Lowen A. C. 2010. Animal models for influenza virus pathogenesis and transmission. Viruses 2:1530–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DiMenna L. J., Ertl H. C. 2009. Pandemic influenza vaccines. Curr. Top. Microbiol. Immunol. 333:291–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dushoff J., Plotkin J. B., Viboud C., Earn D. J., Simonsen L. 2006. Mortality due to influenza in the United States—an annualized regression approach using multiple-cause mortality data. Am. J. Epidemiol. 163:181–187 [DOI] [PubMed] [Google Scholar]
- 7. Foppa I. M., Hossain M. M. 2008. Revised estimates of influenza-associated excess mortality, United States, 1995 through 2005. Emerg. Themes Epidemiol. 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Girard M. P., Tam J. S., Assossou O. M., Kieny M. P. 2010. The 2009 A (H1N1) influenza virus pandemic: a review. Vaccine 28:4895–4902 [DOI] [PubMed] [Google Scholar]
- 9. Hai R., et al. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hale B. G., et al. 2010. Mutations in the NS1 C-terminal tail do not enhance replication or virulence of the 2009 pandemic H1N1 influenza A virus. J. Gen. Virol. 91:1737–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson N. P., Mueller J. 2002. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76:105–115 [DOI] [PubMed] [Google Scholar]
- 12. Kashyap A. K., et al. 2010. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 6:e1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lambert L. C., Fauci A. S. 2010. Influenza vaccines for the future. New Engl. J. Med. 363:2036–2044 [DOI] [PubMed] [Google Scholar]
- 14. Memoli M. J., et al. 2009. An early ‘classical’ swine H1N1 influenza virus shows similar pathogenicity to the 1918 pandemic virus in ferrets and mice. Virology 393:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Minor P. D. 2010. Vaccines against seasonal and pandemic influenza and the implications of changes in substrates for virus production. Clin. Infect. Dis. 50:560–565 [DOI] [PubMed] [Google Scholar]
- 16. Nabel G. J., Fauci A. S. 2010. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat. Med. 16:1389–1391 [DOI] [PubMed] [Google Scholar]
- 17. Neumann G., Noda T., Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palese P. 2006. Making better influenza virus vaccines? Emerg. Infect. Dis. 12:61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pizzi M. 1950. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum. Biol. 22:151–190 [PubMed] [Google Scholar]
- 20. Ramos I., et al. 2011. Effects of receptor binding specificity of avian influenza virus on the human innate immune response. J. Virol. 85:4421–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith G. J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
- 22. Solórzano A., Ye J., Perez D. R. 2010. Alternative live-attenuated influenza vaccines based on modifications in the polymerase genes protect against epidemic and pandemic flu. J. Virol. 84:4587–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava B., et al. 2009. Host genetic background strongly influences the response to influenza A virus infections. PLoS One 4:e4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thakur A. K., Fezio W. L. 1981. A computer program for estimating LD50 and its confidence limits using modified Behrens-Reed-Muench cumulant method. Drug Chem. Toxicol. 4:297–305 [DOI] [PubMed] [Google Scholar]
- 25. Thompson W. W., et al. 2004. Influenza-associated hospitalizations in the United States. JAMA 292:1333–1340 [DOI] [PubMed] [Google Scholar]
- 26. Thompson W. W., et al. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186 [DOI] [PubMed] [Google Scholar]
- 27. Ye J., et al. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog. 6:e1001145. [DOI] [PMC free article] [PubMed] [Google Scholar]