Abstract
Rift Valley fever (RVF) is a mosquito-borne zoonotic disease caused by a phlebovirus of the family Bunyaviridae, which affects humans and ruminants in Africa and the Middle East. RFV virus (RVFV) possesses a single-stranded tripartite RNA genome of negative/ambisense polarity. The S segment utilizes the ambisense strategy and codes for two proteins, the N nucleoprotein and the nonstructural NSs protein, in opposite orientations. The two open reading frames (ORFs) are separated by an intergenic region (IGR) highly conserved among strains and containing a motif, 5′-GCUGC-3′, present on the genome and antigenome, which was shown previously to play a role in transcription termination (C. G. Albarino, B. H. Bird, and S. T. Nichol, J. Virol. 81:5246-5256, 2007; T. Ikegami, S. Won, C. J. Peters, and S. Makino, J. Virol. 81:8421-8438, 2007). Here, we created recombinant RVFVs with mutations or deletions in the IGR and showed that the substitution of the motif sequence by a series of five A's inactivated transcription termination at the wild-type site but allowed the transcriptase to recognize another site with the consensus sequence present in the opposite ORF. Similar situations were observed for mutants in which the motif was still present in the IGR but located close to the stop codon of the translated ORF, supporting a model in which transcription is coupled to translation and translocating ribosomes abrogate transcription termination. Our data also showed that the signal tolerated some sequence variations, since mutation into 5′-GCAGC-3′ was functional, and 5′-GUAGC-3′ is likely the signal for the termination of the 3′ end of the L mRNA.
INTRODUCTION
Rift Valley fever (RVF) is a mosquito-borne zoonotic viral disease affecting livestock and humans (34). The causative agent, RVF virus (RVFV), was first isolated in the 1930s during a severe outbreak affecting sheep and causing deaths and abortions (11). Initially confined to sub-Saharan regions of Africa, where periodic epidemics and epizootics have occurred, RVF spread to Egypt in 1977 and to the Middle East in 2000, representing a high risk to other regions (reviewed in references 23, 26, and 29). Following unusual climatic conditions, outbreaks have occurred between 2006 and 2008 in Kenya, Tanzania, and Sudan as well as in Madagascar and Mayotte, resulting in numerous human and animal deaths (2, 3, 24, 27, 33). In 2010, serious outbreaks occurred in South Africa and in Mauritania. RVFV is responsible for high fatality rates in sheep and cattle, and during outbreaks, various symptoms in humans, from mild febrile illness to fatal hemorrhagic fever, have been observed (for recent reviews, see references 9 and 29).
RVFV belongs to the genus Phlebovirus of the family Bunyaviridae (28). Like all the members of the family, RVFV possesses a single-stranded tripartite RNA genome of negative/ambisense polarity. The L and M segments code for the L protein and the precursor to the glycoproteins Gn and Gc, respectively, which generates two nonstructural proteins of 78 kDa and 14 kDa, respectively, after cleavage. The S segment utilizes an ambisense strategy and codes for two proteins of opposite polarities, the nucleoprotein N and the nonstructural NSs protein (15). The two open reading frames (ORFs) encoded by the S segment are separated by an intergenic region (IGR) highly conserved in sequence and composed of 81 to 85 nucleotides (nt) for most of the strains studied so far (8).
The virus replicates in many cell types, and after uncoating, the L, M, and S RNAs associated with the nucleoprotein and the polymerase in the form of RNPs are the templates for the synthesis of two types of cRNA molecules, the antigenomes and the mRNAs. The antigenomes serve as templates for the replication, leading to the amplification of the genome, whereas the mRNAs are translated into viral proteins. For the ambisense S segment, the S antigenome also serves as a template for the synthesis of the NSs mRNA. mRNAs have a 5′-capped terminal sequence of cellular origin acquired through a cap-snatching mechanism mediated by the L RNA polymerase, which possesses an endonuclease domain in its N-terminal region (31). In contrast, antigenomes have a 5′ triphosphate ribonucleotide end, which triggers the interferon response through the RIG-I activator (17). Antigenomes and mRNAs also differ at their 3′ ends: the antigenome represents the exact full-length copy of the genome, whereas mRNAs are incomplete transcripts terminating before the end of the template. Moreover, with the exception of the Sin Nombre hantavirus mRNA (18), bunyavirus mRNAs are not polyadenylated at their 3′ ends (28). These data suggest that the transcriptase recognizes a signal of transcription termination during mRNA synthesis but not during genome and antigenome syntheses.
The signals for transcription termination were identified only recently in bunyavirus genomes. In the case of Bunyamwera orthobunyavirus, a specific sequence, 5′-GCUGU-3′, within the 5′ untranslated region of the S segment is the signal for the termination of the bicistronic N/NSs mRNA, and such a sequence is present in the L segment. For orthobunyaviruses like Inkoo, La Crosse, Germiston, and snowshoe hare viruses, the motif exhibits a single-nucleotide deviation (5′-GCUGC-3′) (5). In the case of phleboviruses, the 3′ end of the M mRNA of RVFV was mapped by a nuclease protection assay and was found to terminate some 112 nucleotides before the 5′ end of the template (10). More recently, Albarino et al. (1) and Ikegami et al. (20) identified a signal of 6 to 8 nucleotides, 5′-(G/A)CUGC1–3-3′, containing the core sequence 5′-GCUGC-3′, which is conserved in the M and S segments of RVFV strains and several sandfly fever viruses. With regard to the termination in the L segment of RVFV, those two reports noted the absence of a consensus motif sequence in the 5′ noncoding region of this genome segment but did not agree on the identification of the mRNA termination signal. Albarino et al. showed that the L mRNA terminates like the antigenome as a runoff transcript, while Ikegami et al. found that the L mRNA terminates some 20 to 40 nucleotides before the 5′ end of the template in the vicinity of a stable stem structure formed by two complementary 13-nt sequences in the 5′ noncoding region.
Here, we have revisited the transcription termination in the RVFV L and S segments. For the L mRNA, we carried out 3′ rapid amplification of cDNA ends (RACE) analysis, cloned the PCR products, and sequenced individual clones, and for the S segment, we created recombinant RVFVs bearing mutations in their IGRs by reverse genetics and analyzed the 3′ ends of the viral mRNAs by 3′ RACE. Interestingly, we found that in cells infected with RVFV mutants altered within the transcription termination signal present in the IGR, the transcriptase continued to transcribe the template until it reached an upstream motif contained in the ORF with the opposite polarity. We observed a similar situation with mutant viruses in which the motif was present but close to the stop codon of the ORF contained in the transcribed mRNA. The failure of the transcriptase to recognize the wild-type (wt) motif allowed us to propose a model taking into account that transcription is coupled to translation in RVFV- and other bunyavirus-infected cells (4, 6, 21, 36). In addition, we found that although the conserved motif 5′-GCUGC-3′ plays a major role in transcription termination, in some circumstances induced by mutations in the IGR or naturally found in the L segment, a slightly variant sequence can also be recognized as a transcription termination signal.
