Abstract
We detected a high prevalence (12.5%) of novel avian coronaviruses in aquatic wild birds. Phylogenetic analyses of these coronaviruses suggest that there is a diversity of gammacoronaviruses and deltacoronaviruses circulating in birds. Gammacoronaviruses were found predominantly in Anseriformes birds, whereas deltacoronaviruses could be detected in Ciconiiformes, Pelecaniformes, and Anseriformes birds in this study. We observed that there are frequent interspecies transmissions of gammacoronaviruses between duck species. In contrast, deltacoronaviruses may have more stringent host specificities. Our analysis of these avian viral and host mitochondrial DNA sequences also suggests that some, but not all, coronaviruses may have coevolved with birds from the same order.
TEXT
Coronaviruses are important pathogens of birds and mammals, including humans, and are previously classified into 3 genera. Alphacoronaviruses and betacoronaviruses are found in mammals, whereas gammacoronaviruses are detected primarily in birds. Surveillance of coronaviruses in wild mammals has led to the discovery of a significant number of novel alphacoronaviruses and betacoronaviruses in bats (18, 23, 27). In contrast, relatively little is known about the biology of avian coronaviruses in wildlife. Gough and colleagues identified a parrot coronavirus that is genetically distinct from alpha-, beta-, and gammacoronaviruses in 2006 (10). Additional novel coronaviruses that are genetically similar to the parrot coronavirus were subsequently detected in mammals and terrestrial birds (9, 26). Viruses of this novel lineage have been recently proposed to form a new genus, provisionally named Delta-coronavirus (http://talk.ictvonline.org/files/proposals/taxonomy_proposals_vertebrate1/m/vert02/3257.aspx) (8). Besides, findings from other studies suggested that there is diversity of coronaviruses circulating in wild birds (13, 17). These findings have prompted us to launch wild bird surveillance for avian coronaviruses in Hong Kong and Cambodia.
From November 2009 to January 2010, site visits to the Mai Po Marshes, Hong Kong, were organized biweekly. Isolated fresh bird droppings on pond shores were sampled and stored in individual vials containing viral transport medium (VTM) (15). For samples collected in Cambodia, cloacal swabs from captured free-ranging birds were collected in vials with VTM in 2008. RNA was extracted from these samples and reverse transcribed as described previously (5). cDNA was subjected to a pancoronavirus nested PCR (nPCR) for the RNA-dependent RNA polymerase (RdRp) sequence. Briefly, cDNA was amplified in a first-round PCR (forward primer 5′-GGKTGGGAYTAYCCKAARTG-3′ and reverse primer 5′-TGYTGTSWRCARAAYTCRTG-3′; 40 cycles of 94°C for 20 s, 48°C for 30 s, and 72°C for 50 s). The PCR product was then amplified in a second-round PCR under amplification condition identical to those of the first-round PCR, except that a new set of primers was used in the assay (forward primer 5′-GGTTGGGACTATCCTAAGTGTGA-3′, reverse primer 5′-CCATCATCAGATAGAATCATCAT-3′). The final PCR products (440 bp) were analyzed by sequencing.
The bird species studied in Cambodia were identified immediately upon capture by expert ornithologists (20). A well-validated nPCR assay targeting cDNA of the mitochondrial COX1 sequence was used to identify the host species of representative Hong Kong samples (4, 24). Furthermore, a similar nPCR assay using alternative primers for the COX1 sequence was used for result reconfirmations (first round, forward primer 5′-GAYATRGCKTTYCCKCGKATRAA-3′ and reverse primer 5′-ATKGCYCAKACYATKCCYATRTA-3′; second round, forward primer 5′-GCKTTTCCKCGKATRAAYAAYAT-3′ and reverse primer 5′-CCYATRTAKCCRAAKGGYTCYTT-3′). The deduced mitochondrial DNA (mtDNA) sequences were used in a blast search of the GenBank and BOLD databases (19) for host identification.
