Abstract
We analyzed xenotropic murine leukemia virus-related virus (XMRV) integration site sequences previously identified from human prostate tissues for single-nucleotide polymorphisms (SNPs) to discriminate between patient and potential cell line sources of the proviruses. The SNPs of two integration sites were identical to those in cell lines but not the patients, whereas the data on the remaining 12 integration sites were inconclusive. Our results provide direct evidence for contamination during analysis of XMRV integration sites.
TEXT
Xenotropic murine leukemia virus (MLV)-related virus (XMRV) was initially identified as a human gammaretrovirus associated with RNase L-deficient prostate cancer (18). XMRV and other MLV-related viruses have also been implicated in chronic fatigue syndrome (CFS) (8, 9). However, the association of XMRV with human diseases has been in doubt, as many subsequent studies failed to detect the virus (7, 15; for reviews, see references 13 and 16). A major concern in the detection of XMRV by PCR in human tissues is murine DNA contamination, both in sample preparation (4, 10, 12) and in commercial reagents and kits (2, 17). In particular, the observed high incidences of XMRV and other MLV-related viruses in CFS are likely due to the presence of contaminating MLV sequences (7). The significance of XMRV as a human pathogen has further been challenged by the recent finding that XMRV in 22Rv1 cells is the result of recombination between two endogenous retroviruses in mice (11).
Integration of proviral DNA into the host cell genome is a defining feature of retroviral replication. We previously used a linker-mediated PCR method (5) to sequence and map a total of 14 unique provirus integration sites in human prostate DNA from nine different prostate cancer patients (5, 6), supporting XMRV as a bona fide human retrovirus. A recent BLAST analysis showed that two of the patient-derived integration sites are identical to integration sites found in experimental infections of the prostate cancer cell line DU145 (3). Although independent integration events at identical genomic locations have been reported in MLV-induced lymphomas in mice (14) and in HIV-infected T cells in vitro (20), such events are rare and generally reflect a selection process. Given the assumption that the existence of identical integration sites in two different samples is highly unlikely, this suggests that the patient-derived sites might have been the result of PCR contamination. In light of this finding and the controversy surrounding XMRV as a human pathogen, we sought to interrogate our previously acquired integration site data to determine if the integration sites originated in the patient or were the result of laboratory contamination from infected cell lines.
Since XMRV was detected in only a small percentage of stromal and hematopoietic cells, rather than in the clonally expanding, cancerous epithelial cells (18), it is doubtful that the proviral sites reported previously (5, 6) can be recloned. Therefore, we performed an analysis of single-nucleotide polymorphisms (SNPs) in each of the 14 patient-derived integration sites compared to the respective alleles in the corresponding patients and the prostate cancer cell lines LNCaP, DU145, and 22Rv1. These cell lines were selected due to their extensive utilization in molecular studies of XMRV. Each patient-derived integration site is characterized by a region of the XMRV right long terminal repeat followed by a segment of human genomic sequence of variable length, which depends on the location of integration and neighboring restriction enzyme sites in the human genome. Based on the sequence information of each integration site, we amplified and sequenced a region flanking each site (see Table S1 in the supplemental material) in genomic DNA extracted from the cell lines and in prostate tissues from the nine patients analyzed previously (5; also, see the supplemental methods). A minimum of six clones per sample was analyzed to resolve heterozygous alleles. The integration site genomic sequence was then aligned with the corresponding patient and cell line sequences to determine if SNPs were present.
Of the 14 patient-derived integration sites, 11 did not exhibit SNPs (Table 1). Each of these 11 integration site sequences was identical to those found in the patient and in the prostate cell lines, therefore providing no basis for differentiating between candidate sources. This included the patient-derived integration site (GenBank no. EU981810) previously found to be identical to a site (GenBank no. EU981678) cloned from acutely infected DU145 cells (3, 5). Since it is probable that a provirus present in a tumor sample can contaminate an acutely infected cell line and vice versa, we cannot conclude at present that these two identical sites were due to contamination of the tumor sample by XMRV-infected cell lines or vice versa.
Table 1.
Summary of SNPs identified in patient-derived integration sitesa
| SNP status | Sample | Chromosome location of integration site | GenBank no. | SNPs | No. of clones sequenced |
|---|---|---|---|---|---|
| SNPs present, informative | VP432 | 1; 204400002 | EU981801 | rs11075704 (C/T), rs72789205 (A/G) | 8 |
| VP363 | 16; 69090908 | EU981808b | rs9661807 (A/G), rs9660554 (A/G) | 11 | |
| SNPs present, noninformative | VP432 | 6; 111278735 | EU981800 | rs6902336 (A/G), rs9487562 (C/G) | 8 |
| No SNPs present, noninformative | VP268 | 11; 72504631 | EU981802 | 8 | |
| VP283 | 3; 197122283 | EU981803 | 6 | ||
| VP283 | 19; 11254762 | EU981804 | 8 | ||
| VP338 | 12; 46824702 | EU981805 | 7 | ||
| VP433 | 15; 65283282 | EU981806 | 8 | ||
| VP234 | 17; 58591644 | EU981807 | 8 | ||
| VP433 | 14; 31733396 | EU981809 | 6 | ||
| VP268 | 16; 68121168 | EU981810b | 6 | ||
| VP268 | 7; 28723080 | EU981811 | 6 | ||
| VP29 | 3; 73200877 | EU981812 | 6 | ||
| VP229 | 16; 67973893 | EU981813 | 6 |
Integration sites are designated by patient identification numbers, chromosome locations, and GenBank accession numbers. Chromosome locations were mapped to the human genome build hg19 and are represented by the chromosome numbers followed by the nucleotide positions. Additional characteristics of the integration site can be found in reference 5. Identified SNPs are indicated by the RefSNP accession numbers, followed by known allele nucleotides in parentheses. To resolve heterozygosity, a minimum of six clones per sample were sequenced.
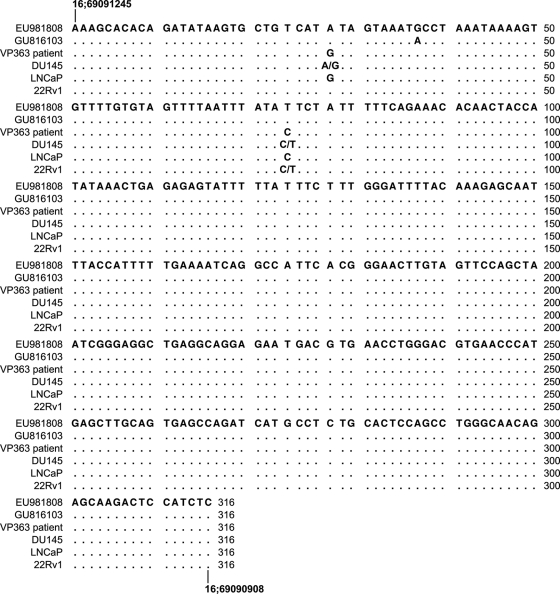
We observed SNPs in three of the integration sites (GenBank no. EU981808, EU981801, and EU981800). Alignment of patient VP363 and cell line genomic sequences with that of the integration site EU981808 showed that two SNP alleles (refSNP no. rs11075704 and rs72789205) are consistent with alleles found in DU145 and 22Rv1 genomes but not in the patient (Fig. 1). The SNP rs11075704 allele contains an A in the cloned integration site, heterozygous (A/G) in the DU145 cell line, and A in 22Rv1, whereas it is homozygous for G in VP363. The SNP rs72789205 allele contains a T in the integration site and heterozygous (C/T) in DU145 and 22Rv1 DNA, compared to C in the patient. EU981808 is identical to an integration site, GU816103, found in an XMRV-infected DU145 clonal cell line (3, 6). Therefore, our SNP results provide direct evidence that this integration site is the result of contamination from infected DU145 cells.
Fig. 1.
Alignment of EU981808 with VP363 patient and prostate cancer cell line genomic DNA. The genomic sequence of EU981808 (patient-derived integration site) is compared to that of GU816103 (DU145-derived integration site), the patient VP363, and the cell lines DU145, LNCaP, and 22Rv1. The locations of this sequence in the human genome build hg19 are shown at the beginning and the end of the sequence as the chromosome number followed by the nucleotide position.
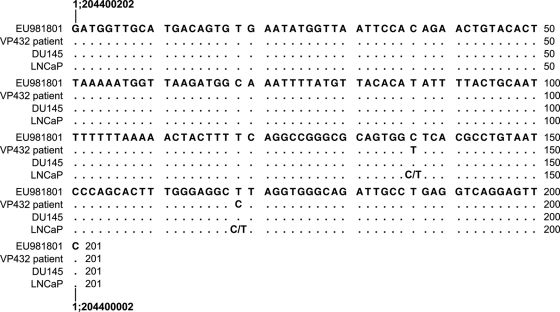
The patient-derived integration site EU981801 also exhibits two SNP alleles (refSNP no. rs9661807 and rs9660554) that differ from those found in the patient sample VP432 (Fig. 2). The SNPs rs9661807 and rs9660554 contain C and T, respectively, in the integration site sequence and in DU145 and heterozygous in LNCaP, whereas the patient VP432 is homozygous for T and C at those sites, respectively (Fig. 2), indicating that this site is also due to cell line-based contamination. Additionally, we identified two SNPs (refSNP no. rs6902336 and rs9487562) in the integration site EU981800 that are identical to those found in the cell lines DU145 and LNCaP (Fig. 3). However, the exact origin of this site could not be determined due to the heterozygosity of the patient VP432 (G/A and C/G, respectively) at these two SNPs.
Fig. 2.
Alignment of EU981801 with VP432 patient and prostate cancer cell line genomic DNA. The genomic sequence of EU981801 (patient-derived integration site) is compared to that of patient VP432 and the cell lines DU145 and LNCaP. The annotations are identical to those in Fig. 1.
Fig. 3.
Alignment of EU981800 with VP432 patient and prostate cancer cell line genomic DNA. The genomic sequence of EU981800 (patient-derived integration site) is compared to that of patient VP432 and the cell lines DU145 and LNCaP. The annotations are identical to those in Fig. 1.
While the majority of integration sites lack SNPs and are therefore noninformative, our analyses indicate that the proviruses designated EU981808 (VP363) and EU981801 (VP432) originated not from the patient samples but from XMRV-infected cell lines. At the time of the original study on XMRV integration sites (1, 5, 6), the UCLA laboratory where the integration site mapping was performed did not culture prostate cancer cell lines nor handle mouse strains, making it unlikely that XMRV or XMRV-like sequences would have been present. However, in addition to the work with the human prostate samples, we performed analyses of XMRV integration sites in genomic DNA isolated from experimentally infected DU145 cells and clones (5, 6). We therefore believe that the integration sites EU981808 and EU981801 were artifactual and possibly a consequence of concurrent work with infected DU145 genomic DNA. Due to the lack of distinguishing SNPs in the integration sites, we cannot confirm or refute the authenticity of the remaining 12 integration sites at this time. It should be noted that there are patient-derived integration sites (GenBank no. EU981807, EU981810, and EU981811) that were cloned at the UCLA laboratory prior to any work with infected DU145 cells.
The advent of new technologies for pathogen detection has led to the identification of many candidate retroviruses thought to be involved in human disease (19). A number of these claims were later invalidated, while several remain the focus of some debate, undoubtedly due to difficulties in confirming whether the retrovirus in question represents a genuine human infection. To validate XMRV as a human retrovirus, it is important that future work on cloning XMRV integration sites from human tissues be performed free of contamination and subjected to subsequent verification, such as the SNP analysis described here.
Supplementary Material
Acknowledgments
We thank John Hackett, Jr., at Abbott Diagnostics for discussions and Gaelle Muller-Greven and Christina Gaughan at the Cleveland Clinic for excellent technical assistance.
This work was supported by the Charlotte Geyer Foundation and the Mal and Lea Bank to R.H.S., and by grant W81XWH-10-1-0268 from the U.S. Department of Defense Prostate Cancer Research Program to S.A.C. Patents are licensed to Abbott Laboratories (J.D.G. and R.H.S.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Dong B., et al. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Erlwein O., et al. 2011. DNA extraction columns contaminated with murine sequences. PLoS One 6:e23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garson J. A., Kellam P., Towers G. J. 2011. Analysis of XMRV integration sites from human prostate cancer tissues suggests PCR contamination rather than genuine human infection. Retrovirology 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hue S., et al. 2010. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology 7:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim S., et al. 2008. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J. Virol. 82:9964–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim S., et al. 2010. Fidelity of target site duplication and sequence preference during integration of xenotropic murine leukemia virus-related virus. PLoS One 5:e10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knox K., et al. 2011. No evidence of murine-like gammaretroviruses in CFS patients previously identified as XMRV-infected. Science 333:94–97 [DOI] [PubMed] [Google Scholar]
- 8. Lo S. C., et al. 2010. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc. Natl. Acad. Sci. U. S. A. 107:15874–15879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Lombardi V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585–589 [DOI] [PubMed] [Google Scholar]
- 10. Oakes B., et al. 2010. Contamination of human DNA samples with mouse DNA can lead to false detection of XMRV-like sequences. Retrovirology 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paprotka T., et al. 2011. Recombinant origin of the retrovirus XMRV. Science 333:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robinson M. J., et al. 2010. Mouse DNA contamination in human tissue tested for XMRV. Retrovirology 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rusmevichientong A., Chow S. A. 2010. Biology and pathophysiology of the new human retrovirus XMRV and its association with human disease. Immunol. Res. 48:27–39 [DOI] [PubMed] [Google Scholar]
- 14. Selten G., et al. 1986. The primary structure of the putative oncogene pim-1 shows extensive homology with protein kinases. Cell 46:603–611 [DOI] [PubMed] [Google Scholar]
- 15. Shin C. H., et al. 2011. Absence of XMRV and other MLV-related viruses in patients with chronic fatigue syndrome. J. Virol. 85:7195–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silverman R. H., Nguyen C., Weight C. J., Klein E. A. 2010. The human retrovirus XMRV in prostate cancer and chronic fatigue syndrome. Nat. Rev. Urol. 7:392–402 [DOI] [PubMed] [Google Scholar]
- 17. Tuke P. W., Tettmar K. I., Tamuri A., Stoye J. P., Tedder R. S. 2011. PCR master mixes harbour murine DNA sequences. Caveat emptor! PLoS One 6:e19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urisman A., et al. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNase L variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Voisset C., Weiss R. A., Griffiths D. J. 2008. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol. Mol. Biol. Rev. 72:157–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang G. P., Ciuffi A., Leipzig J., Berry C. C., Bushman F. D. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.