MATERIALS AND METHODS
Cells and viruses.
Subconfluent monolayers of Vero E6 cells were infected with RVFV ZH548 or recombinant viruses at a multiplicity of infection (MOI) of 0.01 in Dulbecco's maintenance medium (Gibco-BRL) supplemented with 2% fetal calf serum and antibiotics. The medium was collected when a cytopathic effect was observed, at approximately 72 h postinfection (p.i.), and stored at −80°C after clarification. Virus was titrated by plaque assay by infecting monolayers of Vero E6 cells with serial dilutions (10−1 to 10−7) of virus suspensions. Cells were incubated under an agarose layer for 4 to 5 days at 37°C, and plaques were counted after staining with crystal violet. All the manipulations involving RVFV were performed in a biosafety level 3 (BSL-3) containment laboratory.
Baby hamster kidney cells stably expressing T7 RNA polymerase (BHK/T7-9 cells), kindly provided by Naoto Ito (Gifu, Japan) (22), were grown in minimal essential medium (MEM; Gibco-BRL) supplemented with 5% fetal calf serum, 1× tryptose phosphate broth, 10 IU of penicillin, and 10 μg of streptomycin per ml.
Plasmid construction.
For the construction of plasmids with mutated IGRs, pPolI-SZH548 was used as a template for the PCR amplification of two fragments overlapping in the IGR and representing the 5′ or the 3′ half of the genome. The primers annealing in the IGR contained the expected mutation(s). The two PCR products were hybridized with each other, elongated, and amplified into the full-length S segment by PCR using the 5′- and 3′-terminal primers Pol1mSag5′ and Pol1mSag3′, as described previously (7).
Plasmids pPolI-SZH548-IGR20ag and pPolI-SZH548-IGR25ag, in which the IGR in the S segment possesses only the first 20 and 25 nt of the IGR in the genomic sense, respectively, were constructed by PCR with primers IR20/25zh-g/Pol1mSag5′ and Pol1mSag3′/IR20/25zh-ag. Plasmid pPolI-SZH548-IGRpenta51-55, in which the 5′-GCUGC-3′ motif was duplicated at positions 51 to 55, was constructed with primer set Pol1mSag5′/5′heptaIRzhg3′ and Pol1mSag3′/3′heptaIRzhg5′. Primers Pol1mSag3′/pentaN:U-g and Pol1mSag5′/pentaN:U-ag were used to construct plasmid pPolI-SZH548PU9-13ag, in which the 5′-GCUGC-3′ motif at positions 9 to 13 of the IGR was mutated into 5 uridine residues. Similarly, primers Pol1mSag3′/pentaN:A-2g and Pol1mSag5′/pentaN:A-2ag were used to construct plasmid pPolI-SZH548-IGR-PA9-13ag, in which the 5′-GCUGC-3′ motif was replaced by a stretch of 5 adenine residues. Primers Pol1mSag3′/5PentaIRgmuT3 and Pol1mSag5′/3PentaIRgmuT5 allowed the construction of plasmid pPolI-SZH548-IGR-PU:A11ag, in which the U at position 11 of the IGR was mutated into A. All plasmids were sequenced to verify that they had the expected mutations.
Virus rescue.
Recombinant ZH548 viruses carrying mutations in the IGR sequence were rescued by cotransfecting subconfluent monolayers of BHK/T7-9 cells with 0.5 μg of pTM1-NMP12, 0.5 μg of pTM1-LMP12, 1 μg of pPolI-LZH548, 1 μg of pPolI-MZH548, and 1 μg of pPolI-SZH548- or pPolI-SZH548-derived plasmids by use of Fugene 6 transfecting agent (Roche) as described previously (7). After 4 to 5 days posttransfection, the culture supernatants were collected, clarified, and titrated. Viral stocks were produced by the infection of Vero E6 cells at a low MOI (0.01).
RNA extraction.
Vero E6 cells were infected with ZH548 or recombinant virus at an MOI of 3 to 5 and incubated for 6 h at 37°C, and total cellular RNAs were extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The aqueous phase was precipitated with propan-2-ol. The precipitate was centrifuged for 30 min at 17,000 × g, and after washing with 70% ethanol and drying, the pellet was dissolved in 30 μl of RNase-free distilled water.
Northern blotting.
Eight micrograms of total RNA was denatured in 10 mM MOPS (morpholinepropanesulfonic acid) buffer with 1 M glyoxal (Merck) and 50% dimethyl sulfoxide (DMSO). MOPS buffer contained 20 mM 3-(N-morpholino)propanesulfonic acid, 5 mM sodium acetate, and 1 mM EDTA at pH 7. The RNA samples were separated by electrophoresis at 50 V in 1.5% agarose gels in MOPS buffer. RNAs were transferred onto Hybond-N membranes (Amersham Pharmacia Biotec). Blotting was performed as described previously (14). Briefly, the N and NSs probes were synthesized in vitro from linearized plasmids using either T7 or SP6 RNA polymerase in the presence of [α-32P]CTP (3,000 Ci/mmol; Perkin-Elmer). After TRIzol extraction, the probes were purified by filtration through Sephadex Quick Spin columns (Roche).
3′ RACE.
Five micrograms of total RNA extracted from infected cells were used for polyadenylation in a 20-μl reaction mixture under the conditions of the poly(A) polymerase tailing kit (Epicentre Biotechnologies). Polyadenylation was carried out for 5 min at 37°C. RNA was purified and used for reverse transcriptase PCR (RT-PCR) reactions designed to detect the 3′ ends of the mRNAs. Reverse transcription was carried out with the oligo(dT) primer 3′RACE-AP (Invitrogen) and avian myeloblastosis virus (AMV) RT (Promega) and was followed by PCR using LATaq (Takara); 3′RACE-AP and Lzh6060ag (5′-CTATGATTGCTCATCC-3′), Pol1Sag5′/Nsca (5′-CCTTACCTCTAATCAAC-3′), or Pol1Sag3′/NSsg619-638 (5′-CGTTCGGCTTCTGCAAGCAGC-3′), specific for the L, N, or NSs mRNA, respectively.
The resulting RT-PCR products were purified by agarose electrophoresis. The DNA bands with the correct sizes were harvested and purified with the QIAquick purification kit (Qiagen). Purified DNA products were sequenced directly according to standard protocols or cloned into the pCRII-Topo plasmid by using the Topo-TA cloning kit (Invitrogen), and individual plasmids were sequenced.
RESULTS
Mutations in the 5′-GCUGC-3′ motif of the S segment of the IGR resulted in mRNA terminating at an alternate termination signal.
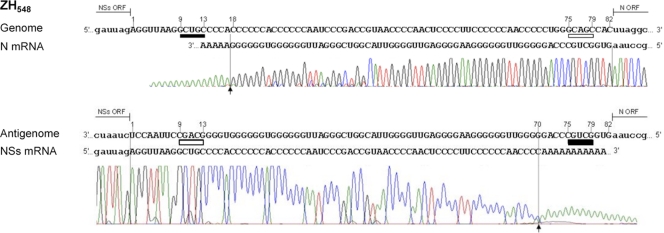
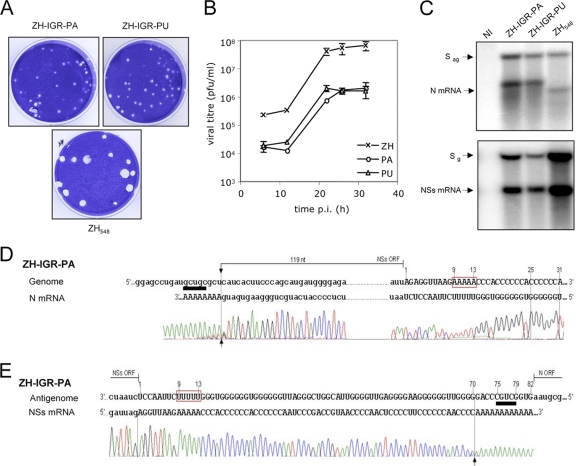
Since reverse genetics is a powerful tool to decipher functional roles of viral sequences, we made use of this methodology to create recombinant viruses carrying mutated IGR in the S segment using the ZH548-based plasmids described previously (7). The IGR sequence contains the 5′-GCUGC-3′ motif and its reverse complementary sequence 5′-GCAGC-3′, located at positions 9 to 13 and 75 to 79, respectively, on the genomic strand, with numbering of the IGR from positions 1 to 82 on the genome (Fig. 1). Characterization of the 3′ ends of the N and NSs mRNAs extracted from ZH548-infected Vero E6 cells was carried out by 3′ RACE after in vitro polyadenylation with poly(A) polymerase and amplification of the 3′-terminal region of the mRNA by RT-PCR using oligo(dT) and a primer specific for the viral sequence. The PCR products were then sequenced (Fig. 1). The 3′ end of the N mRNA was located 4 nucleotides upstream of the 5′-GCUGC-3′ sequence situated at positions 9 to 13 in the IGR of the genome template. The NSs mRNA terminated at positions 70 to 71, again some 4 to 5 nucleotides upstream of the 5′-GCUGC-3′ motif of the antigenome, which is the template for the synthesis of this mRNA. These termination sites correspond to the positions determined previously for the N and NSs mRNAs of other RVFV strains (ZH501 and MP12) (1, 20). In accordance with data described previously by Albarino et al., we noted that the transcriptase terminated very precisely at a major site visualized on the chromatogram as it preceded the poly(A) tract added in vitro. To ascertain the role of the 5′-GCUGC-3′ motif in transcription termination, we created a recombinant RVFV in which the 5′-GCUGC-3′ sequence in the IGR was mutated into a stretch of 5 A's or U's. These viruses, called ZH-IGR-PA and ZH-IGR-PU, were rescued and produced small plaques (Fig. 2A), and their growth kinetics were similarly affected compared to those of wt ZH548 (Fig. 2B): the mutants grew to titers approximately 100-fold lower than those of wt ZH548 (2 × 106 to 3 × 106 PFU per ml for each virus). Analysis of the mRNAs by Northern blotting showed that for both viruses, the N mRNA migrated more slowly than did the wt one (Fig. 2C, top). The larger size of the mRNA was confirmed by 3′ RACE, undertaken only for ZH-IGR-PA, which demonstrated that during N mRNA synthesis, the transcriptase traversed the IGR and continued transcribing up to the next 5′-GCUGC-3′ motif encountered within the NSs ORF at positions 706 to 710 (Fig. 2D). As for the wt virus, the termination occurred 3 to 4 nucleotides upstream of the signal. Thus, the N mRNA of this recombinant RVFV has an extension of 136 nt at its 3′ end compared to the wt one (see the schematic representation of the genomes, antigenomes, and mRNAs of the different mutants in Fig. 6). Interestingly, this sequence is encountered at several places in the genome and antigenome of the S segment, precisely at one location in the NSs ORF, and at several places in the N sequence (Table 1; also summarized in Fig. 6).
Fig. 1.
Mapping of the 3′ ends of RVFV N and NSs mRNAs of ZH548. Shown are data for 3′ RACE analysis of the N and NSs mRNAs of ZH548 expressed in cells collected at 6 h p.i. The sequences of the genome or antigenome template and the N or NSs mRNA sequences are aligned. The intergenic region (uppercase type) is identified from positions 1 to 82, and the motifs 5′-GCUGC-3′ and 5′-GCAGC-3′ are underlined in black and white, respectively. The N and NSs ORFs, including the stop codon, are represented by lowercase type. The arrows indicate the beginning of the poly(A) added in vitro.
Fig. 2.
The 5′-GCUGC-3′ motif in the IGR at positions 9 to 13 is responsible for N mRNA termination. (A) Plaques in Vero E6 cells formed by wild-type recombinant ZH548 and mutated ZH-IGR-PA and ZH-IGR-PU. (B) Viral growth in Vero cells infected by wt and mutant ZH at an MOI of 2. (C) N and NSs mRNAs from ZH548 and recombinant viruses mutated in the intergenic region of the S segment analyzed by Northern blotting. Total RNAs from Vero E6 cells uninfected or infected with recombinant viruses were harvested at 6 h p.i. and used for Northern blotting. The probes detected the N (left) or NSs (right) mRNA. NI, not infected; g, genome; ag, antigenome. (D and E) 3′ RACE analysis of the N (D) and NSs (E) mRNAs from Vero E6 cells infected with RVFV ZH-IGR-PA. The N and NSs ORFs are indicated by lowercase type, and the intergenic region is represented by uppercase type. The 3′ ends of the RNAs monitored by the beginning of the polyadenylation added in vitro are shown by arrows. The 5′-GCUGC-3′ motif and its complementary sequence are underlined in black and white, respectively. The mutations in the IGR are highlighted.
Fig. 6.
Schematic representation of the S segment of wild-type and mutated ZH viruses and their N and NSs mRNAs. The genome, antigenome, N, and NSs mRNAs of ZH548 and mutant RVFVs are represented. The 5′ cap structure of the viral mRNAs is indicated. The 5′-GCUGC-3′ and 5′-GCAGC-3′ motifs are located on the genome and antigenome, shown by black and white boxes. The distance between the transcription termination signal and the codon stop is shown. The overlapping sequences at the 3′ ends of N and NSs mRNAs are also indicated.
Table 1.
Positions of termination motifs in the RVFV ZH548 S segment
| Motifa | Positions or location in: |
|
|---|---|---|
| ZH548 S segment | ORF/IGR | |
| 5′-GCUGC-3′ | 706–710 | NSs |
| 841–845 | IGR | |
| 919–923 | N | |
| 943–947 | N | |
| 1045–1049 | N | |
| 1058–1062 | N | |
| 5′-GCAGC-3′ | 632–636 | NSs |
| 907–911 | IGR | |
| 955–959 | N | |
| 1072–1076 | N | |
| 1130–1134 | N | |
| 1327–1331 | N | |
| 1615–1619 | N | |
The positions refer to the locations in the genome orientation. Because the two motifs are reverse complementary, 5′-GCAGC-3′ will be read as the consensus motif on the complementary strand.
With regard to the NSs mRNA, the termination motif in the IGR of the antigenome was not modified. As expected, the Northern blot did not reveal any difference from wt ZH548 (Fig. 2C, bottom), and the 3′ end of the RNA molecule determined by 3′ RACE was the same as that of the wt virus (Fig. 2E).
Recombinant virus ZH-IGR20/25 expresses an N mRNA terminating within the NSs ORF but not at the wild-type motif.
The above-described results pointed out the essential role of the 5′-GCUGC-3′ motif for transcription termination but did not exclude that cis-acting sequences might also be important. To address this question, we attempted to create infectious RVFV with progressive deletions in the 3′-terminal region of the IGR, conserving the N mRNA termination motif at positions 9 to 13 preceded by the natural sequence (Fig. 3A). The recombinant viruses ZH-IGR20 to ZH-IGR25, in which the IGR possesses the first 20 to 25 bases with 1 nucleotide increment per virus, were all viable. However, they formed small plaques (only ZH-IGR20 and -IGR25 are shown in Fig. 3B) and were strongly attenuated in their growth throughout the infection period at a high or low MOI (Fig. 3C), reaching titers of 8.6 × 105 and 9.5 × 105 PFU per ml. mRNAs from Vero cells infected with these mutants were analyzed by Northern blotting and 3′ RACE. The N mRNA synthesized by all the recombinant viruses migrated as a major band slightly more slowly than did the N mRNA from ZH548 (Fig. 3D, top). Another minor and diffuse band migrating faster than the wt one was also visible. The 3′-terminal sequence of the N mRNA was determined for ZH-IGR20 and -IGR25. After the poly(A) addition and amplification of the 3′ end of the N mRNA by RT-PCR, the DNA product was found to migrate as a single band. The chromatograms revealed the position of the poly(A) addition upstream of the 5′-GCUGC-3′ motif within the NSs ORF (Fig. 3E), indicating that the transcriptase did not recognize the expected signal located at positions 9 to 13 but continued transcribing up to the next 5′-GCUGC-3′ motif encountered within the NSs ORF. The 3′ end of the mRNA was thus extended by 136 nt compared to the wt one, as it was for ZH-IGR-PA. The positions of the motifs recognized or ignored and the termination of the mRNAs are illustrated in Fig. 6. Again, the termination occurred 3 to 4 nucleotides upstream of the signal. The same results were obtained for both ZH-IGR20 and -IGR25 (Fig. 6). Most likely, the sequence data obtained here correspond to the major molecule visible in the Northern blot (Fig. 3D, top). However, it could be that the wt termination signal was occasionally recognized, leading to an mRNA with a shorter 3′ end, which was likely detected as the additional molecule migrating faster in the Northern blot. However, we were not able to amplify it by RT-PCR, probably because the molecule was present in limited amounts.
Fig. 3.
Rescue of recombinant ZH virus carrying a deleted IGR. (A) Schematic representation of the deletions in the S segment. NC, noncoding region. (B) Plaques in Vero E6 cells formed by wild-type recombinant ZH548 and mutant strains ZH-IGR25 and ZH-IGR20. (C) Viral growth in Vero cells infected by wt and mutant ZH strains at an MOI of 2. (D) Expression of N and NSs mRNAs from the wild-type virus and the recombinant virus carrying the deleted intergenic region. Total RNA from Vero E6 cells uninfected or infected with recombinant viruses ZH-IGR25 to -IGR20 were harvested at 6 h p.i. and used for Northern blotting. The probes detected the N (top) or NSs (bottom) mRNA. (E and F) 3′ RACE analysis of the N (E) and NSs (F) mRNAs from Vero E6 cells infected with RVFV ZH-IGR25 (top) and ZH-IGR20 (bottom). The N and NSs ORFs are indicated by lowercase type, and the intergenic region is represented by uppercase type. The 3′ ends of the RNAs monitored by the beginning of the polyadenylation added in vitro are shown by arrows. The 5′-GCUGC-3′ motif and its complementary sequence are underlined in black and white, respectively.
The NSs mRNA of recombinant ZH-IRG20/25 terminates within the N ORF.
We then exploited these mutant viruses to investigate the transcription termination of the NSs mRNA, since the wt termination motif was deleted. By Northern blot analysis, the NSs mRNA appeared as a predominant RNA molecule migrating more slowly than its wt counterpart (Fig. 3D, bottom). Other RNA molecules with a diffuse pattern were also visible in the Northern blot. However, 3′ RACE, which was carried out for ZH-IGR20 and ZH-IGR25, led to the synthesis of only one PCR product, probably representing the major band. Sequencing showed that for both mutated viruses, the NSs mRNA terminated some 3 nucleotides upstream of a 5′-GCUGC-3′ motif present at positions 1072 to 1076 on the antigenome template within the N ORF (Fig. 3F). Interestingly, this is the second 5′-GCUGC-3′ motif encountered by the L protein copying the N sequence on the antigenome, with the first one being located at positions 955 to 959 (Table 1 and see Fig. 6). Upon closer observation of the Northern blot (Fig. 3D, bottom), minor bands were visible, migrating faster or slightly slower, suggesting that transcription termination occurred at other sites, in particular at the first consensus sequence at positions 955 to 959 in the N ORF or even the one at positions 1130 to 1134, which would lead to the longest 3′ end (Table 1). The 3′ ends of these molecules could not be amplified.
Similar to the data obtained with the N mRNA of ZH-IGR-PA, the results upon the termination of the NSs mRNA showed that when the natural transcription termination motif is altered, the transcriptase transcribed until it encountered an upstream termination motif present in the opposite ORF. Thus, for these mutants, the transcriptase terminated before the end of the genome template (for N mRNA synthesis) or antigenome template (for NSs mRNA synthesis), recognizing a transcription termination signal present in the opposite ORF, which is normally nonfunctional, as it is never encountered by the transcriptase (see Fig. 6). Interestingly, such sequences exist in both ORFs, as if the signal for termination had to be rescued if the normal termination is deficient (Table 1).
The 5′-GCAGC-3′ motif is recognized as a termination signal in recombinant RVFV with a mutated IGR.
The mutant virus ZH-IGR-PA allowed us to ascertain that the 5′-GCUGC-3′ motif is the termination signal recognized in the IGR of the genomic and antigenomic templates. However, the data obtained with ZH-IGR20/25 upon the termination of the N mRNA raised the question of why the motif was not recognized by the transcriptase in the deleted IGR. In these viruses, the stop codon of the N ORF was located at a short distance (i.e., 7 or 12 nucleotides) from the transcription termination motif, compared to the wt situation, where the distance was 69 nucleotides. This strongly suggested to us that, as described previously for Bunyamwera virus (4), with transcription being coupled to translation (4, 6, 21, 36), the presence of ribosomes on the nascent mRNA interferes with transcription termination, and a minimal distance between the stop codon and the transcription termination signal might be required for the transcriptase to recognize the termination motif and detach from the template. For Bunyamwera virus, it was shown that the translocation of the ribosomes on the nascent mRNA abrogated transcription termination.
To address this issue, we created ZH-IGRpenta51-55, in which a second motif was introduced into the IGR upstream of the wt one, by mutating the existing sequence at positions 51 to 55 into 5′-GCUGC-3′ (see the IGR sequence in Fig. 4 and the schematic representation in Fig. 6). We chose this location because the preceding sequence has a strong similarity with the sequence preceding the wt motif. This upstream termination motif is located 28 nucleotides from the stop codon of the N ORF, compared to 69 nt in the wt virus.
Fig. 4.
Presence of alternate termination motifs in the S segment. (A) Plaques in Vero E6 cells formed by wild-type recombinant ZH548 and the ZH-IGRpenta51-55 and -U:A11 mutants. (B) Viral growth in Vero cells infected by wt and mutant ZH strains at an MOI of 2. (C) N and NSs mRNAs from ZH548 and recombinant viruses mutated in the intergenic region analyzed by Northern blotting. Total RNAs from Vero E6 cells uninfected or infected with recombinant viruses were harvested at 6 h p.i. and used for Northern blotting. The probes detected the N (left) or NSs (right) mRNA. (D and E) 3′ RACE analysis of the N (D) and NSs (E) mRNAs from Vero E6 cells infected with RVFV ZH-IGRpenta51-55 (top) and ZH-IGR-U:A11 (bottom). The N and NSs ORFs are indicated by lowercase type, and the intergenic region is represented by uppercase type. The 3′ ends of the RNAs monitored by the beginning of the polyadenylation added in vitro are shown by arrows. The 5′-GCUGC-3′ motif and its complementary sequence are underlined in black and white, respectively. The mutations are highlighted.
The recombinant virus rZH-IGRpenta51-55 was rescued, and its virus growth curve and titer reached at the plateau (2.5 × 107 PFU per ml) were similar to those obtained with wt ZH548 (Fig. 4B). The plaques were also similar to the wt ones (Fig. 4A). When analyzed by Northern blotting, the N and NSs mRNAs appeared to migrate like those expressed by the wt virus (Fig. 4C). The 3′ end of the N mRNA was determined by 3′ RACE. The chromatogram shown in Fig. 4D (top) indicated that the new motif was not recognized, and the N mRNA terminated like the wt one, 3 to 4 nucleotides upstream of the motif present at positions 9 to 13.
This mutation did not introduce a novel 5′-GCUGC-3′ motif in the antigenomic strand and therefore was not expected to affect the expression of NSs. Unexpectedly, the chromatogram of the NSs mRNA terminal sequence revealed that the 3′ end was located near position 48 in the IGR (Fig. 4E, top), just before the 5′-GCAGC-3′ sequence newly introduced into the antigenomic strand of this mutated template and corresponding to the sequence reverse complementary to the 5′-GCUGC-3′ motif introduced at positions 51 to 55 (see Fig. 6). The transcriptase recognized this signal as it did for the wt motif, stopping transcription 2 or 3 nucleotides before it encountered the motif.
To ascertain the role of the 5′-GCAGC-3′ motif as a transcription termination signal, we created a recombinant ZH strain in which the U of the termination motif at position 11 in the IGR was changed into A. The ZH-IGR-U:A11 virus was rescued, formed small plaques (Fig. 4A), and was severely affected in its growth kinetics (Fig. 4B). RNAs from cells infected with this virus were analyzed by Northern blotting and were shown to migrate like the wt ones (Fig. 4C). The 3′ end of the N mRNA of this mutant virus ZH-IGR-U:A11 was determined by 3′ RACE, which showed that the termination occurred as in the wt virus (Fig. 4D, bottom, and see also Fig. 6), indicating that the 5′-GCAGC-3′ motif was recognized exactly like the wt one.
Since the U-to-A mutation in the genome introduced a new “bona fide” motif in the antigenome template, we questioned whether this new sequence is utilized as a termination motif for the NSs mRNA or whether it is too close to the stop codon (9 nucleotides). The 3′ end of the NSs mRNA of this mutant ZH strain was determined by 3′ RACE, which showed that the newly introduced 5′-GCUGC-3′ motif was not recognized but that transcription terminated at the wt signal (Fig. 4E, bottom).
The variant motif 5′-GUAGC-3′ is likely functional for the termination of the L mRNA.
The motif 5′-GCUGC-3′ is conserved in the 3′ untranslated region of RVFV M and in the IGR of the S genome and antigenome (1, 10, 20) but is absent in the L segment. To clarify the mechanism of transcription termination in the L segment, we tried to determine the 3′ end of the L mRNA from ZH548-infected cells by using the 3′ RACE methodology. Since the L segment is less abundant in infected cells, cells were harvested at 10 h p.i. in order to let the mRNA accumulate. After in vitro polyadenylation, the 3′-terminal region of the L antigenome/mRNA was amplified by RT-PCR using oligo(dT) and a primer located within the L sequence. When analyzed by agarose gel electrophoresis, the amplified DNA migrated as a single band (not shown), which was eluted from the gel and sequenced. The chromatogram of the product clearly showed a sequence ending at the exact 3′-terminal 3′-UGUGUUUC… consensus sequence of the template followed by an added poly(A) tract (Fig. 5A). Moreover, one could also notice that another poly(A) sequence was entangled with the first one, starting at positions 28 to 29 from the 3′ end. This suggests the existence of two PCR products that could not be separated, one corresponding to the full-length L segment antigenome and the other one, 28 or 29 nucleotides shorter, representing the L mRNA. The termination occurred just one or two nucleotides upstream of a 5′-GUAGC-3′ sequence present on the L template, which exhibited similarities with the consensus motif sequence. Identical sequences were not found elsewhere in the genome, either in the IGR of the S segment or in the 5′ noncoding region of the M segment.
Fig. 5.
Mapping of the 3′ end of the L mRNA. (A) 3′ RACE analysis of L mRNA from ZH548-infected cells collected at 10 h p.i. The 5′ noncoding sequence of the L segment is shown in the genomic sense from positions 1 to 90. The chromatogram of the RT-PCR product corresponding to the 3′ ends of the L antigenome/mRNA is presented. The initial position of the internal in vitro polyadenylation sequence is indicated by an arrow at position 28. (B) Sequence obtained from individual PCR products after cloning into pCRII-Topo plasmids. The sequences of the antigenome were obtained from 37 plasmids. The poly(A) added in vitro is not shown. The sequences highlighted in red indicate possible transcription termination motifs. (C) Alignment of the sequences surrounding the transcription termination site in the IGR of the genome (g), antigenome (ag), N, NSs, M, and L ORFs.
To ascertain the existence of two L RNA molecules with different 3′ extremities, we cloned the PCR amplification products obtained after in vitro polyadenylation into the pCRII-Topo vector, and the individual sequences in the recombinant plasmids were determined (Fig. 5B). Among 45 plasmids harboring the L viral sequence, 37 terminated exactly at the phlebovirus consensus sequence 3′-UGUGUUU… and thus represented the L antigenome. Among the remaining 8 plasmids, 6 terminated at positions 27 to 30 from the 3′ end just upstream of the sequence 5′-GUAGC-3′. Two additional clones were also found to terminate at positions 71 and 94, upstream of a sequence resembling the transcription termination motif (Fig. 5B). These data confirmed the existence of the L mRNA ending before the 3′ end of the genome template and highlighted possible variations in the sequence motif recognized by the transcriptase.
DISCUSSION
Most of the cellular or viral mRNAs are polyadenylated at their 3′ ends through posttranscriptional modifications that contribute to nuclear export, translation initiation, and RNA stability (12). Among the negative-stranded RNA viruses, only the Arenaviridae and Bunyaviridae have no poly(A) at the 3′ ends of their mRNAs. For the bisegmented arenaviruses, transcription termination is not dependent on a sequence motif, but the viral transcriptase stops in the vicinity of a stem-loop structure present in the IGR sequences (25, 30). A similar mechanism seems to occur for the ambisense S segment of tospoviruses, which contains a hairpin structure (35). In this paper, we demonstrated that RVFV transcriptase recognizes a sequence motif as a terminator of transcription.
Transcription termination occurs at a 5′-GCUGC-3′ motif present in the IGR or within the opposite ORF.
Using recombinant viruses, we demonstrated that the transcription termination signal consists of a sequence, 5′-GCUGC-3′, which is recognized by the transcriptase at the end of the IGR in the S genome and antigenome. We also found that in some mutants in which the signal in the IGR is abrogated, the termination occurred at a similar motif present in the opposite ORF. Similar recombinant RVFVs were created previously by Albarino et al. (1), but those authors reported that the N or NSs mRNAs of recombinant viruses with a deletion of the transcription termination signals in the IGR were runoff transcripts, which is in contrast with our present data. It seems unlikely that the difference could be due to the strains used, because the alternate motifs are conserved. Instead, the conditions under which RT-PCR for 3′ RACE analysis was performed likely favored the synthesis of the full-length molecule. Similarly, Ikegami et al. (20) did not observe termination within the opposite ORF, but they utilized minigenomes rather than rescued recombinant viruses in their study. It is therefore worthwhile to report our initial experiments based on the use of minigenomes. Although the mRNAs expressed by a minigenome with a wt IGR reproduced exactly the transcription termination that occurs in RVFV-infected cells, the mRNAs expressed from minigenomes with a deleted IGR (S-CAT-IR 20/25) did not behave like those expressed by ZH-IGR20/25 carrying the same deletion. In this context, we found that the NSs mRNA terminated as a runoff transcript (not shown), which retrospectively was not surprising, since the chloramphenicol acetyl transferase (CAT) ORF does not contain such a motif sequence. The same applies to the minigenomes containing Renilla luciferase (20), as the sequence is also deprived of the termination motif.
The 5′-GCAGC-3′ sequence is an alternate transcription termination signal.
The termination of the NSs mRNA in ZH-IGRpenta51-55-infected cells revealed a novel motif, 5′-GCAGC-3′. This was confirmed by creating ZH-IGR-U:A11 and showing that the sequence is recognized by the transcriptase like the authentic one during the synthesis of the N mRNA. There is indirect evidence for the use of the 5′-GCAGC-3′ termination motif in the Punta Toro virus S segment (13, 19): although the mapping of the 3′ ends of the N and NSs mRNAs was not precisely established, the two mRNAs did not overlap, suggesting that the unique 5′-GCUGC-3′ motif in the genome (and 5′-GCAGC-3′ in the antigenome) located just at the tip of the stem-loop in the IGR is utilized as a terminator on the genome and antigenome templates.
Most of the mutations in the IGR affected virus growth, as most of the mutants produced small plaques and had reduced yields, especially for ZH-IGR20/25, in which the deletion reduced virus production by almost 100-fold. How does the deletion affect virus growth? A schematic representation of the N and NSs mRNAs as well as the genomes and antigenomes is presented in Fig. 6. A striking feature is that in most of the mutants, the 3′ noncoding region is extended compared to the wt ones. For instance, while the 3′ ends of wt N and NSs mRNAs overlap on 52 nucleotides, the overlap is increased to 185, 293, and 298 nucleotides in ZH-IGR-PA, ZH-IGR20, and ZH-IGR25, respectively. While this allows annealing between the two mRNAs and likely induces a higher level of stability, the 3′ ends, including part of the ORF (the N ORF for ZH-IGR20/25 or the NSs ORF for ZH-IGR-PA) are in a double-stranded form, which may impede the translation of the protein. If N (and possibly NSs) is produced in limited amounts, this could impact the virus yield. However, factors other than the overlap of the 3′ ends might be involved in attenuation, because ZH-IGR-U:A11 has overlapping 3′ ends, like wt ZH548, but is attenuated in its growth. Interestingly, in ZH-IGRpenta51-55, the two mRNAs overlap only on 30 nucleotides, and the virus is similar to the wt in its ability to replicate. It should be noted that N and NSs mRNAs ending in the opposite ORFs are not unusual for phleboviruses: they have been reported previously for Uukuniemi and Toscana viruses (16, 32).
In previous studies, the motif sequence was delineated to 5′-(G/A)CUGC1–3-3′ (1) or 5′-GCUGC-3′ (20). Here, we compared and aligned the motifs and flanking sequences present in the IGR or the opposite ORFs as well as in the L and M noncoding regions (Fig. 5C). While the nucleotides surrounding the motif are variable, our data indicated that the pentanucleotide 5′-GCUGC-3′ tolerates some variation at position 3, with U being replaced by A (“A” version), and in the L segment, the “A” version of the motif contains an additional change at position 2, where C can be replaced by U. Similar variations occur among other phleboviruses in which the 5′-GCUGC-3′ signal is not strictly conserved but could be 5′-ACUGC-3′ in Toscana virus or Uukuniemi virus (1). Altogether, these data show that transcription termination is conserved not only among phleboviruses but also among orthobunyaviruses, which utilize 5′-GCUGC-3′ or some variants as a transcription termination signal (5).
Coupling of transcription and translation affects transcription termination.
The transcription termination of the N mRNAs of ZH-IGR20/25 indicated that the first termination signal encountered in the IGR was not recognized. A logical hypothesis could be that additional elements, such as the stretches of C's or G's preceding the motif, are required for termination, in addition to the consensus motif. However, our data showed that termination could occur at motifs which are not preceded by G's or C's: the recombinant viruses ZH-IGR20 and -IGR25 terminated N or NSs mRNA synthesis within the opposite ORFs, which are not G or C rich and have no homology with each other. This strongly suggested to us that the sequence preceding the signal is not crucial for the transcriptase to terminate.
It has been known for a long time that the efficient transcription of bunyaviruses must be coupled to translation (4, 6, 21, 36). Although the molecular basis of the mechanism remains unknown, early work on the in vitro transcription of Germiston virus showed that the viral transcriptase produced runoff transcripts in the presence of ribosomes and edeine, an inhibitor of protein synthesis which prevents ribosomal complex formation but allows the scanning of the 40S subunit until the 3′ end of the mRNA (36). These data strongly suggested that the presence of 40S ribosomal subunits prevents transcription termination and, reciprocally, that the transcription termination signal is recognized only in regions depleted of ribosomes. This would explain why the transcription termination sequences present in the ORFs (Table 1) are normally not functional and that only incomplete transcripts terminating at spurious sites are synthesized in the presence of inhibitors of translation such as cycloheximide or puromycin (21; E. Lara and M. Bouloy, unpublished data).
More recently, Barr proposed that the movement of ribosomes disrupts RNA-RNA interactions between the nascent strand and the template and prevents the transcription termination signals from functioning (4). The signaling ability of the motif was affected both by the presence of an in-frame stop codon and by the distance between the termination signal and the stop codon, with a too-short distance leading to inefficient transcription termination. In accordance with this hypothesis, the 3′ ends of all the mRNAs in phlebovirus-infected cells are relatively long, varying from 55 to 56 nt for Toscana virus M and NSs mRNAs to approximately 220 nt for Punta Toro virus N mRNA.
Mechanistically, two nonexclusive models can be elaborated. It could be that close interactions exist between ribosomes and transcriptase. Therefore, the termination motifs encountered within the ORFs or too close to the stop codon were ignored because of the synergetic effect of the ribosome pushing the polymerase. Under normal conditions, the constraint existing between the ribosomes and the transcriptase downstream of the stop codon is alleviated, since the ribosomes have been downloaded. However, a minimal distance between the stop codon and the transcription termination signal might be required for the transcriptase to slow down and for ribosomes to detach from their mRNAs. According to the second model, similar to the one proposed by Barr (4), the RNA interactions between the nascent strand and the template (usually disrupted by the scanning ribosomes in the ORF) must be stable enough to prevent the polymerase from moving further; the longer the distance between the stop codon and the signal, the more stable the interaction. Figure 6 represents the location of the 3′ ends of the N and NSs mRNAs and the consensus motifs (“U or A version”) encountered and recognized or ignored on the genome or antigenome template. Clearly, a distance of 9, 7, or 12 nucleotides between the first motif encountered (and not recognized) and the stop codon of the ZH-IGR-U:A11 NSs mRNA or the ZH-IGR20 or ZH-IGR25 N mRNA, respectively, did not allow the termination of the NSs mRNA (motif in the “A” version) or the N mRNA (motif in the “U” version). Similarly, the alternate motif 5′-GCAGC-3′ seems also to function only when it is away from the stop codon of the translated ORF and the termination signal. As examples, the 5′-GCAGC-3′ sequences encountered at positions 75 to 79 on the genome template during N mRNA synthesis, located 3 nucleotides from the N stop codon or encountered at positions 9 to 13 on the antigenome during NSs mRNA synthesis, are not recognized naturally. It is difficult to determine what should be the minimal distance between the stop codon and the termination motif because the context of the sequence upstream of the motif may have an effect on the rate of the polymerase or the stability of the RNA interactions. It may be shorter when termination occurs in the IGR, where the C's or G's may slow down the polymerase or induce more stable interactions, than those with termination occurring in the N or NSs ORF. In wt ZH548, the distances for wt N and NSs mRNAs are 69 and 75 nucleotides, respectively, between the stop codon and the signal in the IGR. For the mutants, the shortest distance that is functional with a version “A” termination motif in the IGR is 51 nucleotides for the NSs mRNA of ZH-IGRpenta51-55. In contrast, the version “U” motif located in the N ORF at positions 955 to 959 and 60 or 65 nucleotides distant from the stop codon of the NSs mRNA of ZH-IGR20 or ZH-IGR25, respectively, was not recognized, but termination occurred at positions 1072 to 1076, which are 177 or 182 nt away from the NSs stop codon, respectively. The version “A” motif was encountered, but not recognized, at four positions by the transcriptase transiting within the N ORF during the synthesis of the NSs mRNA of ZH-IGR20/25 (Fig. 6). The distance between the stop codon is, however, comparable to what was observed with the wt motif 5′-GCUGC-3′, suggesting that the 5′-GCAGC-3′ motif may not be as efficient as the consensus one.
An intriguing question is how the transcriptase senses this upstream motif and stops elongation before it is transcribed. It could be that the polymerase, being oligomeric (37), occupies a rather large volume and that the downstream signal sequence is recognized by the closest monomer or the oligomer, leading to its release from the template. The exact molecular mechanism and the mode of recognition of the motif by the transcriptase remain undetermined and could be a subject for further work.
ACKNOWLEDGMENTS
We are thankful to Marie Flamand for critical reading of the manuscript and fruitful comments. We gratefully acknowledge excellent technical assistance of Carole Tamietti all along during this study.
E.L. was a recipient of a fellowship from la Délégation à l'Armement (DGA) and the Pasteur Institute (bourse Weizmann). This work was supported by funds from the NIH (grant 7-U01-AI66327), from the Agence Nationale de la Recherche (ANR-08-MIE-022), and from the Pasteur Institute.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Albarino C. G., Bird B. H., Nichol S. T. 2007. A shared transcription termination signal on negative and ambisense RNA genome segments of Rift Valley fever, sandfly fever Sicilian, and Toscana viruses. J. Virol. 81:5246–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andriamandimby S. F., et al. 2010. Rift Valley fever during rainy seasons, Madagascar, 2008 and 2009. Emerg. Infect. Dis. 16:963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anyangu A. S., et al. 2010. Risk factors for severe Rift Valley fever infection in Kenya, 2007. Am. J. Trop. Med. Hyg. 83:14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barr J. N. 2007. Bunyavirus mRNA synthesis is coupled to translation to prevent premature transcription termination. RNA 13:731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barr J. N., Rodgers J. W., Wertz G. W. 2006. Identification of the Bunyamwera bunyavirus transcription termination signal. J. Gen. Virol. 87:189–198 [DOI] [PubMed] [Google Scholar]
- 6. Bellocq C., Kolakofsky D. 1987. Translational requirement for La Crosse virus S-mRNA synthesis: a possible mechanism. J. Virol. 61:3960–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Billecocq A., et al. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bird B. H., Khristova M. L., Rollin P. E., Ksiazek T. G., Nichol S. T. 2007. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J. Virol. 81:2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bouloy M., Flick R. 2009. Reverse genetics technology for Rift Valley fever virus: current and future applications for the development of therapeutics and vaccines. Antiviral Res. 84:101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collett M. S. 1986. Messenger RNA of the M segment RNA of Rift Valley fever virus. Virology 151:151–156 [DOI] [PubMed] [Google Scholar]
- 11. Daubney R. J., Hudson J. R., Garnham P. C. 1931. Enzootic hepatitis of Rift Valley fever. An underdescribed virus disease of sheep, cattle and man from East Africa. J. Pathol. Bacteriol. 34:545–579 [Google Scholar]
- 12. Edmonds M. 2002. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 71:285–389 [DOI] [PubMed] [Google Scholar]
- 13. Emery V. C., Bishop D. H. L. 1987. Characterization of Punta Toro S mRNA species and identification of an inverted complementary sequence in the intergenic region of Punta Toro phlebovirus ambisense S RNA that is involved in mRNA transcription termination. Virology 156:1–11 [DOI] [PubMed] [Google Scholar]
- 14. Gauliard N., Billecocq A., Flick R., Bouloy M. 2006. Rift Valley fever virus noncoding regions of L, M and S segments regulate RNA synthesis. Virology 351:170–179 [DOI] [PubMed] [Google Scholar]
- 15. Giorgi C., et al. 1991. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology 180:738–753 [DOI] [PubMed] [Google Scholar]
- 16. Gro M. C., Di Bonito P., Accardi L., Giorgi C. 1992. Analysis of 3′ and 5′ ends of N and NSs messenger RNAs of Toscana phlebovirus. Virology 191:435–438 [DOI] [PubMed] [Google Scholar]
- 17. Habjan M., et al. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hutchinson K. L., Peters C. J., Nichol S. T. 1996. Sin Nombre virus mRNA synthesis. Virology 224:139–149 [DOI] [PubMed] [Google Scholar]
- 19. Ihara T., Matsuura Y., Bishop D. H. 1985. Analyses of the mRNA transcription processes of Punta Toro phlebovirus (Bunyaviridae). Virology 147:317–325 [DOI] [PubMed] [Google Scholar]
- 20. Ikegami T., Won S., Peters C. J., Makino S. 2007. Characterization of Rift Valley fever virus transcriptional terminations. J. Virol. 81:8421–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ikegami T., Won S., Peters C. J., Makino S. 2005. Rift Valley fever virus NSs mRNA is transcribed from an incoming anti-viral-sense S RNA segment. J. Virol. 79:12106–12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito N., et al. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47:613–617 [DOI] [PubMed] [Google Scholar]
- 23. LaBeaud A. D., Kazura J. W., King C. H. 2010. Advances in Rift Valley fever research: insights for disease prevention. Curr. Opin. Infect. Dis. 23:403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LaBeaud A. D., et al. 2008. Interepidemic Rift Valley fever virus seropositivity, northeastern Kenya. Emerg. Infect. Dis. 14:1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lopez N., Franze-Fernandez M. T. 2007. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res. 124:237–244 [DOI] [PubMed] [Google Scholar]
- 26. Meegan J. M., Bailey C. J. 1988. Rift Valley fever, vol. IV CRC Press Inc., Boca Raton, FL [Google Scholar]
- 27. Mohamed M., et al. 2010. Epidemiologic and clinical aspects of a Rift Valley fever outbreak in humans in Tanzania, 2007. Am. J. Trop. Med. Hyg. 83:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nichol S. T., et al. 2005. The Bunyaviridae, p. 695–716 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 29. Pepin M., Bouloy M., Bird B. H., Kemp A., Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 41:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pinschewer D. D., Perez M., de la Torre J. C. 2005. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J. Virol. 79:4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reguera J., Weber F., Cusack S. 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 6:e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simons J. F., Pettersson R. F. 1991. Host-derived 5′ ends and overlapping complementary 3′ ends of the two mRNAs transcribed from the ambisense S segment of Uukuniemi virus. J. Virol. 65:4741–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sissoko D., et al. 2009. Rift Valley fever, Mayotte, 2007-2008. Emerg. Infect. Dis. 15:568–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swanepoel R., Coetzer J. A. W. 2004. Rift Valley fever, p. 1037–1070 In Coetzer J., Tustin R. (ed.), Infectious diseases of livestock, vol. 2 Oxford University Press, Cape Town, South Africa [Google Scholar]
- 35. van Knippenberg I., Goldbach R., Kormelink R. 2005. Tomato spotted wilt virus S-segment mRNAs have overlapping 3′-ends containing a predicted stem-loop structure and conserved sequence motif. Virus Res. 110:125–131 [DOI] [PubMed] [Google Scholar]
- 36. Vialat P., Bouloy M. 1992. Germiston virus transcriptase requires active 40S ribosomal subunits and utilizes capped cellular RNAs. J. Virol. 66:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zamoto-Niikura A., Terasaki K., Ikegami T., Peters C. J., Makino S. 2009. Rift Valley fever virus L protein forms a biologically active oligomer. J. Virol. 83:12779–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]