A total of 658 samples collected in Hong Kong were tested. Ninety-nine (15.0%) of these samples were reverse transcription (RT)-PCR positive for coronavirus. Positive samples were detected on each visit, with positivity rates ranging from 13.2% to 22.6%. Host mtDNA was used to determine the host species of all the positive samples. In order to get an overview of the species diversity of our sample collections, 94 randomly selected coronavirus-negative specimens were also subjected to the mtDNA barcoding assays. Comparable host mtDNA detection rates were found in coronavirus-positive (81.8%) and coronavirus-negative (74.5%) samples. Species that were found to be positive for coronaviruses are summarized in Table 1. The range of species that were coronavirus positive was broadly similar to that of coronavirus-negative ones (Table 1) (P > 0.05, t test). None of the mtDNA positive samples were found to have ambiguous results in the barcoding assays.
Table 1.
Detection of coronavirus in avian samples
| Location and species | Total no. (no. positive/negative) | No. (%) positive for: |
|
|---|---|---|---|
| Gammacoronavirus | Deltacoronavirus | ||
| Hong Konga | |||
| Common teal (Anas crecca) | 18 (10/8) | 9 (50.0) | 1 (5.6) |
| Northern shoveler (Anas clypeata) | 31 (24/7) | 22 (71.0) | 2 (6.5) |
| Eurasian wigeon (Anas penelope) | 24 (10/14) | 9 (37.5) | 1 (4.2) |
| Northern pintail (Anas acuta) | 38 (17/21) | 16 (42.1) | 1 (2.6) |
| Tufted duck (Aythya fuligula) | 1 (1/0) | 1 (100) | 0 |
| American wigeon (Anas americana) | 1 (1/0) | 0 | 1 (100) |
| Gray heron (Ardea cinerea) | 10 (4/6) | 0 | 4 (40.0) |
| Night heron (Nycticorax nycticorax) | 1 (0/1) | 0 | 0 |
| Great cormorant (Phalacrocorax carbo) | 24 (13/11) | 0 | 13 (54.2) |
| Black-faced spoonbill (Platalea minor) | 3 (1/2) | 0 | 1 (33.3) |
| Cambodia | |||
| Pond heron (Ardeola bacchus/speciosa) | 123 (16/107) | 0 | 16 (13.0) |
| Lesser whistling duck (Dendrocygna javanica) | 33 (1/32) | 1 (3.0) | 0 |
| Ruddy-breasted crake (Porzana fusca) | 80 (80/0) | 0 | 0 |
Only samples that were informative in the DNA barcoding analysis are listed.
Cloacal swabs (n = 263) were collected from pond herons, lesser whistling ducks, and ruddy-breasted crakes in Cambodia. Coronavirus-positive reactions were detected in 13.0% (16/123) of pond herons and 3.0% (1/33) of lesser whistling ducks (Table 1).
