Abstract
Recombinant strains of replication-competent rhesus monkey rhadinovirus (RRV) were constructed in which strong promoter/enhancer elements were used to drive expression of simian immunodeficiency virus (SIV) Env or Gag or a Rev-Tat-Nef fusion protein. Cultured rhesus monkey fibroblasts infected with each recombinant strain were shown to express the expected protein. Three RRV-negative and two RRV-positive rhesus monkeys were inoculated intravenously with a mixture of these three recombinant RRVs. Expression of SIV Gag was readily detected in lymph node biopsy specimens taken at 3 weeks postimmunization. Impressive anti-SIV cellular immune responses were elicited on the basis of major histocompatibility complex (MHC) tetramer staining and gamma interferon enzyme-linked immunospot (ELISPOT) assays. Responses were much greater in magnitude in the monkeys that were initially RRV negative but were still readily detected in the two monkeys that were naturally infected with RRV at the time of immunization. By 3 weeks postimmunization, responses measured by MHC tetramer staining in the two Mamu-A*01+ RRV-negative monkeys reached 9.3% and 13.1% of all CD8+ T cells in peripheral blood to the Gag CM9 epitope and 2.3% and 7.3% of all CD8+ T cells in peripheral blood to the Tat SL8 epitope. Virus-specific CD8+ T cell responses persisted at high levels up to the time of challenge at 18 weeks postimmunization, and responding cells maintained an effector memory phenotype. Despite the ability of the RRVenv recombinant to express high levels of Env in cultured cells, and despite the appearance of strong anti-RRV antibody responses in immunized monkeys, anti-Env antibody responses were below our ability to detect them. Immunized monkeys, together with three unimmunized controls, were challenged intravenously with 10 monkey infectious doses of SIVmac239. All five immunized monkeys and all three controls became infected with SIV, but peak viral loads were 1.2 to 3.0 log10 units lower and chronic-phase viral loads were 1.0 to 3.0 log10 units lower in immunized animals than the geometric mean of unimmunized controls. These differences were statistically significant. Anti-Env antibody responses following challenge indicated an anamnestic response in the vaccinated monkeys. These findings further demonstrate the potential of recombinant herpesviruses as preventive vaccines for AIDS. We hypothesize that this live, replication-competent, persistent herpesvirus vector could match, or come close to matching, live attenuated strains of SIV in the degree of protection if the difficulty with elicitation of anti-Env antibody responses can be overcome.
INTRODUCTION
The difficulties in finding an effective vaccine against human immunodeficiency virus type 1 (HIV-1)/AIDS revolve principally around the properties of HIV-1 itself. Once HIV-1 establishes an initial infection, it has an uncanny ability to replicate persistently and relentlessly despite apparently strong virus-specific humoral and cellular immune responses. It is now clear that HIV, as well as its monkey counterpart simian immunodeficiency virus (SIV), has evolved specific strategies to evade intrinsic, innate, and adaptive immunity (16, 33). For adaptive immunity, the evasion strategies are multifaceted. These retroviruses have error-prone reverse transcriptases to replicate their genetic information and are able to quickly select for immune escape variants (both humoral and cellular) that are able to replicate efficiently in the face of ongoing immune responses (8, 9, 24, 53). The viruses have constructed their envelope glycoprotein spikes in a way that makes it difficult for antibodies to access and difficult for them to neutralize viral infectivity (10, 22, 49, 57). One of the viral gene products, Nef, downregulates major histocompatibility complex (MHC) class I molecules from the surfaces of infected cells, making these infected cells less susceptible targets of virus-specific CD8+ T cells (13, 58, 60). There is enormous sequence variability in HIV-1 circulating in the population, and it is not clear how a vaccine will be able to provide protection against this great diversity of sequences. Finally, the main target cell for viral replication is the CD4+ T lymphocyte, a major orchestrator of effective immune responses.
One vaccine approach that has worked reasonably well—much better than other vaccine approaches, in fact—against SIV in monkeys is live attenuated deletion mutant strains of virus (14, 31, 68). While live attenuated strains of SIV with Nef deleted have protected well against homologous challenge or challenge by strains closely matched in sequence, even live attenuated SIV has not performed so well against challenge by heterologous strains not closely matched in sequence (52, 67). This inability to protect well against sequence-mismatched strains of SIV may be analogous to the inability of infection by one HIV-1 strain to routinely protect against superinfection by a different circulating strain of HIV-1 (5, 50).
Through complex genetic engineering, strains of SIV that are capable of a single round of infection and a single round of virus production but are incapable of further spread to other cells have been constructed (19, 20). Repeated administrations of such “single-cycle SIV” can provide some level of protection against subsequent challenge by homologous SIV, but the degree of protection is nowhere near as great as that observed with live attenuated strains (30). One possible explanation for this observation is that anamnestic immune responses may simply not be sufficient to respond in time to blunt replication of a wild-type challenge strain, even one that is totally matched in sequence.
Herpesviruses provide one potential means to examine the importance of persistent antigen expression for the degree of protection. Herpesviruses have large double-stranded DNA genomes that can stably accommodate large amounts of inserted foreign DNA. They persist for life, and immune responses to them in terms of effector cells persist at readily detectable levels for life (59). Eight distinct human herpesviruses have been identified, and each falls into one of the distinct subgroups: alphaherpesvirus, betaherpesvirus, gamma-1 herpesvirus, and gamma-2 herpesvirus. Herpesviruses from different subgroups can be quite distinctly different, targeting different types of cells for primary replication and different types of cells for persistence, and they even carry different compositions of genes. To date, the use of recombinant herpesvirus vectors to express SIV antigens in monkeys has been reported for the alphaherpesvirus herpes simplex virus (HSV) (35, 47) and for the betaherpesvirus cytomegalovirus (CMV) (26).
Here, we report the construction and performance of recombinants of the gamma-2 herpesvirus of rhesus monkeys, rhesus monkey rhadinovirus (RRV) (2, 17).
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney cells (293T) were maintained on Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 2 mM l-glutamine, and penicillin-streptomycin (50 IU and 50 μg/ml, respectively) at 37°C in a humidified incubator with 5% CO2. Rhesus macaque skin fibroblasts (RF) were maintained in DH20 (DMEM supplemented with 20% fetal calf serum, 2 mM l-glutamine, penicillin-streptomycin [50 IU and 50 μg/ml, respectively], and 10 mM HEPES) at 37°C in a humidified incubator with 5% CO2.
Cosmid cloning.
Procedures for insertion of foreign sequences upstream of the RRV R1 reading frame in the leftmost cosmid clone, ah28ΔA/H, have been described previously (6). Specific features of the recombinants used for the current study are illustrated in Fig. 1 and described here. For insertion of the expression-optimized SIV envelope gene driven by the elongation factor 1 promoter (EF1-SIVenv-eo), complementary oligonucleotides, 5′-CTAGTTGTTTAAACGGGGCGCCGGA-3′ and 5′-CTAGTCCGGCGCCCCGTTTAAACAA-3′, were annealed at 55°C and phosphorylated using T4 polynucleotide kinase, forming an SpeI-PmeI-SpeI adaptomer. The adaptomer featured a cut SpeI site at each end flanking a central PmeI site. For insertion of the codon-optimized SIV gag gene driven by the CMV promoter (CMV-SIVgag) and the SIV rev-tat-nef fusion construct driven by a simian virus 40 (SV40) promoter (SV40-SIVRTN), complementary oligonucleotides, 5′-CTAGTGGCTAGGGATAACAGGGTAATA-3′ and 5′-CTAGTATTACCCTGTTATCCCTAGCCA-3′, were annealed and phosphorylated as before to form an SpeI-ISceI-SpeI adaptomer. The adaptomer featured a cut SpeI site at each end flanking a central ISceI site. The ah28ΔA/H cosmid was linearized at base pair 206 with SpeI and dephosphorylated using calf intestinal phosphatase (CIP). Subsequently, the linearized ah28ΔA/H cosmid was ligated to the SpeI-PmeI-SpeI or SpeI-ISceI-SpeI adaptomer, yielding ah28ΔA/H-PmeI or ah28ΔA/H-ISceI, respectively.
Fig. 1.
Schematic representation of recombinant RRV-SIV constructions. The site of insertion into the leftmost RRV cosmid clone is as described by Bilello et al. (6). The transcriptional elongation factor 1 promoter region was used to drive expression of a codon-optimized SIVmac239 gp160 envelope sequence. The CMV immediate-early promoter was used to drive expression of a codon-optimized SIVmac239 Gag sequence, and the SV40 promoter was used to drive expression of a Rev-Tat-Nef fusion protein.
Each SIV expression insert was designed to be noncomplementary to the others in order to avoid recombination events when subsequent SIV-recombinant RRV viruses were used to coinfect monkeys. To generate the ah28ΔA/H EF1-SIVenv cosmid (Fig. 1), expression-optimized SIVenv sequences were excised from a modified p64s S23T plasmid (obtained from E. Yuste, New England Primate Research Center [NEPRC], Southborough, MA) and ligated into pEF1 p(A), a pEF1-mycHisA plasmid (Invitrogen) that was altered to contain (i) an HSV thymidine kinase poly(A) sequence, HSVtk p(A), downstream from the XbaI site within the plasmid and (ii) an additional PmeI restriction endonuclease site upstream from the EF1 promoter. Briefly, the pEF1-mycHisA plasmid was digested with NotI and XbaI and ligated to an adaptomer containing the HSVtk p(A) sequence flanked by NotI and XbaI. This adaptomer was formed in the same manner described above using complementary oligonucleotides, 5′-GGCCGCAATAAAAAGACAGAATAAAT-3′ and 5′-CTAGATTTATTCTGTCTTTTTATTGC-3′. To insert the PmeI restriction endonuclease site upstream from the EF1 promoter, an adaptomer containing the PmeI restriction site flanked by MluI restriction sites was formed in the same manner as described above using complementary oligonucleotides, 5′-CGCGTTGTTTAAACGGGGCGCCGGA-3′ and 5′-CGCGTCCGGCGCCCCGTTTAAACAA-3′. The pEF1-mycHisA plasmid was digested with MluI and ligated to this adaptomer. The p64s S23T plasmid was modified to contain a KpnI restriction endonuclease recognition site by the ligation of a EcoRI-KpnI-EcoRI adaptomer into the EcoRI site just upstream from the expression-optimized SIVenv gene. This adaptomer was formed in the same manner as described above using complementary oligonucleotides, 5′-AATTCCGCGGATCCGCGGGGTACCG-3′ and 5′-AATTCGGTACCCCGCGGATCCGCGG-3′. Finally, pEF1 p(A) and the modified p64s S23T were digested with KpnI and gel extracted. Following dephosphorylation of pEF1 p(A) with CIP (NEB), the two products were ligated together to make the pEF1-64s plasmid. The ah28ΔA/H-PmeI cosmid was digested with PmeI, dephosphorylated with CIP, and gel extracted using the QiaExII kit (Qiagen). The expression-optimized SIV env gene driven by the EF1 promoter was excised from the pEF1-64s plasmid by digestion with PmeI, gel extracted, and ligated to the ah28ΔA/H-PmeI fragment to generate the ah28ΔA/H EF1-SIVenv cosmid.
To generate the ah28ΔA/H SV40-RTN cosmid (Fig. 1), the SIV rev-tat-nef (RTN) sequence was excised from the pcDNA/RTN plasmid (the kind gift of David Knipe, Harvard Medical School) by digestion with BamHI and ligated into a modified pSG5 plasmid that was digested with BamHI and dephosphorylated using CIP. The pSG5 plasmid (Stratagene) was modified to contain the SV40 promoter, a multicloning site containing a single BamHI restriction endonuclease site, and the SV40 poly(A) sequence flanked by ISceI restriction endonuclease recognition sites, giving rise to the pSG5-RTN-B plasmid. The ISceI site upstream from the SIV-RTN sequence was generated by QuikChange (Agilent Technologies) mutagenesis following the manufacturer's protocol using the following oligonucleotides: 5′-CGGCCAGTGAATTGTCGACTAGTGAGGCGGAAAGAACCAGCTG-3′ and 5′-CAGCTGGTTCTTTCCGCCTCACTAGTCGACAATTCACTGGCCG-3′. The ISceI site downstream from SIV-RTN was created by insertion of a BglII-ISceI-BglII adaptomer formed as described above using complementary oligonucleotides, 5′-GATCTGGCTAGGGATAACAGGGTAATA-3′ and 5′-GATCTATTACCCTGTTATCCCTAGCCA-3′. The ah28ΔA/H-ISceI cosmid was digested with ISceI, dephosphorylated with CIP, and gel extracted using the QiaExII kit (Qiagen). The SIV-RTN sequence driven by the SV40 promoter was excised from the modified pSG5 plasmid by digestion with ISceI, gel extracted, and ligated to the ah28ΔA/H-ISceI fragment to generate the ah28ΔA/H SV40-RTN cosmid. Insertion of the SV40 SIV-RTN fragment occurred in the antisense orientation of the ah28ΔA/H ISceI cosmid relative to the R1 reading frame in this cosmid.
To generate the ah28ΔA/H CMV-SIVgag cosmid (Fig. 1), the codon-optimized SIV gag sequence was excised from the pTkdGag plasmid and inserted into a modified pcDNA3.1+ (Invitrogen, Carlsbad, CA) plasmid containing the CMV immediate-early promoter (CMV-IE) and the bovine growth hormone (BGH) poly(A) sequence. An ISceI restriction endonuclease sequence was introduced both 5′ and 3′ of the CMV-IE promoter-SIV gag-BGH poly(A) sequence in a manner similar to that described above, giving rise to the pcDNA/gag plasmid. The codon-optimized SIV-gag sequence driven by the CMV promoter was excised from pcDNA/gag by digestion with ISceI, gel extracted, and ligated to the ah28ΔA/H-ISceI fragment to generate the ah28ΔA/H CMV-SIVgag cosmid.
DNA sequencing.
Cosmid and plasmid constructs were sequenced with a CEQ 8000 Genetic Analysis System using a dye terminator cycle-sequencing kit as specified by the manufacturer (Beckman Coulter, Fullerton, CA).
Cotransfection and virus preparation.
Prior to transfection, overlapping cosmids necessary to reconstruct the RRV26-95 genome, including either the ah28ΔA/H EF1-SIVenv, ah28ΔA/H CMV-SIVgag, or ah28ΔA/H SV40-RTN cosmid, were digested overnight with the ICeuI homing endonuclease to remove the RRV26-95 sequence from the pSuperCos 1 backbone vector as described by Bilello et al. (6). The cosmid DNA was precipitated by adding 3 volumes of 5% 3 M sodium acetate/95% ethanol and incubating it for >1 h at −20°C. The DNA was then pelleted by centrifugation for 10 min at maximum speed in a microcentrifuge. The pellets were washed in 70% ethanol, dried, and rehydrated in H2O. One day postseeding, 293T cells (4.5 × 105 cells/well in 6-well plates) were transfected with different combinations of digested overlapping cosmids (0.4 μg each cosmid) using Transfectin reagent (Bio-Rad Laboratories, Hercules, CA) following a scaled-down procedure. As a positive control, 0.25 μg of whole viral RRV DNA isolated from column-purified RRV26-95 was transfected in the same manner. At 5 days posttransfection, cell-free culture supernatant was collected and stored at 4°C. To amplify recombinant stocks generated in 293T cells, fresh rhesus monkey fibroblast cultures were inoculated with 1 ml of the supernatant collected from the 293T transfections. The inoculated fibroblast cultures were passaged until the emergence of viral plaques was observed in the cultures, and then the cultures were maintained without splitting until complete lysis of the fibroblast monolayer occurred. High-titer recombinant RRV stocks were subsequently generated in fresh rhesus fibroblast cultures.
Isolation and analysis of RRV DNA.
For each RRV, supernatant collected following complete lysis of RRV-infected rhesus fibroblasts was subjected to low-speed centrifugation to remove cellular debris. The supernatant was then filtered through a 0.45-μm-pore-size filter to remove any additional debris. The filtered supernatant was then centrifuged for 3 h at 45,000 × g in a Sorvall Type 19 rotor to pellet the virus. The crude virus was resuspended in Tris-EDTA (TE) buffer and lysed by adding 0.1 volume 1% N-lauroylsarcosine and proteinase K and incubating it at 60°C for 1 h. The mixture was extracted twice with phenol-chloroform, followed by four chloroform washes. The DNA was recovered by precipitation with 2.5 volumes 5% 3 M sodium acetate/95% ethanol, rinsed in 80% ethanol, and resuspended in Tris-EDTA buffer. Viral DNA was digested with restriction endonucleases, separated on a 0.5% agarose electrophoretic gel, and stained with ethidium bromide.
Plaque assay.
The titers of parental RRV26-95 and recombinant RRV stocks were determined as previously described (18). Briefly, cell-free culture supernatant was collected following complete lysis of RRV-infected rhesus fibroblasts. Fresh fibroblasts were seeded into 12-well plates at 2 × 105 cells/well. The following day, 10-fold serial dilutions of the virus-containing supernatant were made in DH20. The media were removed from the RF cultures and replaced with 200 μl of diluted virus/well. The cultures were then incubated for 1 h at 37°C with gentle rocking every 15 min. After 1 h, 2 ml of Hank's buffered saline solution (HBSS) was added to each well and subsequently aspirated. Two milliliters of overlay medium (a 1:1 ratio of 2× DMEM and 1.5% methylcellulose [Sigma, St. Louis, MO] supplemented with 2% fetal calf serum) was applied, and the cultures were incubated at 37°C and 5% CO2 for 1 week. The overlay medium was aspirated, and a staining solution (0.8% crystal violet in 50% ethanol) was applied for 10 min. Each well was washed 5 times with distilled water, and the number of plaques at each dilution of inoculum was determined.
Transfection of positive-control plasmids.
One day postseeding, 293T cells (4.5 × 105 cells/well in 6-well plates) were transfected with either the pEF1-64s (for SIV env), pcDNA/gag, or SG5-RTN-B (for SIV rev-tat-nef) expression plasmid using Transfectin reagent (Bio-Rad Laboratories, Hercules, CA) following a scaled-down procedure.
Immunoblotting.
At 48 h posttransfection or 4 days postinfection, cultures were rinsed with phosphate-buffered saline (PBS) and lysed with RIPA buffer (Boston BioProducts, Boston, MA). The lysates were sonicated at 20% for 10 s, and the debris was spun down at maximum speed in a microcentrifuge for 1 min. These proteins or those obtained by lysis of SIVmac239 isolates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked overnight at room temperature in 5% milk in phosphate-buffered saline containing 0.1% Tween 20 (PBS-T; Sigma, St. Louis, MO). Primary antibody to SIVenv gp41/gp160 (KK41 from the NIH AIDS reagent repository), SIVenv gp120 (3.11H; a kind gift from James Robinson, Tulane University), SIVgag (N27), or SIV-nef (from the NIH AIDS reagent repository) diluted in 5% milk in PBS-T was applied and rocked at room temperature for 1 h. After successive washings in PBS-T, the blots were incubated in secondary anti-mouse (Santa Cruz) diluted in 5% milk in PBS-T. The blots were washed in PBS-T, and antibody binding was detected using the SuperSignal West Pico chemiluminescent reagent (Pierce) and a Fuji LAS3000 chemiluminescence imager (Bio-Rad).
Antibody detection by ELISA.
Purification of RRV, coating of enzyme-linked immunosorbent assay (ELISA) plates, and detection of anti-RRV antibody responses by ELISA have been described previously (45). Detection of anti-SIV antibody responses by ELISA used procedures described by Desrosiers et al. (15) except that SIVmac239 gp120 protein was used for the coating of ELISA plates.
IFN-γ ELISPOT assays.
SIV-specific T cell responses were enumerated using an enzyme-linked immunospot (ELISPOT) assay for detection of macaque gamma interferon (IFN-γ) (Mabtech, Mariemont, OH) using standard methodologies as previously described (1, 63). Briefly, freshly isolated peripheral blood mononuclear cells (PBMC) in R10 medium were plated in duplicate at 3 × 105 cells/well and incubated with overlapping peptide pools corresponding to the SIVmac239 Gag, Env, Rev, Tat, and Nef sequences at a final concentration of 2 μg/ml for individual peptides. The numbers of spots (representing IFN-γ-producing T cells) were detected in a colorimetric assay for bound IFN-γ and were counted using an automated ELISPOT plate reader (Zellnet Consulting, New York, NY). To determine the absolute numbers of SIV-specific IFN-γ-producing T cells per 106 PBMC, the number of background spots in medium-only control wells was subtracted from the number of spots in peptide-stimulated wells. Concanavalin A-stimulated PBMC served as a positive control. The peptides were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, or were synthesized by the Massachusetts General Hospital Peptide Core Facility, Boston, MA.
Viral load measurement.
Plasma viral loads were determined by real-time PCR methods as described previously (12). For each sample, 1.5 ml of plasma was processed to yield a threshold limit of detection of 10 Gag RNA copy equivalents per ml.
Flow cytometric analysis.
Staining and phenotyping of SIV tetramer binding cells was performed as previously described (56, 61) with modifications as noted below. Briefly, 200 μl of whole blood was incubated with titrated amounts of tetramers in 100 μl PBS with 2% fetal bovine serum (FBS) at 37°C and 5% CO2 for 60 min in the dark and then stained with the monoclonal antibodies indicated below. For demonstration of the effector memory phenotype, PBMC (1 × 106 to 2 × 106) were incubated with tetramers for 45 min in the dark prior to staining with monoclonal antibodies. Mamu-A*01 class I tetramers conjugated to phycoerythrin (PE) and complexed with the A*01 Gag181-189CM9 (4) and A*01 Tat28-35SL8 (3, 38) epitopes were kindly provided by Nancy Wilson and David Watkins (Wisconsin National Primate Research Center, Madison WI). For intracellular staining, samples were fixed with Caltag Fix and Perm Solution A (Burlingame, CA) for 10 min at room temperature, washed, incubated with intracellular antibodies in Caltag Fix and Perm Solution B, and then processed as described above. Samples were analyzed on a Becton Dickinson (BD) FACSCalibur or BD LSR-II flow cytometer (BD Immunocytometry Systems, San Jose, CA). Anti-mouse IgG(κ/κ)-negative polystyrene beads (BD CompBeads; BD, CA) or PBMC stained separately with individual monoclonal antibodies used in the test samples were used for electronic compensation. Gates for the expression of perforin, CCR7, and CD127 were established using fluorescence-minus-one (FMO) controls (54).
The following fluorescently labeled monoclonal antibodies were obtained from BD Biosciences (La Jolla, CA): CD3-fluorescein isothiocyanate (FITC) (SP34-2), CD3-allophycocyanin (APC)-Cy7 (SP34-2), CD4-peridinin chlorophyll protein complex (PerCP) (L-200), CD8a-PerCP, CD8a-Alexa 700 (RPA-T8), and CD28-PE-Texas Red (L293). Perforin-FITC (Pf-344) was purchased from Mabtech (Cincinnati, OH). CCR7 (150503; R&D Systems, Minneapolis, MN) was used as a custom Pacific Blue conjugate prepared at the NEPRC. All antibodies were tested for cross-reactivity and titrated on rhesus macaque PBMC to determine optimal staining concentrations and conditions.
Immunohistochemistry.
Immunohistochemistry was performed on lymph node biopsy specimens obtained 3 and 9 weeks following vaccination to identify Gag-expressing cells using an avidin-biotin complex immunostain technique, as previously described (28). Briefly, immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue sections, which were deparaffinized in xylene and rehydrated through graded ethanol to distilled water. Incubation in 3% H2O2 was used to block endogenous peroxidase activity and was followed by 60 min of incubation with the primary monoclonal antibody directed against SIVp27 (clone 183-H12-5C; NIH AIDS Research and Reference Reagent Program). The sections were sequentially treated with a biotinylated secondary antibody and horseradish peroxidase-conjugated streptavidin. The chromogenic substrate 3,3′-diaminobenzidine (DAB) (Dako Corp.) was used to localize antigen-antibody complexes. Tissue sections were counterstained with Mayer's hematoxylin (Sigma-Aldrich, St. Louis, MO), cleared, and coverslipped with permanent mounting medium. An isotype-matched irrelevant control antibody was used on all sections to control for nonspecific staining.
Animal studies.
Rhesus macaques (Macaca mulatta) of Indian ancestry were specific pathogen free of B virus, simian T-lymphotropic virus, simian retrovirus, and SIV. The animals were housed at the NEPRC and maintained in accordance with the Guide for the Care and Use of Laboratory Animals of the Institute of Laboratory Resources, National Research Council. The facility is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International, and all work was approved by Harvard Medical School's Standing Committee on Animals. The RRV serostatus was evaluated by ELISA prior to the initiation of the study as previously described (17). In the vaccine phase of the study, five animals were intravenously inoculated with a mixture of RRV-SIV recombinants (1 × 105 PFU of each recombinant), and one control animal was inoculated with 3 × 105 PFU of the parental cloned RRV26-95. The animals were followed prospectively with sequential blood draws and lymph node biopsies performed at 3 and 9 weeks. At 18 weeks, these animals and two additional seropositive controls were challenged intravenously (i.v.) with 10 infectious doses of SIVmac239. Euthanasia criteria were developed prior to the initiation of the study and were carried out in accordance with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
Statistical analyses.
Geometric means and associated 95% confidence intervals (CI) of peak viral loads were computed for vaccinated and control groups. Between-group differences in log10-transformed peak viral loads were tested using a two-sided t test. Between-group differences in geometric means were also expressed as a ratio of the vaccinated group to the control group, as well as percent reduction. Geometric means of set point viral loads (weeks 6 to 32) for the vaccinated and control groups were estimated using a linear mixed model with a random intercept for each subject. The data were centered at week 16 postchallenge (the midpoint between weeks 6 and 32). The model accounts for the correlations among the repeated measurements on the same experimental subject. Based on the same model, differences in set point values were estimated and tested for statistical significance using the Wald test. The estimated difference was then expressed as a ratio and as percent reduction.
RESULTS
Construction of RRV recombinants.
A genetic system that employs overlapping cosmid clones of RRV has been described (6). A unique ISceI site in the leftmost cosmid clone (ah28) was used as the site of insertion of SIV gene expression cassettes (6) (Fig. 1). The promoter region of cellular transcriptional elongation factor 1 (EF1) was used to drive expression of a codon-optimized (expression-optimized) version of the env gene of SIVmac239 (27, 51). The CMV immediate-early promoter was used to drive expression of an expression-optimized version of the SIVmac239 gag gene (27, 51), and the SV40 promoter was used to drive expression of a rev-tat-nef fusion construct (64). Replication-competent recombinant RRV strains were generated by transfection of overlapping cosmid clones and expanded on early-passage rhesus monkey fibroblast cultures as described previously (6).
Permissive rhesus fibroblasts were infected in culture with each recombinant RRV, and cell lysates were prepared and analyzed for the presence of the appropriate SIV gene product by immunoblotting using appropriate monoclonal antibodies to individual products (Fig. 2). The recombinant RRV-SIVenv made the expected SIV Env products gp160, gp120, and gp41. These Env proteins were readily detected and were as abundant as in 293T cells transfected with the expression plasmid containing the codon-optimized sequences (Fig. 2A). The RRV-SIVgag recombinant expressed a Gag protein of the expected 55 kDa as the principal product, as well as apparent proteolytic breakdown products (Fig. 2B). The RRV-SIVrev-tat-nef recombinant expressed a protein product of approximately 60 kDa, the predicted mass of the Rev-Tat-Nef fusion protein, as well as lower-molecular-weight (MW) bands that are likely proteolytic breakdown products (Fig. 2C). The predicted mass of the Rev-Tat-Nef fusion protein is 56 kDa.
Fig. 2.
Demonstration of expression of the desired SIV proteins in rhesus fibroblast cultures infected with RRV-SIV recombinants. Cell lysates were prepared, proteins were separated by SDS-polyacrylamide gel electrophoresis, and the separated proteins were Western blotted to a filter and reacted with SIV-specific antibodies. (A) RRV-env1 and RRV-env2 represent lysates from cells infected with two independently derived stocks of RRV-SIVenv. RRV26-95 and RRV-GFP are also infected-cell lysates. pEF1-64s is a cell lysate from 293T cells transfected with the expression plasmid for SIVmac239 Env. In the top gel, blots were reacted with an antibody specific for the Env surface subunit (gp120), and in the lower gel, blots were reacted with a monoclonal antibody that recognizes the Env transmembrane subunit (gp41). (B) RRV-gagA and RRV-gagB represent lysates from cells infected with two independently derived stocks of RRV-SIVgag. pcDNA/gag is a cell lysate from 293T cells transfected with the expression plasmid for codon-optimized SIV gag. (C) Western blots for detection of Rev-Tat-Nef fusion protein. RRV-RTN-2, -3, -5, and -6 represent lysates from cells infected with four independently derived stocks of RRV-SIVrev-tat-nef. SG5-RTN-B is a cell lysate from 293T cells transfected with the expression plasmid for the Rev-Tat-Nef fusion protein.
RRV virions were purified, DNA was isolated, and the virion DNA was subjected to restriction endonuclease analysis (Fig. 3). The restriction endonuclease BamHI was particularly useful for discriminating left-end fragments in recombinant versus parental DNA. In each case, a left-end BamHI fragment present in the parental strain was lost and replaced in the recombinant strains by a higher-MW band of the expected size for the newly inserted fragment (Fig. 3). Digestion of RRV-SIV recombinant virus DNA with SphI or SpeI restriction endonuclease also showed changes in fragmentation patterns indicative of the inserted SIV expression cassettes. These restriction endonuclease analyses revealed no additional changes to the composition of the virion DNA other than the recombinant DNA fragment that was introduced.
Fig. 3.
Restriction endonuclease analysis of recombinant RRV-SIV strains confirms the expected genetic changes. DNA was extracted from concentrated virions and digested with the indicated restriction endonucleases, and the fragmented DNA was analyzed by agarose gel electrophoresis. U, undigested; E, EcoRI; B, BamHI. Fragments with altered mobility due to the insertion of an SIV expression cassette are marked with arrowheads.
Vaccine phase.
Five rhesus monkeys were enrolled in a vaccine study for evaluation of these RRV-SIV recombinants. Three of the monkeys were RRV negative at the time of enrollment, and two were intentionally selected as naturally infected and RRV positive (Table 1). The use of two RRV-positive monkeys allowed us to subsequently evaluate the impact of prior RRV infection on the take of the recombinant RRV vaccine strains. Four of the five monkeys were Mamu-A*01 positive to allow convenient use of MHC tetramers for the evaluation of virus-specific CD8+ T cell responses to defined epitopes (Table 1). A mixture of the three RRV-SIV recombinant strains was made to contain equivalent numbers of PFU, and the mixture was inoculated intravenously into each of the five test animals on the day of vaccination (DOV) (Table 1). One of the three control monkeys (309-04) was experimentally infected with the parental cloned RRV26-95 at this time, and the other two control monkeys (390-93 and 328-04) were naturally RRV positive (Table 1).
Table 1.
Characteristics of rhesus monkeys used in vaccine trials
| Monkey | Inoculum (DOV)a | Mamu A*01 | Mamu A*02 | Mamu B*17 | Mamu B*08 | RRV status |
|
|---|---|---|---|---|---|---|---|
| DOV | DOCb | ||||||
| 166-91 | RRV-SIV | +d | + | − | − | − | + |
| 175-91 | RRV-SIV | + | − | − | − | − | + |
| 440-92 | RRV-SIV | + | − | − | − | + | + |
| 128-04 | RRV-SIV | − | − | − | − | − | + |
| 247-04 | RRV-SIV | + | − | − | − | + | + |
| 309-04 | RRV | − | − | − | − | − | + |
| 390-93 | Nonec | + | + | − | − | + | + |
| 328-04 | Nonec | + | + | − | − | + | + |
DOV, day of vaccination. 440-92 and 247-04 were already RRV positive as a consequence of natural RRV infection at the time of RRV-SIV administration.
DOC, day of challenge.
Monkey was naturally RRV positive at the time of enrollment.
+, positive; −, negative.
Immunohistochemistry was used to localize Gag-expressing cells in peripheral lymph node biopsy specimens obtained from vaccinated animals. Small to moderate numbers of Gag-expressing cells were identified (Table 2 and Fig. 4) in RRV-seronegative animals receiving the RRV-SIV vaccine at 3 weeks postinoculation. Rare Gag-positive cells were also observed in one of the two RRV-positive monkeys (440-92) that received RRV-SIV. Cytoplasmic staining was observed primarily in cells in the parafollicular cortex. No staining was observed in animals receiving the cloned or uncloned RRV or in irrelevant antibody control sections. Staining was absent in biopsy specimens taken 9 weeks following inoculation.
Table 2.
Localization of Gag-expressing cells by immunohistochemistry
| Animal | Vaccine | Prior RRV status | IHC Gag staininga |
|---|---|---|---|
| 166-91 | RRV-SIV | Negative | Positive (+) |
| 175-91 | RRV-SIV | Negative | Positive (++) |
| 128-04 | RRV-SIV | Negative | Positive (+) |
| 247-04 | RRV-SIV | Positive | Negative |
| 232-04 | Uncloned RRV | Negative | Negative |
| 309-04 | Cloned RRV | Negative | Negative |
| 440-92 | RRV-SIV | Positive | Positive (+) |
+, small, and ++, moderate numbers of Gag-positive cells. IHC, immunohistochemistry.
Fig. 4.
Detection of SIVgag in peripheral lymph node biopsy specimens by immunohistochemistry. (A) SIV Gag-expressing cells in a lymph node biopsy specimen from animal 175-91 (inset: positive cell) inguinal lymph node; DAB chromogen. (B) Irrelevant antibody control.
SIV-specific T cell responses after inoculation with RRV-SIV recombinants were followed using IFN-γ ELISPOT assays and MHC class I tetramers. IFN-γ ELISPOT assays were carried out using overlapping peptide pools corresponding to the Gag, Env, Rev, Tat, and Nef open reading frames. In the RRV-naive animals 166-91, 175-91, and 128-04, robust ELISPOT responses to Gag were observed 3 weeks after infection (>1,000 spot-forming cells [SFC] per 106 PBMC) with little apparent decay at 9 weeks postinoculation (Fig. 5A). Lower-level responses to Gag were observed in the RRV-seropositive animals, and they decreased at 9 weeks postinoculation. Clearly positive, although low-level, responses to Tat, Rev, and Env were also observed, which were higher in RRV-seropositive animals, while no significant responses to Nef were observed in either group of vaccinated animals. The lowest ELISPOT responses were observed in an RRV-seropositive monkey (440-92), but the other RRV-seropositive monkey, 247-04, showed robust responses to Gag and detectable responses to Tat and Env, as well (Fig. 5A).
Fig. 5.
SIV-specific IFN-γ ELISPOT responses in RRV-SIV-vaccinated macaques (Mm). PBMC were obtained at the indicated time points, and SIV-specific responses to the indicated SIV peptide pool were calculated after subtraction of spots obtained in wells incubated with R10 medium alone. (A) Vaccine phase. (B) Challenge phase. (C) Challenge phase for the unvaccinated control monkeys. *, monkeys that were already RRV positive at the time of RRV-SIV vaccination.
Analysis of SIV-specific CD8+ T cell responses using MHC tetramers to the immunodominant Mamu A*01-restricted Gag181-189CM9 (4) and Tat28-35SL8 (3, 38) epitopes revealed a similar pattern of immune response after inoculation with recombinant RRV. Representative flow cytometric data for Mamu-A*01 Gag CM9 tetramer binding cells are shown in Fig. 6 for the RRV-naive animal 166-91. A low frequency (0.25% of CD8+ T cells) of Gag CM9 tetramer binding cells was first observed at 2 weeks postinoculation, rapidly increased to >9% by 3 weeks postinoculation, and was sustained at relatively high levels thereafter until the time of challenge at week 18 (Fig. 7A). A similar pattern was observed in the other RRV-naive animal, 175-91. These sustained high-level Gag-specific responses in RRV-naive animals with only minor decreases over periods of >10 weeks are strongly suggestive of ongoing antigenic stimulation. Gag-specific tetramer responses in RRV-seropositive animals had lower peaks and, in animal 247-04, a more accentuated decay. A similar pattern was observed for Tat SL8-specific responses in both RRV-naïve and RRV-seropositive animals, although the overall frequencies of Tat-specific responses in RRV-naive and RRV-seropositive animals were lower than for Gag-specific cells (Fig. 7B).
Fig. 6.
Frequency of Mamu-A*01 Gag CM9 tetramer binding cells in macaque 166-91. PBMC were obtained at the indicated times postinoculation (PI) from monkey 166-91 (RRV naive). The frequencies of tetramer binding cells in CD3+ CD8+ lymphocytes are shown. DOI, day of inoculation.
Fig. 7.
Summary of Gag CM9 and Tat SL8 tetramer binding cells in RRV-inoculated animals. Macaques 166-91 and 175-91 were RRV naive; macaques 440-92 and 247-04 were RRV seropositive. Macaques 390-93 and 328-04 were unvaccinated controls that were inoculated with SIV at week 19. All of these macaques expressed the Mamu-A*01 allele. *, monkeys that were already RRV positive at the time of RRV-SIV vaccination.
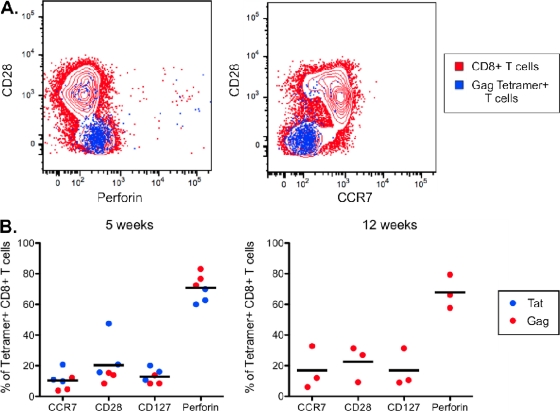
One of the distinguishing characteristics of persistent viral infections is their ability to induce long-lived effector memory CD8+ T cell responses (23). Phenotypic analysis using Gag and Tat tetramer binding cells in RRV-vaccinated animals revealed a characteristic effector memory phenotype. Analysis of both Gag and Tat tetramer binding cells at 5 weeks after inoculation revealed that SIV-specific cells were predominantly CD28− CCR7− CD127− and perforin positive, with no significant evolution between 5 and 12 weeks (Fig. 8A and B).
Fig. 8.
SIV-specific CD8+ T cells in RRV-vaccinated animals display an effector memory phenotype. (A) Representative flow cytometry data from monkey 175-91 showing the phenotype of Gag tetramer binding cells (blue) compared with the bulk CD8+ T cell population (red) at 12 weeks postinoculation. (B) Summary of the phenotype of Gag and Tat tetramer binding cells in RRV-vaccinated animals. The frequency of Tat SL8 tetramer binding cells at 12 weeks was too low to provide reliable phenotypic analysis. Samples from animal 440-92 were also excluded due to too few positive events.
Serum samples taken on the DOV and over the subsequent weeks were used to evaluate antibody responses to RRV by whole-lysed-virus ELISA (Fig. 9). The two RRV-positive monkeys, 440-92 and 247-04, as expected, already had strong antibody reactivity to RRV proteins on the DOV. The other three test monkeys (166-91, 175-91, and 128-04) and the RRV-inoculated control monkey for this study (309-04), as well as one additional control (232-04), also as expected were RRV negative on the DOV, but all convincingly seroconverted to strong anti-RRV antibody reactivity over the subsequent weeks (Fig. 9A). Analysis of serial dilutions of serum demonstrated that by 12 weeks the six seroconverting monkeys had achieved anti-RRV antibody titers similar to those of monkeys 440-92 and 247-04, which were naturally infected with RRV (Fig. 9B).
Fig. 9.
Antibody responses to RRV during the vaccine phase. Antibodies to RRV were measured by ELISA using whole lysed virions for detection, as previously described (17). Monkeys 440-92 and 247-04 were already RRV positive at the time of enrollment. The straight red line without data points represents the reactivity of a positive control (140-83) at a straight 1:10 dilution of serum. The black dashed line represents the reactivity of a negative control (288-94) at a straight 1:10 dilution of serum. (A) A straight 1:10 dilution of serum was used at the indicated weeks after vaccine administration. (B) Serial dilutions of serum taken at week 12 after vaccine administration were tested for reactivity to RRV virions. *, monkeys that were already RRV positive at the time of RRV-SIV vaccination.
Despite the ready detection of SIV Env protein in tissue culture cells infected with the RRV-SIVenv recombinant and despite the strong take of RRV-SIV when inoculated into the RRV-negative monkeys, we were unable to detect anti-Env antibody responses in the inoculated monkeys. The methods used included standard and high-sensitivity ELISA to gp120, Western blotting, and the appearance of neutralizing activity in serum to the very neutralization-sensitive laboratory-adapted SIVmac251 (46).
Challenge phase.
At 18 weeks, the five immunized monkeys and the three immunized controls were challenged intravenously with 10 infectious doses of cloned SIVmac239. The preparation and titration of this stock has been described previously (40), and it has been used extensively by a number of different laboratories for controlled-dose challenges (11, 20, 21, 29, 41, 47). On the basis of SIV RNA burdens in plasma (Fig. 10) and seroconversion (Fig. 11), all 5 immunized monkeys and all 3 controls became infected with SIV.
Fig. 10.
SIV RNA loads in plasma following challenge. *, monkeys that were already RRV positive at the time of RRV-SIV vaccination.
Fig. 11.
Antibody responses to SIV gp120 following SIV challenge. Antibodies were measured by ELISA at a straight 1:200 dilution of serum. *, monkeys that were already RRV positive at the time of RRV-SIV vaccination.
Viral loads in vaccinated monkeys at peak height (0.49 × 106 copies of viral RNA per ml of plasma) were 1.85 log10 units lower than those of control monkeys (35.3 × 106 copies of viral RNA per ml of plasma). This 70-fold (98.6%) reduction was statistically significant using a two-sided two-independent-group t test for log10(viral loads) (P = 0.006). Viral loads in vaccinated monkeys during the chronic phase of infection, weeks 6 to 32 postchallenge (0.25 × 105 copies of viral RNA per ml of plasma), were on average 1.80 log10 units lower than those of control monkeys (15.5 × 105 copies of viral RNA per ml of plasma). This 63-fold (98.4%) reduction was statistically significant using the Wald test (P < 0.001) based on a linear mixed model. Similar findings were obtained when weeks 6 through 12 were analyzed. The viral loads in the control animals in this study were not significantly different from those in a number of other studies from a number of different laboratories that used the same stock of SIVmac239 (26, 30, 34, 35, 65). The two monkeys that were RRV+ at the time of vaccination (440-92 and 247-04) fared no worse than the other vaccinated monkeys following SIV challenge. There was no apparent correlation of vaccine phase immune response measurements with postchallenge viral loads.
Analysis of anti-SIVenv antibody responses postchallenge measured by ELISA revealed responses in all five vaccinated monkeys and all three control monkeys (Fig. 11 and data not shown). However, the anti-Env responses in the vaccinated monkeys rose more quickly and to higher levels than in the control monkeys, suggestive of an anamnestic response. Cellular responses to SIV antigens also increased to various degrees postchallenge on the basis of ELISPOT assays (Fig. 5B and C) and MHC tetramer binding (Fig. 7).
DISCUSSION
Except for live attenuated strains of SIV, which have typically apparent provided apparent sterilizing immunity against challenge by homologous or closely matched strains, the vast majority of vaccine challenge experiments in monkeys have not shown an effect on the acquisition of SIV infection. Protective effects afforded by vaccination have typically been measured by the extent to which viral loads have been lowered following challenge. The extent of viral load reduction afforded by recombinant RRV-SIV in the experiments described here compares favorably with maximal viral load reductions that have been reported in the literature. A number of investigators using a variety of vaccine approaches have reported maximal SIV load reductions in the 1.5 to 2.0 log10 unit range (geometric mean) (30, 42, 55, 65). Of course, many studies have not achieved this degree of viral load reduction. Perhaps the greatest SIV load reductions have been observed recently by Manrique et al. (44). Using recombinant MVA (modified vaccinia virus Ankara, a poxvirus) expressing SIV Gag, Pol, and Env proteins, augmented by DNA-mediated cytokine expression, these investigators reported an approximate 3 log10 unit reduction in viral loads following repeated low-dose mucosal challenge with SIVmac251 (44).
Hansen et al. (26) have described the construction and performance of replication-competent CMV-SIV recombinants using the betaherpesvirus from rhesus monkeys. Recombinants expressing Gag, Env, and a Rev-Tat-Nef fusion protein were used, as we did here for our RRV-SIV recombinants. More recently, subsequent to the original submission of our manuscript, Hansen et al. published a more detailed account of immune responses and protective efficacy with their rhCMV-SIV recombinants (25). Interestingly, despite adequate expression of Env in cells infected in culture, few or no anti-Env antibody responses were observed by Hansen et al. (26), similar to our experience here with the RRV-SIVenv recombinant. Cellular responses with rhCMV-SIV were persistent and had an effector memory phenotype, also similar to what we report here. However, the immunodominant A*01-restricted Gag-CM9 and Tat-SL8 epitopes were not immunodominant in the context of rhCMV-SIV immunization. Although significant reductions in chronic-phase viral loads in rhCMV-SIV-vaccinated monkeys were not observed by Hansen et al. (25, 26), a significant fraction of the vaccinated monkeys did not exhibit progressive systemic dissemination following repeated SIVmac239 mucosal exposure.
Virus-specific memory T cells can be categorized into central memory and effector memory subsets (39). Classically, central memory T cells are more quiescent than effector memory cells, have increased proliferative capacity, and retain the ability to secrete interleukin 2 (IL-2). Characteristic expression markers of central memory cells include CD28, CCR7, and CD127. Central memory T cells are enriched in secondary lymphoid tissues and relatively absent from mucosal effector sites. In contrast, effector memory cells are CD28− CCR7− CD127−, generally express large amounts of perforin, and are highly enriched in effector mucosal sites. Hansen et al. reported preferential induction of SIV-specific effector memory CD8+ T cells by their recombinant CMV-SIV strains (26). Although there was some animal-to-animal variation in our study, SIV-specific CD8+ T cells induced by recombinant RRV similarly were predominantly a classic effector memory phenotype (CD28− CCR7− CD127−and perforin positive).
A number of factors could potentially contribute to the poor elicitation of anti-Env antibodies in our study. Despite the occurrence of very high anti-Env antibody levels during the course of HIV-1 and SIV infections (7), the Env protein may actually not be that immunogenic (37). The high level of anti-Env antibody responses in natural infection may have more to do with the prolonged, continuous exposure to high levels of antigen than it does with the inherent immunogenicity of the protein. Envelope protein is covered with a glycan shield and is locked into a tight trimer configuration; others have noticed a low inherent immunogenicity of the natural gp160 Env protein when expressed from DNA or other forms of vectored expression (30, 36, 43, 66). It is also possible that the temporally unregulated expression of Env in the construct used may be problematic for the recombinant virus in terms of replication kinetics, yield per cell, toxicity, or ability to be limited by the host immune response. Additionally, we do not know whether Env traffics properly to the cell surface in an RRV-infected cell that is making a variety of other RRV glycoproteins.
It has been speculated that optimal levels of vaccine protection may be achieved when both cellular and humoral responses are optimal and can act in concert (62). Live attenuated strains of SIV persist in monkeys and elicit mature antibody responses and persistent cellular responses (23, 32). Our hope in doing these experiments with replication-competent RRV recombinants was that we could match the characteristics of the anti-SIV immune response achieved by live attenuated strains of SIV. Although the anti-SIV CD8+ cellular responses that we achieved with RRV-SIV are impressive in their magnitude and durability, we have certainly failed on the humoral side. Others have presented evidence for improved efficacy with other vaccine approaches when anti-Env responses were included in the vaccine regimen (48, 69). If the problem of anti-Env antibody responses with RRV-SIV can be resolved, there will still be reason to hope that we can match, or come close to matching, the degree of protection observed with live attenuated SIV.
ACKNOWLEDGMENTS
We thank Jennifer Morgan, Hannah Sanford, Jacqueline Gillis, Angela Carville, and Mike Piatak for assistance with certain aspects of these experiments. We also thank David Knipe, George Pavlakis, David Watkins, and Eloisa Yuste for providing reagents.
This work was supported by NIH grants R01 AI63928 (R.C.D.), RR0168 for support of the New England Primate Research Center, and 5T32AI07245 (J.P.B.). The work was also funded in part with federal funds from the National Cancer Institute under contract HHSN261200800001E (J.D.L.).
Footnotes
Published ahead of print on 7 September 2011.
REFERENCES
- 1. Abdel-Motal U. M., et al. 2005. Kinetics of expansion of SIV Gag-specific CD8+ T lymphocytes following challenge of vaccinated macaques. Virology 333:226–238 [DOI] [PubMed] [Google Scholar]
- 2. Alexander L., et al. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen T. M., et al. 2001. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J. Virol. 75:738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allen T. M., et al. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 160:6062–6071 [PubMed] [Google Scholar]
- 5. Altfeld M., et al. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434–439 [DOI] [PubMed] [Google Scholar]
- 6. Bilello J. P., Morgan J. S., Damania B., Lang S. M., Desrosiers R. C. 2006. A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization. J. