Abstract
The trimeric envelope glycoprotein (Env) spikes displayed on the surfaces of simian immunodeficiency virus (SIV) and human immunodeficiency virus type 1 (HIV-1) virions are composed of three heterodimers of the viral glycoproteins gp120 and gp41. Although binding of gp120 to cell surface CD4 and a chemokine receptor is known to elicit conformational changes in gp120 and gp41, changes in quaternary structure of the trimer have only recently been elucidated. For the HIV-1 BaL isolate, CD4 attachment results in a striking rearrangement of the trimer from a “closed” to an “open” conformation. The effect of CD4 on SIV trimers, however, has not been described. Using cryo-electron tomography, we have now determined molecular architectures of the soluble CD4 (sCD4)-bound states of SIV Env trimers for three different strains (SIVmneE11S, SIVmac239, and SIV CP-MAC). In marked contrast to HIV-1 BaL, SIVmneE11S and SIVmac239 Env showed only minor conformational changes following sCD4 binding. In SIV CP-MAC, where trimeric Env displays a constitutively “open” conformation similar to that seen for HIV-1 BaL Env in the sCD4-complexed state, we show that there are no significant further changes in conformation upon the binding of either sCD4 or 7D3 antibody. The density maps also show that 7D3 and 17b antibodies target epitopes on gp120 that are on opposites sides of the coreceptor binding site. These results provide new insights into the structural diversity of SIV Env and show that there are strain-dependent variations in the orientation of sCD4 bound to trimeric SIV Env.
INTRODUCTION
The discovery that simian immunodeficiency virus (SIV) causes an AIDS-like disease in primates has provided a relevant animal model to study human immunodeficiency virus type 1 (HIV-1) and progression to AIDS (12, 37). SIV and HIV-1 are similar in many of their biological properties and display significant similarities in sequence and viral genome organization (30). SIV infection also results in many of the key pathogenic manifestations of HIV, including CD4+ T-cell depletion, opportunistic infections, and encephalopathy (7, 12, 15, 37, 40, 50). The rhesus macaque (Macaca mulatta), which is commonly employed for modeling AIDS pathogenesis because of its ease of availability and shorter time course for disease onset, provides a good model for the study of HIV-1 pathogenesis under relatively well-controlled experimental conditions (13, 15, 30).
HIV-1 and SIV utilize surface envelope glycoproteins (Env) to enter target cells by binding cellular surface receptors, CD4, and CCR5/CXCR4 (11, 14, 21, 49, 57). On virions, the envelope glycoproteins are found in trimers composed of heterodimers of the viral transmembrane glycoprotein (gp41) and surface glycoprotein (gp120) (59). Macaque SIV and HIV-1 envelope glycoproteins have ∼35% sequence identity and 70% similarity with disulfide bonds in gp120 and gp41, which are largely conserved (2, 6). Viral entry by both SIV and HIV-1 is thought to occur by binding of gp120 to cellular CD4 and coreceptor proteins, followed by exposure of the gp41 fusion peptide domain, thereby initiating fusion between viral and cellular membranes (22).
Previously, molecular architectures of trimeric Env from both HIV-1 and SIV were determined by using cryo-electron tomography (38, 58). These studies showed that the overall molecular architectures of trimeric Env displayed on HIV-1 BaL, SIVmac239, and SIVmneE11S are similar, revealing a propeller-shaped quaternary structure with gp120 blades connected at the apex (V1/V2) and base (gp41) (58). Despite these structural similarities between HIV-1 and SIV Env, numerous studies highlight functional differences in Env upon binding CD4. Striking differences between HIV-1 and SIV have been reported in terms of their sensitivity to neutralization by soluble CD4 (sCD4). Functional studies have shown that sCD4 binding to SIV Env results in a stable, activated state (53) with limited exposure to the gp41 N-terminal heptad repeat bound by C34 (23). In contrast, results reporting the effects of incubating sCD4 with HIV-1 Env vary widely, ranging from activation to neutralization, inactivation, and gp120 shedding (5, 10, 26, 27, 41–44, 51, 55). Cryo-electron tomographic structural studies have begun to also provide insights into quaternary structural changes in trimeric Env that occur with CD4 binding (38, 58). Upon binding to sCD4 and the coreceptor binding site antibody 17b, a large conformational change is observed for HIV-1 BaL Env in which each of the gp120 protomers is reoriented leading to an opening at the apex of the spike. Interestingly, trimeric Env displayed on SIV CP-MAC, a strain that can mediate viral entry into target cells even in the absence of cell surface CD4, is already in the same open conformation even in the absence of sCD4 binding (58).
As binding CD4 is a necessary step for viral entry in both wild-type HIV and SIV, we have carried out cryo-electron tomographic experiments aimed at characterizing the sCD4-bound states of trimeric Env displayed on different SIV strains. These strains differ in neutralization sensitivity, onset of pathogenic manifestations, and antigenicity. SIVmac239 is highly resistant to neutralization, with infection typically resulting in rapid progression to simian AIDS (29). In contrast, SIV CP-MAC is highly antigenic and neutralization sensitive (18, 36), while SIVmneE11S is thought to lie somewhere in between these two extremes (S. L. Hu and J. Overbaugh, personal communication). In addition, to obtain insights into the possible diversity in the binding of neutralizing antibodies that bind gp120 at the coreceptor binding site, we investigated the structure of SIV CP-MAC bound to 7D3, a potent neutralizing antibody that does not affect CD4 binding to the virus but instead prevents the interaction of Env with coreceptor CCR5 (18).
MATERIALS AND METHODS
Sample preparation.
SIVmneE11S and SIVmac239 viruses were produced by infection of Hut78 and SupT1 cells, respectively, treated with 2,2′dithiodipyridine (Aldrithiol-2 [AT-2]) and purified by sucrose gradient centrifugation to obtain concentrated preparations (typically ∼1011 virions/ml) that retained functionally intact Env as previously described (58). Human soluble CD4 protein containing only the first two domains (sCD4-183) was obtained from the NIH AIDS Reference and Research Reagent Program. sCD4 was added at a concentration of 30 μM and incubated with virus for a minimum of 30 min at 4°C, unless stated otherwise. Protein A gold particles (10 nm) (CMC-UMC, Utrecht, The Netherlands) were mixed in with all samples to provide fiducial markers for tilt-series alignment. 7D3 (4 μM) was purified as described previously (58) and incubated for 30 min at 4°C with virus and sCD4 (30 μM). Holey carbon-coated Multi A 200 mesh grids for electron microscopy were used from Quantifoil GmbH (Jena, Germany) and glow discharged immediately prior to specimen preparation. Samples were deposited on the grids at room temperature and transferred to the chamber of a Mark III Vitrobot (FEI Company, OR) maintained at 25°C and 100% humidity. Grids were then blotted for 6 s and plunged into an ethane slurry cooled by liquid nitrogen.
Data collection and three-dimensional (3D) reconstruction.
Tilt series were collected on an energy-filtered (GIF 2002; Gatan, Inc.) Tecnai G2 Polara transmission electron microscope (FEI, The Netherlands) operated at 200 kV, with an extraction voltage of 3.95 kV, a gun lens of 5, and a nominal magnification setting of 34,000× resulting in a pixel size of 4.1 Å at the specimen plane, with aperture settings of 100 and 70 μm for the objective and condenser 2, respectively. The settings for the energy filter were set for a slit width of 20 eV. The specimen was maintained at −193°C, and the average ice thickness was estimated to be between 100 and 200 nm. Data collection was carried out over a tilt range spanning ±60 to 65° by using a linear tilt scheme with tilt increments ranging from 1 to 2° and a nominal underfocus of 2.5 μm. Samples were imaged by using a dose distribution factor of 1.5 (i.e., ratio of dose between images at zero tilt angle and the highest tilt angle), typically at doses of ∼1 to 2 el/Å2 for each image. Tilt series were aligned by using fiducial based alignment as implemented in IMOD (34), and tomograms were reconstructed by using R-weighted back projection. The number of tomograms utilized to obtain the maps for the CD4-bound complexes of SIVmneE11S, SIVmac239, 7D3-bound SIVmneE11S, CP-MAC, and 7D3-bound CP-MAC were 69, 43, 29, 61, and 54, respectively. Virion centroids were identified manually, and subtomograms (480 by 480 by 480 voxels) containing only the virions were selected. The numbers of virions utilized to obtain the maps for the CD4-bound complexes of SIVmneE11S, SIVmac239, 7D3-bound SIVmneE11S, CP-MAC, and 7D3-bound CP-MAC were 354, 426, 329, 217, and 308, respectively.
Particle picking.
For purposes of the selection of Env spikes, virion subtomograms were down-sampled by a factor of 4, denoised by using edge-enhancing anisotropic diffusion as implemented in IMOD, and subjected to unsupervised membrane segmentation by using an energy-based 3D approach (3). In order to identify the locations of spikes in an automated manner, a scalar value was associated with every point on the segmented virion surface corresponding to the cross-correlation value between an external 3D template and the image data immediately outside the membrane. Spikes were identified at the locations corresponding to the local maxima of this function that were above a given threshold. A cylindrically symmetric phantom was used as a template for the search; the same template was used for all maps presented here and for those presented in reference 58.
Classification and averaging.
Subvolumes (100 by 100 by 100 voxels) corresponding to reconstructions of individual spikes (without denoising or binning) were cut from the virion subtomograms at the automatically extracted positions. The orientations of the long axes of the spikes were determined by using the normal to the automatically segmented membrane at the location of each spike, providing initial estimates for two of the three Euler angles. The remaining in-plane rotation was initially randomized to prevent any possible bias in subsequent alignments. After application of the Euler angles, subvolumes were translationally aligned to their cylindrically averaged global average to ensure they all shared the same center of mass. The 10% of subvolumes that correlated most poorly with the updated global average were left out of subsequent steps in data analysis. Spike volumes were aligned and classified without using external references and with proper accounting of the missing wedge by using the framework described in the work of Bartesaghi et al. (4). Subvolume alignments were progressively refined at each iteration and spike volumes repeatedly clustered into 10 classes. Early stages of classification clearly showed classes with inherent 3-fold symmetry, and typically at the fourth iteration, 3-fold symmetry was imposed. At each round, the classes that showed the most clearly delineated features in all regions of the spike (typically ∼50 to 60%) were selected and combined to be used as reference for the next round. The number of Env subvolumes utilized to obtain the maps for the CD4-bound complexes of SIVmneE11S, SIVmac239, 7D3-bound SIVmneE11S, CP-MAC, and 7D3-bound CP-MAC were 4,826, 4,561, 930, 1,844, and 2,808 spikes, respectively. Final maps were obtained after ∼5 to 12 refinement rounds and included contributions from ∼50% of subvolumes in each data set. The resolutions of the maps were determined from plots of the frequency dependence of the Fourier shell coefficient between randomly selected halves of the data and were ∼25 Å in each case.
Coordinate fitting.
Steepest-ascent local optimization, as implemented in UCSF Chimera, was utilized for fitting coordinates into density maps (45). Coordinates were initially placed in random orientations, and local maxima of the sum of pointwise products between the coordinates and the map were determined. Fitting was performed to convergence by performing multiples of 100 steepest-ascent steps. Atomic coordinates were fit by generating a map simulated from the atomic coordinates at 20 Å. The fits shown below (see Fig. 3) were carried out by using 1GC1 coordinates. These coordinates contain a truncated version (residues 119 to 129 and 194 to 202) of the V1/V2 loop region (spanning residues 119 to 202). To eliminate any bias in the fits from the inclusion of the partial complement of loop residues, loop residues were excluded for purposes of coordinate fitting, although their inclusion did not alter the fits significantly. The use of deposited gp120 X-ray coordinates resulting from other ternary complexes with CD4 and antibodies also did not result in noticeably different fits. However, the 2BF1 coordinates for unliganded, monomeric SIV gp120 did not fit the SIV maps, as in the case of the HIV-1 BaL density map described previously (38, 58). For fitting Env with 7D3 density, 17b Fab X-ray coordinates (heavy and light chains from 1GC1.pdb) were used to represent 7D3. Difference density maps (see next section) were calculated by subtracting sCD4-bound SIV CP-MAC map from the 7D3/sCD4-bound CP-MAC. 7D3 (represented by 17b Fab X-ray coordinates) was placed into the difference density map corresponding to 7D3 and then further refined by using steepest-ascent local optimization as described above.
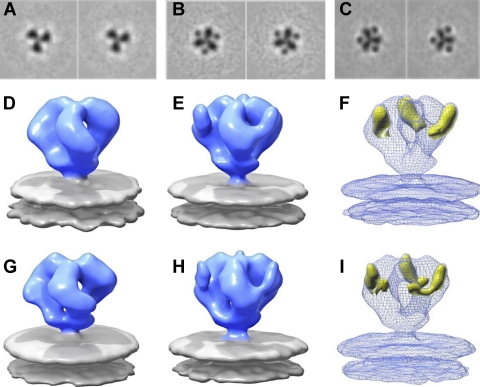
Fig. 3.
Coordinate fits of gp120 and sCD4 into native and sCD4-bound SIV Env maps. (A and B) Perspective view of native SIVmneE11S (A) and SIVmac239 (B) Env fit with gp120 coordinates (1GC1.pdb) (58). Env is displayed as a transparent blue isosurface, with gp120 coordinates shown as gray space-filling model with V1/V2, V3, and sCD4-binding site shown in red, green, and yellow, respectively. Red spheres indicate the locations of the V1/V2 loops missing from the coordinates. (C and D) Rigid body fitting with X-ray coordinate subset containing gp120 and sCD4 coordinates (PDB ID:1GC1) into sCD4-bound SIVmneE11S (C) and SIVmac239 (D) Env density maps. Env and gp120 are displayed as in A and B, with sCD4 shown in yellow ribbons. (E and F) Top views of the density maps and coordinate fits shown as in C and D for sCD4-bound SIVmne E11S (E) and SIVmac239 (F) Env, respectively. The small opening at the apex of the trimer can be visualized by the displacement in the estimated portions of the V1/V2 loops (red spheres) compared to the native state.
Difference maps.
Densities corresponding to the Env component of each map were masked in order to exclude any contributions from the membrane and background regions in the analysis. Next, the isolated spike densities were normalized and difference maps were calculated in USCF Chimera (45). The native maps were subtracted from the sCD4-bound maps, and difference maps were thresholded to show the greatest positive density differences, corresponding to the likely location of bound sCD4, with the difference density peak threshold enclosing a volume of ∼32,530 Å3 or 26 kDa, which is similar in mass to that reported for sCD4-183 (∼26 kDa) (24, 39). The volume of negative difference density peaks was <1% of the positive peaks.
RESULTS
Structural analysis of sCD4-bound Env on SIVmneE11S and SIVmac239.
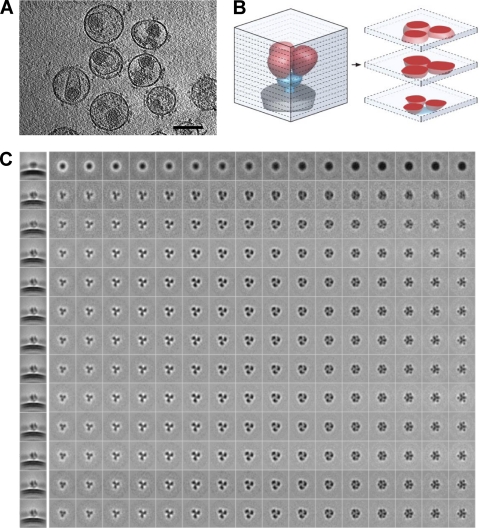
Inspection of tomographic slices from individual SIVmneE11S virions incubated with sCD4 show clearly delineated spikes on the viral membrane surface (Fig. 1A). Automatically selected spike volumes were subjected to 12 rounds of alignment, classification, and 3D averaging. Improvements in spike alignment and classification result in progressively clearer views of the structure, as illustrated in Fig. 1C, where each row presents a series of successive slices through the averaged density map at a particular iteration, as illustrated in Fig. 1B, which provides a schematic of tomographic slices through the spike. During iterative refinement, the density from bound sCD4, which is largely smeared in the initial map (Fig. 1C, first row, final column), becomes distinct by the final refinement cycle, as evidenced by the map slice taken from the height where the density from the bound sCD4 is especially prominent (Fig. 1C, last row, final column).
Fig. 1.
Cryo-electron tomography of trimeric SIV Env. (A) Slice (2.4 nm thick) through a reconstructed, cryo-electron tomogram of SIVmac239 virions incubated with sCD4 (30 μM). Env spikes on the surface of the viral membrane as well as the inner viral core are visible. Bar, 100 nm. (B) Schematic illustrating relationship between the 3D structure of the sCD4-Env complex and the slices through the tomographic reconstruction shown in C, with gp120 in red, gp41 in blue, and viral membrane in gray. (C) Slices through the tomographic reconstruction of sCD4-bound SIVmac239 Env with successive rounds of refinement shown in each row. The first column on the left shows a side-projection view through the density map before refinement (first row) and through the 12 successive rounds of refinement. The second and successive columns show slices every 4.1 Å starting at the base (density corresponding to the transmembrane glycoprotein) through Env to the apex (density corresponding to gp120 and sCD4) in the last column on the right. sCD4 density can be visualized from the second iteration onwards (third row from the top) as punctate dots in the slices at the apex of the spike.
We have previously shown that density maps for native trimeric Env from SIVmneE11S and SIVmac239 strains are composed of a propeller-shaped region with three gp120 blades connected both at the apex and at the base, corresponding to the gp120/gp41 interface (58). The density maps for these two Env variants are similar in shape and closely match that reported previously for native HIV-1 BaL Env (38). Inspection of apical slices from the density maps for sCD4-bound trimeric Env from both SIVmneE11S and SIVmac239 clearly show the additional density from the bound sCD4 (Fig. 2A to C). The additional density from sCD4 is visualized in 3D in the isosurface representation of the density maps (Fig. 2D to I). The structures of Env-sCD4 complexes formed on SIVmneE11S and SIVmac239 viruses are similar in structure, but they also include a density that is attached to each gp120 monomer and which protrudes outward and points upward away from the top of the spike (Fig. 2E and H). Subtraction of the density map for native trimeric Env from their respective sCD4-bound counterpart maps yields 3D difference density maps that both have large positive difference peaks situated where each CD4 is predicted to bind, with minimal quaternary structure changes in the rest of the spike (Fig. 2F and I). Thus, sCD4 binding to trimeric Env in these two SIV strains does not result in the large opening in trimeric Env seen previously for HIV-1 BaL (38).
Fig. 2.
Comparison of trimeric SIV Env molecular architecture in native and sCD4-bound states. (A to C) Consecutive slices illustrating the density of the map at the apex of the spike in native SIVmneE11S Env (A), sCD4-bound SIVmneE11S Env (B), and sCD4-bound SIVmac239 Env (C). The densities from gp120 and sCD4 are clearly delineated. (D and E) Density map of SIVmneE11S Env displayed as an isosurface (blue and gray represent Env and viral membrane, respectively) in native (D) (58) and in sCD4-bound (E) conformations. (F) Superposition of density map from sCD4-Env complex on SIVmneE11S (shown in mesh) with positive difference density map (yellow isosurface) obtained by subtracting the native density map from the sCD4-bound density map. (G to I) Native, sCD4-bound, and superposition of liganded density maps with difference density maps for trimeric SIVmac239 Env, as in D to F for SIVmneE11S. Although both positive and difference densities were calculated for the respective maps, the positive difference density overwhelms the negative difference density peaks and are not visible at the threshold displayed. The mass contained within the displayed difference density threshold was estimated by procedures implemented in the EMAN software (39) and is ∼78 kDa, roughly matching the approximate molecular mass of 3 sCD4 molecules (24). No difference in structure was observed when sCD4 incubation was carried out at 37°C instead of 4°C.
Molecular interpretation of these density maps to confirm the placement of sCD4 and to create working models of the interaction was achieved by fitting the maps by using coordinates from the structure reported for the complex formed between HIV-1 gp120, sCD4, and 17b (35) (Protein Data Bank [PDB] ID: 1GC1), since there are no available X-ray structures for either SIV or HIV-1 trimer-associated gp120 complexed with sCD4 alone. Three copies of the coordinates for the gp120 monomer, or gp120 and sCD4 (subsets of PDB ID: 1GC1), readily fit into the density maps of native and sCD4-bound SIV Env trimers (Fig. 3B and D), respectively. Comparison of the molecular structures of the native (Fig. 3A and B) and sCD4-bound (Fig. 3C and D) SIV Env trimers indicates that in both SIVmneE11S and SIVmac239 trimers, the orientation of sCD4 is such that the D2 domain points toward the target cell membrane. The coordinate fits additionally indicate a rearrangement of the gp120 monomers in the sCD4-bound state that is best described as a small opening of the trimer at the apex of the spike. This opening, which can be seen by comparing the change in locations of the V1/V2 loops (Fig. 3E and F), is much smaller than the reorientation of gp120 that occurs in the sCD4/17b bound state of the HIV-1 BaL Env trimer (38).
Binding of trimeric HIV-1gp120 to either sCD4 alone or to sCD4 in combination with the coreceptor mimic 17b results in an opening of the trimeric Env structure (28, 38). To our knowledge, no coreceptor binding site antibodies elicited against SIVmneE11S or SIVmac239 Env have been reported yet. However, a coreceptor binding site antibody, 7D3 (18), has been reported for SIV CP-MAC Env (36, 58). We analyzed the structure of SIVmneE11S Env in the presence of sCD4 and 7D3 antibody but obtained the same maps as those obtained in the absence of the antibody (Fig. 3), implying that there was no significant binding of the antibody to this virus. In contrast, when the same experiment was carried out with SIV CP-MAC, significant binding was observed, as determined by visualization of the additional density corresponding to the antibody (see next section).
Structural analysis of sCD4-bound SIV CP-MAC.
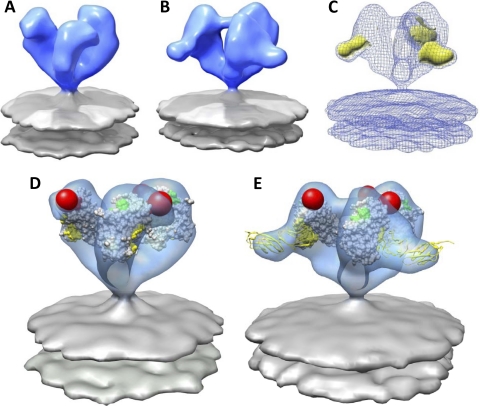
In the CD4-independent strain SIV CP-MAC, the native trimeric spikes on virions display a constitutively open conformation (Fig. 4A and D) (58). This conformation resembles the open state that is observed with HIV-1 BaL Env only after CD4 binding to the gp120 trimer (38). Determination of the structure of the SIV CP-MAC Env-sCD4 complex (Fig. 4B) shows the presence of an extra density at the outer rim of the sCD4-bound spike as compared to the native spike. The location of this extra density coincides with the location of the main peak in the difference map between the sCD4-bound and the native Env (Fig. 4C). Using a subset of the 1GC1.pdb X-ray coordinates, fitting gp120 or gp120-sCD4 into the native SIV CP-MAC or sCD4-bound SIV CP-MAC density maps, respectively, shows that both structures and coordinate fits are similar (Fig. 4D and E). Both display a structure for trimeric gp120 that is closely comparable to the open conformation seen for sCD4-complexed HIV-1 BaL Env (38), in which there are large reorientations of each gp120 protomer and a significant outward displacement of the V1/V2 loop. In addition, we compared the structures of trimeric SIV CP-MAC Env bound to the 7D3 antibody in the absence (Fig. 5A) (58) and presence (Fig. 5B) of sCD4. As previously described, 7D3 was elicited against SIV CP-MAC Env expressed as native trimers on the surfaces of infected cells and targets a conformational epitope overlapping the CCR5 coreceptor binding site (18). The presence of bound sCD4 did not influence the ability of 7D3 to bind gp120 and also did not alter the orientation or arrangement of trimeric gp120. The 7D3 antibody binds near the apex and points away from the 3-fold axis of the spike, while the two domains of sCD4 are pointed equatorially in a direction that is roughly parallel to that of the viral membrane. Together, these results show that in the case of SIV CP-MAC, where trimeric gp120 resides in a fully open conformation, the presence of bound 7D3, bound sCD4, or both 7D3 and sCD4 does not result in significant further changes in the quaternary conformation of trimeric gp120.
Fig. 4.
Molecular architecture and coordinate fits of sCD4-bound SIV CP-MAC Env. (A and B) Native (A) (58) and sCD4-bound (B) SIV CP-MAC Env density map obtained from 3D classification and averaging rendered as a blue isosurface with gray lipid bilayer. (C) Greatest positive difference density (yellow isosurface) when subtracting the native SIV CP-MAC Env (A) density map from that of sCD4-bound trimeric Env (B) density map, shown here as a blue mesh. (D) Fit of gp120 subset of 1GC1 coordinates into native SIV CP-MAC Env density map. The gp120 protomers are rotated outward compared to the conformation observed for native SIVmac239 and SIVmneE11S Env trimers. (E) Fit of gp120 and sCD4 components of 1GC1 coordinates into the sCD4-bound SIV CP-MAC Env density map. Coordinates of gp120 are displayed as in Fig. 3, with sCD4 shown as yellow ribbons.
Fig. 5.
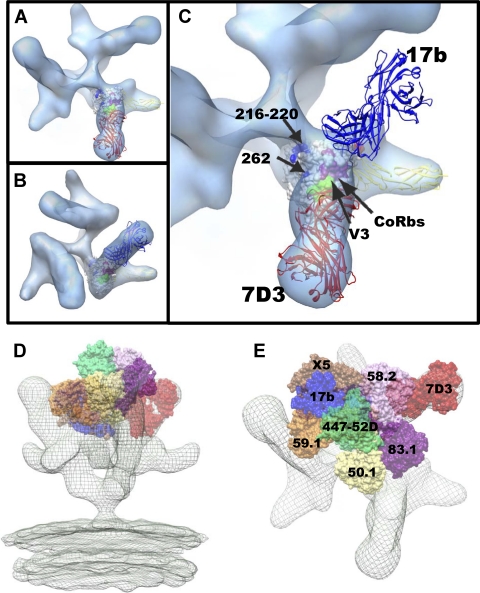
Molecular architecture of CCR5-binding site antibody (7D3) complexes on SIV CP-MAC Env with fitted gp120 coordinates. (A and B) Perspective views of 7D3-bound (red ribbons) SIV CP-MAC Env shown in blue transparent isosurface rendering in the absence (A) (58) and presence (B) of sCD4 (yellow ribbons). Insets show the same averaged maps as in the main figure panel, but without the fitted coordinates. Coordinate fits of a subset of gp120 X-ray coordinates from 1GC1 (A) or gp120 and sCD4 (B) rendered as in Fig. 3. Residues highlighted in magenta and blue indicate gp120 residues involved in coreceptor binding (47) and gp120 residues which have complete sequence conservation between HIV-1 and SIV, respectively. The 17b component of 1GC1 coordinates were used to model the 7D3 Fab fragment (red ribbons), since no crystal structure is available for 7D3 Fab, and it was then fitted into difference density calculated by subtracting the native SIV CP-MAC Env density from that of the antibody-bound map (see Materials and Methods for details).
Comparison of coreceptor binding site and V3- loop antibodies.
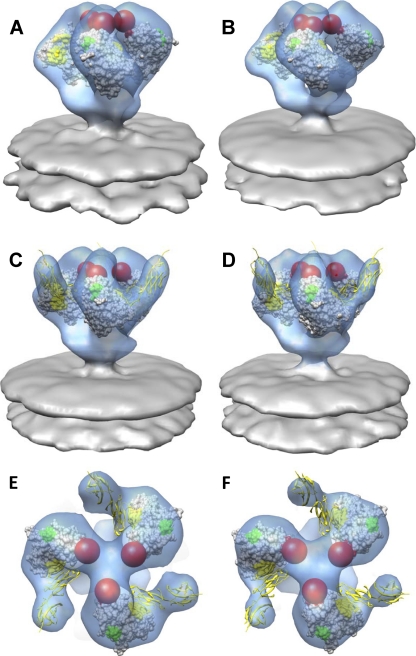
It is interesting to compare the location and orientation of the 7D3 and sCD4 bound to trimeric SIV CP-MAC Env with the location of 17b and sCD4 bound to trimeric HIV-1 BaL Env. Inspection of the top views of these complexes (Fig. 6A and B) shows that there is a dramatic difference between the angle of approach of 7D3 and that of 17b, even though both antibodies recognize residues in the coreceptor binding site and in the vicinity of the V3 loop. The binding of both antibodies is such that they point away from the central 3-fold axis but differ by over 110° in their approach angle to gp120 (Fig. 6C). Using the published structures of the various V3-loop binding antibodies and the molecular structure of the open conformation of the gp120 trimer, it is possible to obtain an impression of the variations in approach angle and situation of different antibodies that bind to gp120 (Fig. 6D and E). First, we utilized the X-ray coordinate subset of gp120-sCD4 (1GC1.pdb) to fit them to the density map for SIV CP-MAC Env complexed to both 7D3 and sCD4. Next, we superimposed the X-ray coordinates of gp120 containing the V3 loop (2B4C.pdb) to the coordinates, and then coordinates for antibodies with bound V3 peptides derived from HIV-1 gp120 were superimposed onto the V3-containing gp120 (2B4C.pdb) as described previously (32). These antibodies are shown as space-filling models in the context of the ternary complex of SIV CP-MAC Env trimer bound to sCD4 and 7D3 (Fig. 6D and E). The complexes with 17b and 7D3 are the only two examples where molecular architectures are directly determined for the complexes of these antibodies on trimeric Env. By combining the crystallographic and tomographic results, this way of visualizing the likely locations of coreceptor binding site antibodies provides a glimpse of the diversity of gp120-antibody interactions that can be accommodated on the surfaces of HIV-1 and SIV (Fig. 6D and E).
Fig. 6.
Comparison of the binding site footprint for coreceptor binding site (CoRbs) and V3-loop targeting antibodies. (A) Top view of trimeric SIV CP-MAC Env complexed with coreceptor binding site antibody 7D3 in the presence of bound sCD4. (B) Top view of trimeric HIV-1 BaL Env complexed with coreceptor binding site antibody 17b in the presence of bound sCD4 (38). Fitted X-ray coordinates for the coreceptor binding site antibodies 7D3 and 17b are shown as red (A) and blue (B) ribbons, respectively. As in Fig. 5, the X-ray coordinates from 17b Fab were used to represent 7D3 for coordinate fitting (see Materials and Methods for details). (C) Magnified top view showing locations of fitted coordinates for gp120, sCD4, and 7D3 (red ribbons) in the SIV Env map shown in A, superimposed with the location of 17b (blue ribbons) based on the corresponding ternary complex with HIV-1 gp120 and sCD4. (D and E) Visualizing locations of V3-loop and coreceptor binding site antibodies in the context of SIV CP-MAC/sCD4/7D3 ternary complex. To better understand how the binding of 7D3 to SIV CP-MAC gp120 compares with the binding of comparable antibodies directed against HIV-1 gp120, we compared the locations of these CD4i (CD4-induced) antibodies after placing them at the apex of the spike by using the complexed V3 peptide as a structural marker for alignment, as suggested in reference 32. Perspective (D) and top (E) views of the sCD4/7D3-bound density map (mesh) superposed with predicted locations of various V3-loop reactive antibodies. Positions were derived by first fitting gp120 X-ray coordinates (1GC1 with 17b shown in blue) without V3 into the density map, aligning the coordinates with V3-loop containing gp120 coordinates (2B4C with X5 shown in brown) and subsequent superposition of V3-loop peptides of V3-binding antibody complexes (59.1, 447-52D, 50.1, 83.1, and 58.2 in orange, green, yellow, purple and pink van der Waals surfaces, respectively).
DISCUSSION
These studies highlight differences observed in quaternary structure upon sCD4-binding Env on HIV-1 BaL and on three SIV strains (SIVmneE11S, SIVmac239, and SIV CP-MAC). HIV-1 BaL Env displays large conformational changes upon sCD4 binding, from a closed, native state to an open, CD4-bound state (28, 38) (Fig. 7). Env from all three strains of SIV displayed minor conformational changes upon sCD4 binding as compared to their respective native conformations (58), which in SIVmneE11S and SIVmac239 are observed as a slight opening at the apex of trimeric Env. Despite similar minor conformational changes in Env with sCD4 binding, these SIV strains have varied pathogenicities, antigenicities, and neutralization sensitivities (8, 18, 20, 29, 48, 56).
Fig. 7.
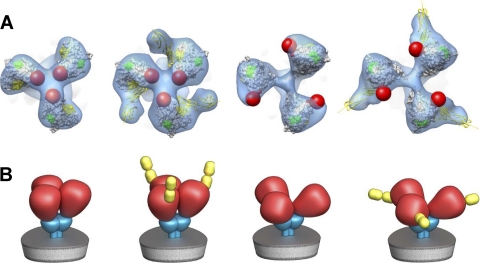
Top and schematic views of trimeric SIV envelope glycoproteins in native and sCD4-bound states. (A) Top views of the density maps (shown as a transparent blue isosurface) with fitted gp120 coordinates for SIVmne, sCD4-bound SIVmne, SIV CP-MAC, and sCD4-bound CP-MAC, shown from left to right. The red spheres indicate the likely locations of the V1/V2 loops, which are not present in the X-ray coordinates. (B) Schematic as in A, representing the essentially closed (first two) and open (last two) states of trimeric Env without and with sCD4 bound, showing gp120, gp41, and sCD4 shown in red, cyan, and yellow, respectively.
Early biochemical studies of the stoichiometry of CD4 binding to SIV Env suggested multimeric binding of sCD4 (17), while more-recent investigations have concluded only one sCD4 molecule binds per gp120 trimer (9, 33). It has also been reported that the stoichiometry of binding to SIV Env is different from the binding to HIV-1, where three sCD4 molecules bind each gp120 trimer (9). Because the structures we report are based on averaging thousands of individual trimers, we cannot determine the absolute sCD4 occupancy or exclude the possibility that a subpopulation of the Env complexes have only one bound sCD4. Nevertheless, the orientation and geometry of binding of sCD4 derived in our structural analysis show that three sCD4 molecules can be bound without steric hindrance to a single SIV trimer in the quaternary conformations displayed by all three SIV strains analyzed.
The architecture of the sCD4-bound state of SIV provides a structural basis to rationalize the striking differences between the sensitivities of SIV and most HIV-1 viruses to neutralization by sCD4 (10, 41, 52). The finding that relatively small quaternary structural changes are induced in trimeric gp120 upon sCD4 binding is consistent with previous reports that sCD4 binding causes local changes in the exposure of V2 and V3 loops but without measurable gp120 dissociation (52). The fact that this sCD4-bound state is stable in SIV, potentially capable of interacting with coreceptor molecules on the target cell membrane, also explains why cell surface CD4 is no longer required for SIV entry in the presence of exogenously added sCD4 (53) and that SIV can remain infectious at higher concentrations of sCD4, under conditions when most HIV-1 viruses are neutralized (10). Reports that sCD4 binding results in the formation of a transient intermediate in HIV-1 strains whose decay correlates with an irreversible loss of infectivity (27) and that sCD4 concentrations too low to result in gp120 dissociation can enhance viral infection (1, 54) are both fully consistent with the proposal that the sCD4 complex with SIV Env may resemble a similar structural intermediate that occurs transiently in HIV-1.
An unresolved question from our present work is whether further conformational changes occur in SIV Env before viral entry into target cells. It could be that a large quaternary conformational change such as that seen with HIV-1 occurs upon contact with the cell and that we have not yet identified conditions to capture this Env intermediate in our experiments. It could also be that the more subtle changes observed for SIVmac239 and SIVmacE11S are sufficient to expose coreceptor binding sites and that further changes may not be necessary for viral entry, since both viruses are capable of infecting human cell lines that express human CD4 and an appropriate coreceptor (19, 25, 31, 46). Yet another possibility is that the large conformational change does occur transiently but that the equilibrium is shifted to favor this state in HIV-1 BaL but not in certain SIV strains, except for strains such as SIV CP-MAC, where the conformation is already in a fully open state.
Another possible origin of the difference in the extents of conformational change observed with sCD4 binding to HIV-1 BaL, as compared to the minor changes upon sCD4 addition to the SIV strains investigated here, is our use of human sCD4 instead of simian sCD4 for the binding studies. However, since these strains are fully capable of infecting cells expressing human CD4 and human CCR5, this is unlikely to explain differences we observed for SIV and HIV-1 with respect to the consequences of sCD4 binding. Although human versus rhesus CD4 sequences are highly conserved (92%), there are amino acid changes in the rhesus CD4 near residues which have been shown to be critical in binding gp120 (35). It remains to be determined whether rhesus macaque CD4 residue mutations contribute to an alternate SIV Env conformation upon binding that is different than that observed with human sCD4.
While the detailed mechanistic implications of the structural differences between HIV-1 and SIV strains will require thorough analysis of a wider spectrum of viruses ranging from neutralization-resistant primary isolates to laboratory-adapted neutralization-sensitive strains, the present studies establish that sCD4 can bind and stabilize distinct conformations of trimeric Env and that cryo-electron tomography can be used to identify strain-dependent variations in the quaternary structures of unliganded and liganded Env. Continuing advances in computational strategies to separate distinct Env conformations from heterogeneous spike populations will provide further possibilities for separating conformational variants and improved prospects for understanding the structural diversity of trimeric Env as displayed on intact viruses.
ACKNOWLEDGMENTS
This work was supported by funds from the Center for Cancer Research at the National Cancer Institute and in part with Federal funds from the National Cancer Institute, NIH, under contract HHSN261200800001E.
We thank Steven Fellini and colleagues for assistance with use of the high-performance computational capabilities of the Biowulf Linux cluster at NIH, Bethesda, MD (http://biowulf.nih.gov); Tseng-Ming Chou, Dan Shi, and Robert Mueller for assistance with electron microscopy; and Ethan Tyler for expert assistance with figures.
Footnotes
Published ahead of print on 21 September 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Allan J. S., Strauss J., Buck D. W. 1990. Enhancement of SIV infection with soluble receptor molecules. Science 247:1084–1088 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Bartesaghi A., Sapiro G., Subramaniam S. 2005. An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms. IEEE Trans. Image Process. 14:1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartesaghi A., et al. 2008. Classification and 3D averaging with missing wedge correction in biological electron tomography. J. Struct. Biol. 162:436–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berger E. A., Lifson J. D., Eiden L. E. 1991. Stimulation of glycoprotein gp120 dissociation from the envelope glycoprotein complex of human immunodeficiency virus type 1 by soluble CD4 and CD4 peptide derivatives: implications for the role of the complementarity-determining region 3-like region in membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 88:8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen B., et al. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 7. Clements J. E., Mankowski J. L., Gama L., Zink M. C. 2008. The accelerated simian immunodeficiency virus macaque model of human immunodeficiency virus-associated neurological disease: from mechanism to treatment. J. Neurovirol. 14:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole K. S., et al. 2001. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology 290:59–73 [DOI] [PubMed] [Google Scholar]
- 9. Crooks E. T., et al. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daar E. S., Li X. L., Moudgil T., Ho D. D. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. U. S. A. 87:6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dalgleish A. G., et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 12. Daniel M. D., et al. 1985. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228:1201–1204 [DOI] [PubMed] [Google Scholar]
- 13. Daniel M. D., et al. 1987. Long-term persistent infection of macaque monkeys with the simian immunodeficiency virus. J. Gen. Virol. 68:3183–3189 [DOI] [PubMed] [Google Scholar]
- 14. Deng H., et al. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666 [DOI] [PubMed] [Google Scholar]
- 15. Desrosiers R. C. 1990. The simian immunodeficiency viruses. Annu. Rev. Immunol. 8:557–578 [DOI] [PubMed] [Google Scholar]
- 16. Douglas N. W., Munro G. H., Daniels R. S. 1997. HIV/SIV glycoproteins: structure-function relationships. J. Mol. Biol. 273:122–149 [DOI] [PubMed] [Google Scholar]
- 17. Earl P. L., Doms R. W., Moss B. 1992. Multimeric CD4 binding exhibited by human and simian immunodeficiency virus envelope protein dimers. J. Virol. 66:5610–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edinger A. L., et al. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Endres M. J., et al. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745–756 [DOI] [PubMed] [Google Scholar]
- 20. Faith S. A., et al. 2010. Induction of antibody-mediated neutralization in SIVmac239 by a naturally acquired V3 mutation. Virology 400:86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng Y., Broder C. C., Kennedy P. E., Berger E. A. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877 [DOI] [PubMed] [Google Scholar]
- 22. Gallo S. A., et al. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36–50 [DOI] [PubMed] [Google Scholar]
- 23. Gallo S. A., Sackett K., Rawat S. S., Shai Y., Blumenthal R. 2004. The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors. J. Mol. Biol. 340:9–14 [DOI] [PubMed] [Google Scholar]
- 24. Garlick R. L., Kirschner R. J., Eckenrode F. M., Tarpley W. G., Tomich C. S. 1990. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res. Hum. Retroviruses 6:465–479 [DOI] [PubMed] [Google Scholar]
- 25. Gazdar A. F., et al. 1980. Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood 55:409–417 [PubMed] [Google Scholar]
- 26. Groenink M., Moore J. P., Broersen S., Schuitemaker H. 1995. Equal levels of gp120 retention and neutralization resistance of phenotypically distinct primary human immunodeficiency virus type 1 variants upon soluble CD4 treatment. J. Virol. 69:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haim H., et al. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris A., et al. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirsch V. M., Johnson P. R. 1994. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 32:183–203 [DOI] [PubMed] [Google Scholar]
- 30. Hirsch V. M., Lifson J. D. 2000. Simian immunodeficiency virus infection of monkeys as a model system for the study of AIDS pathogenesis, treatment, and prevention. Adv. Pharmacol. 49:437–477 [DOI] [PubMed] [Google Scholar]
- 31. Hoxie J. A., et al. 1986. Alterations in T4 (CD4) protein and mRNA synthesis in cells infected with HIV. Science 234:1123–1127 [DOI] [PubMed] [Google Scholar]
- 32. Huang C. C., et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim M., et al. 2001. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J. Biol. Chem. 276:42667–42676 [DOI] [PubMed] [Google Scholar]
- 34. Kremer J. R., Mastronarde D. N., McIntosh J. R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 35. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. LaBranche C. C., et al. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J. Virol. 68:5509–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Letvin N. L., et al. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71–73 [DOI] [PubMed] [Google Scholar]
- 38. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ludtke S. J., Baldwin P. R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 40. Mansfield K. G., Lackner A. A. 1997. Simian immunodeficiency virus-inoculated macaques acquire Mycobacterium avium from potable water during AIDS. J. Infect. Dis. 175:184–187 [DOI] [PubMed] [Google Scholar]
- 41. Moore J. P., McKeating J. A., Norton W. A., Sattentau Q. J. 1991. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implications for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J. Virol. 65:1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moore J. P., McKeating J. A., Weiss R. A., Sattentau Q. J. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142 [DOI] [PubMed] [Google Scholar]
- 43. O'Brien W. A., Mao S. H., Cao Y., Moore J. P. 1994. Macrophage-tropic and T-cell line-adapted chimeric strains of human immunodeficiency virus type 1 differ in their susceptibilities to neutralization by soluble CD4 at different temperatures. J. Virol. 68:5264–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orloff S. L., Kennedy M. S., Belperron A. A., Maddon P. J., McDougal J. S. 1993. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J. Virol. 67:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pettersen E., et al. 2004. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 46. Raport C. J., Gosling J., Schweickart V. L., Gray P. W., Charo I. F. 1996. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J. Biol. Chem. 271:17161–17166 [DOI] [PubMed] [Google Scholar]
- 47. Rizzuto C. D., et al. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953 [DOI] [PubMed] [Google Scholar]
- 48. Rudensey L. M., Kimata J. T., Long E. M., Chackerian B., Overbaugh J. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samson M., et al. 1996. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725 [DOI] [PubMed] [Google Scholar]
- 50. Sasseville V. G., Lackner A. A. 1997. Neuropathogenesis of simian immunodeficiency virus infection in macaque monkeys. J. Neurovirol. 3:1–9 [DOI] [PubMed] [Google Scholar]
- 51. Sattentau Q. J., Moore J. P. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sattentau Q. J., Moore J. P., Vignaux F., Traincard F., Poignard P. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schenten D., et al. 1999. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 73:5373–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith D. H., et al. 1987. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science 238:1704–1707 [DOI] [PubMed] [Google Scholar]
- 55. Stamatatos L., Werner A., Cheng-Mayer C. 1994. Differential regulation of cellular tropism and sensitivity to soluble CD4 neutralization by the envelope gp120 of human immunodeficiency virus type 1. J. Virol. 68:4973–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steckbeck J. D., Cole K. S. 2006. Dissecting the humoral immune response to simian immunodeficiency virus: mechanisms of antibody-mediated virus neutralization. Immunol. Res. 36:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trkola A., et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 58. White T. A., et al. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wyatt R., Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]