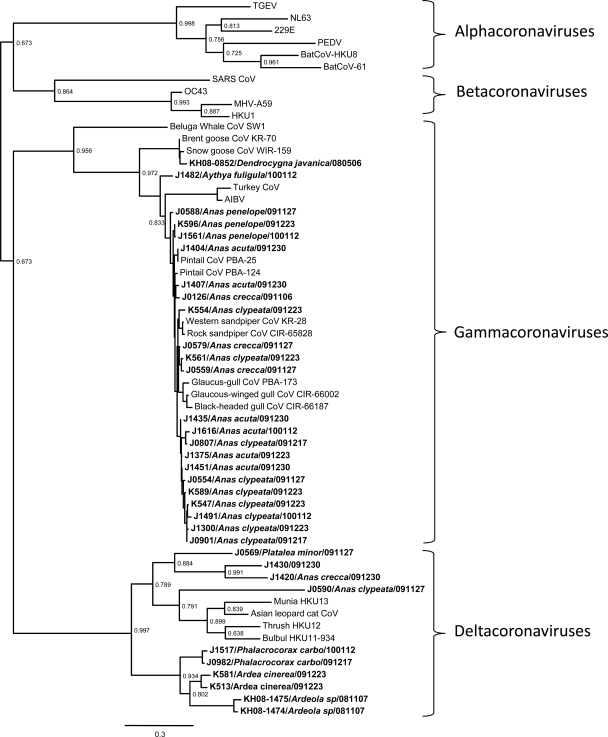
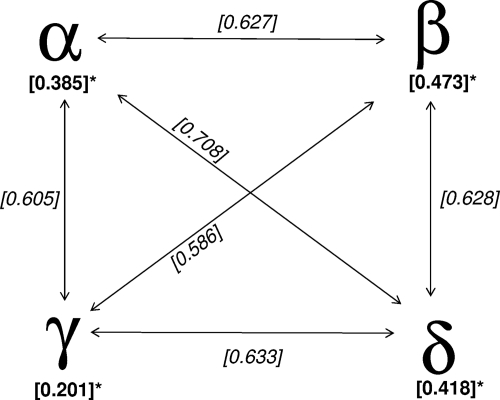
All the coronaviruses identified in this study could be phylogenetically classified as gammacoronaviruses and deltacoronaviruses (Fig. 1; see Fig. S1 in the supplemental material). This observed phylogeny is comparable to the split off between alphacoronaviruses and betacoronaviruses in mammals. To roughly estimate the genetic difference between these partial RdRp sequences, we selected 10 representative viruses from each genus to estimate the genetic distances between these viruses (data not shown). The genetic distance between these two groups of avian coronaviruses is similar to the intergenus distances between coronaviruses from different genera, and this genetic distance is significantly greater than those observed within the same genus (Fig. 2, P < 0.005).
Fig. 1.
Phylogenetic analysis of alphacoronaviruses, betacoronaviruses, gammacoronaviruses, and deltacoronaviruses. Partial viral RdRp sequences (360 bp) were used in this analysis. Sequences were aligned and edited manually by ClustalW (25). The best evolution model describing an alignment was determined by MEGA5 (22). Phylogenetic trees were generated using Phyml with the best substitution model selected, and with aLRT statistics SH-like branch supports (11). The novel avian coronaviruses identified in this study are in bold. The host identities determined by DNA fingerprinting techniques are in brackets. Sample collection dates are indicated (year/month/day). aLRT statistics SH-like branch supports are indicated at the nodes. The GenBank accession numbers of the genes used are as follows: 229E (human coronavirus 229E), AF304460; AIBV (avian infectious bronchitis virus), FJ904722; Asian leopard cat CoV, EF584908; BatCoV 61, AY864196; BatCoV HKU8, DQ249228; beluga whale CoV SW1, EU111742; black-headed gull CoV CIR-66187, GU396686; Brent goose CoV KR-70, GU396676; bulbul HKU11 (Bulbul coronavirus HKU11), FJ376619; glaucous-winged gull CIR-66002, GU396682; glaucous gull CoV PBA-173, GU396674; HKU1 (human coronavirus HKU1), AY597011; MHV-A59 (mouse hepatitis virus), FJ647225; munia HKU13 (munia coronavirus HKU13), FJ376622; NL63 (human coronavirus NL63), AY567487; OC43 (human coronavirus OC43), AY903460; PEDV (porcine epidemic diarrhea coronavirus), AF353511; pintail CoV PBA-124, GU396673; pintail CoV PBA-25, GU396671; rock sandpiper CoV CIR-65828, GU396688; SARS CoV (human severe acute respiratory syndrome coronavirus), JF292915; snow goose CoV WIR-159, GU396690; TGEV (transmissible gastroenteritis coronavirus), DQ811789; thrush HKU12 (thrush coronavirus HKU12), FJ376621; turkey CoV, EU095850; western sandpiper CoV KR-28, GU396675.
Fig. 2.
Genetic distances between alphacoronaviruses, betacoronaviruses, gammacoronaviruses, and deltacoronaviruses. Partial ORF1b sequences of 10 representative viruses from each genus were analyzed (data not shown). The genetic distances were estimated by using the Jukes-Cantor model. The median genetic distances between the viruses studied are shown. An asterisk indicates that all intragenus genetic distances were found to be significantly shorter than the relevant intergenus genetic distances (P < 0.005).
Gammacoronaviruses found in this study were detected in wild waterfowl of orders Anseriformes. These viruses include those from little whistling ducks, tufted ducks, common teals, northern shovelers, Eurasian wigeons, and northern pintails. Previously known representative coronaviruses in this group include avian infectious bronchitis virus, peafowl coronavirus, turkey coronavirus, goose coronaviruses, pintail coronaviruses, gull coronavirus, and a virus collected from a sick beluga whale (2, 16, 17). In contrast, novel deltacoronaviruses found in this study were detected in waterbirds from the orders Ciconiiformes, Pelecaniformes, and Anseriformes. These viruses include those from gray herons, pond herons, great cormorants, black-faced spoonbills, and several duck species (Anas spp.). In addition, the prevalence of deltacoronavirus was found to be much lower than that of gammacoronaviruses in the duck samples studied. Of 63 avian coronaviruses obtained from the Anas spp., only 6 (9.5%) are deltacoronaviruses (Table 1). Previously known members of this group include coronavirus (CoV) HKU11 from bulbuls, CoV HKU12 from thrushes, CoV HKU13 from munias, and Asian leopard cat coronavirus (9, 26).
Our findings demonstrate that wild birds are major reservoirs of a wide range of gamma- and deltacoronaviruses. Some of the species studied were found to have a high prevalence of coronaviruses (Table 1). It is possible that these avian coronaviruses do not cause severe illness in their hosts, thereby allowing themselves to be endemic in some avian populations. Interestingly, gammacoronaviruses which are similar to those discovered in the Bering Strait area in 2005 (17) were also detected in our study. In particular, viruses that have identical partial RdRp sequences were detected in northern pintails at both sites studied (Fig. 1, J1404 and pintail CoV PBA-25), indicating that some avian coronaviruses can persist in a specific bird species and are carried by these migrating birds to other geographic locations (http://www.werc.usgs.gov/ProjectSubWebPage.aspx?SubWebPageID=8&ProjectID=37&List=SubWebPages&Web=Project_37&Title=Movements).
We also noted that not all of our studied species were positive for coronaviruses. Many of these species may have been negative as a result of the small sample sizes. However, we tested a substantial number of samples from ruddy-breasted crakes (n = 80) that were all RT-PCR negative for coronavirus. The ruddy-breasted crake is a permanent-resident bird in Cambodia (20). In contrast, the positivity rates of avian coronavirus in some of our studied migratory species, such as ducks and great cormorants, are extremely high (Table 1). Previous studies of migratory waterbirds and resident terrestrial birds have similar findings (17, 26). It is not known whether these observations are due to the intrinsic biological differences between coronaviruses circulating in these two distinct bird populations and/or the behavioral and physiological differences between these two groups of birds (1).
The phylogenetic topology of gammacoronaviruses is very different from that of deltacoronaviruses. The gammacoronaviruses sampled from ducks, sandpipers, and gulls form a distinct clade (Fig. 1). The RdRp genes from this clade of viruses are genetically closely related, and the overall RdRp gene sequence homology differs by <7%. In this virus cluster, the same avian species may contain viruses that are genetically heterogeneous (e.g., J1404, J1407, J1435, and J1451 collected from northern pintails on the same sampling day), whereas viruses that are genetically very similar could be detected in different duck species (e.g., J0807 and J1616). These findings suggest that there are frequent intergenus and interspecies transmissions of this group of gammacoronaviruses among these avian species. In contrast, viruses in the deltacoronavirus group were found to have distinct phylogenetic branches (e.g., J1517 and J0982 from great cormorants sampled on different dates), indicating that coronaviruses in this group have a more stringent host specificity than those in the gammacoronavirus group. Overall, these results suggest that the ecology of gammacoronaviruses and deltacoronaviruses may be very different.
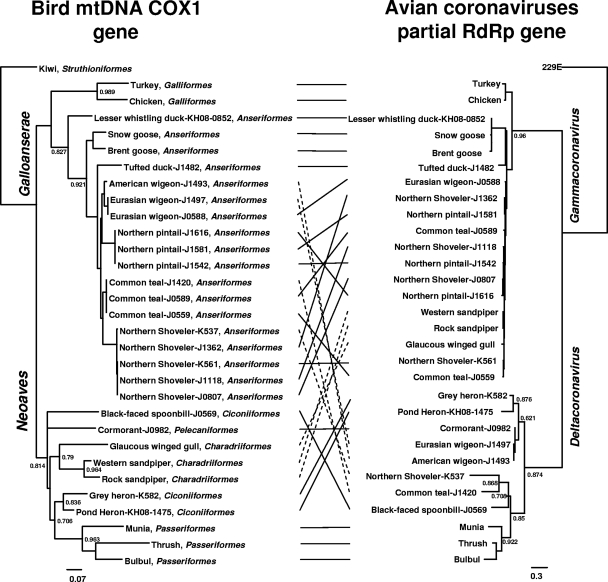
It was previously reported that there is a coevolved virus-host relationship between vespertilionid bats and their coronaviruses (7). We used some of our representative sequences to perform a similar analysis with avian coronaviruses and their hosts (Fig. 3). It was reported that both gammacoronaviruses and deltacoronaviruses could be detected in birds in the superorders Galloanserae and Neoaves (12), with the gamma group found predominantly in Galloanserae birds and the delta group found predominantly in Neoaves birds (see Fig. S1 in the supplemental material [highlighted in blue and green, respectively]). Some coronaviruses and their hosts even fall into the same monophyletic subclade in the corresponding trees (e.g., gammacoronaviruses from ducks and deltacoronaviruses from Passeriformes) (Fig. 3). We also observed that there are some exceptional cases in this study. For example, gammacoronaviruses and deltacoronaviruses were detected in Charadriiformes and Anseriformes, respectively (see Fig. S1 in the supplemental material [highlighted in red]). As our sample set was limited to just a few avian orders, additional surveillance work is needed to reveal the full picture of avian coronavirus ecology. Nonetheless, our findings suggest that at least some avian coronaviruses may have circulated and coevolved with specific orders or subsets of avian hosts. To test this hypothesis, we have also examined additional coronaviruses reported to be detected in different avian species (3, 6, 10, 13, 14, 17, 21, 26). Our preliminary analyses indicate that, except for ducks, which can be infected by both the gamma and delta groups, all the bird orders examined are positive only for either gammacoronaviruses or deltacoronaviruses (data not shown).
Fig. 3.
Phylogenetic correlation between avian coronaviruses and their hosts. A phylogenetic tree of avian host mtDNA COX1 gene (450 bp, left) and viral RdRp gene sequences (360 bp, right) is shown. The order of each bird species studied is shown. Only viral and host sequences from selected coronavirus-positive bird dropping samples in our study were included. Lines between the 2 trees were added to help visualize virus and host sequence congruence (solid lines) or incongruence (broken lines). Accession numbers of the reference host sequences used: Brent goose, DQ433366; bulbul, FJ378536; chicken, GU261717; glaucous-winged gull, HM033523; kiwi, EU525317.1; munia, EF515788; rock sandpiper, GU571303; snow goose, DQ434538; thrush, GQ482858; turkey, EF153719; western sandpiper, AY666261.
Our data suggest that the circulation of coronaviruses in apparently healthy populations of wild aquatic birds is more common than previously recognized. Additional coronavirus surveillance in birds of different orders and a full genome analysis of avian coronaviruses may help us to better understand the evolution of coronaviruses.
Nucleotide sequence accession numbers.
All novel virus gene sequences generated in this study were deposited in GenBank under accession numbers JN788780 to JN788888. The host mtDNA sequences generated in this work were deposited in GenBank under accession numbers JN788889 to JN788908.
Supplementary Material
Acknowledgments
We thank Kai-Chi Chow, Chuk-Kwan Ho, and Yu-On Wu at the University of Hong Kong for sample collection. We thank Bena Smith and Katherine Leung at the Mai Po Nature Reserve, World Wide Fund, Hong Kong, for their assistance in sample collection. We also thank colleagues at the Department of Agriculture, Fisheries, and Conservation, Hong Kong, for facilitating the study.
This project was supported by the National Institutes of Health (NIAID contract HHSN266200700005C) and EMPERIE (grant EU FP7 223498).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 28 September 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Altizer S., Bartel R., Han B. A. 2011. Animal migration and infectious disease risk. Science 331:296–302 [DOI] [PubMed] [Google Scholar]
- 2. Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet. Res. 38:281–297 [DOI] [PubMed] [Google Scholar]
- 3. Cavanagh D. 2005. Coronaviruses in poultry and other birds. Avian Pathol. 34:439–448 [DOI] [PubMed] [Google Scholar]
- 4. Cheung P. P., et al. 2009. Identifying the species-origin of faecal droppings used for avian influenza virus surveillance in wild-birds. J. Clin. Virol. 46:90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chu D. K., et al. 2006. Coronaviruses in bent-winged bats (Miniopterus spp.). J. Gen. Virol. 87:2461–2466 [DOI] [PubMed] [Google Scholar]
- 6. Circella E., et al. 2007. Coronavirus associated with an enteric syndrome on a quail farm. Avian Pathol. 36:251–258 [DOI] [PubMed] [Google Scholar]
- 7. Cui J., et al. 2007. Evolutionary relationships between bat coronaviruses and their hosts. Emerg. Infect. Dis. 13:1526–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Groot R. J., et al. In King A. M. Q. (ed.), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses, in press Elsevier, New York, NY [Google Scholar]
- 9. Dong B. Q., et al. 2007. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in southern China. J. Virol. 81:6920–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gough R. E., Drury S. E., Culver F., Britton P., Cavanagh D. 2006. Isolation of a coronavirus from a green-cheeked Amazon parrot (Amazon viridigenalis Cassin). Avian Pathol. 35:122–126 [DOI] [PubMed] [Google Scholar]
- 11. Guindon S., et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 12. Hackett S. J., et al. 2008. A phylogenomic study of birds reveals their evolutionary history. Science 320:1763–1768 [DOI] [PubMed] [Google Scholar]
- 13. Hughes L. A., et al. 2009. Genetically diverse coronaviruses in wild bird populations of northern England. Emerg. Infect. Dis. 15:1091–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jonassen C. M., et al. 2005. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos). J. Gen. Virol. 86:1597–1607 [DOI] [PubMed] [Google Scholar]
- 15. Leung Y. H., et al. 2007. Poultry drinking water used for avian influenza surveillance. Emerg. Infect. Dis. 13:1380–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mihindukulasuriya K. A., Wu G., St Leger J., Nordhausen R. W., Wang D. 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82:5084–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muradrasoli S., et al. 2010. Prevalence and phylogeny of coronaviruses in wild birds from the Bering Strait area (Beringia). PLoS One 5:e13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poon L. L., et al. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ratnasingham S., Hebert P. D. 2007. The Barcode of Life Data System (http://www.barcodinglife.org) Mol. Ecol. Notes 7:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robson C. 2005. A field guide to the birds of South-East Asia. New Holland Publishers Ltd., London, United Kingdom [Google Scholar]
- 21. Sun L., et al. 2007. A Massachusetts prototype like coronavirus isolated from wild peafowls is pathogenic to chickens. Virus Res. 130:121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K., et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang X. C., et al. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tavares E. S., Baker A. J. 2008. Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol. Biol. 8:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woo P. C., et al. 2009. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 83:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woo P. C., et al. 2006. Molecular diversity of coronaviruses in bats. Virology 351:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.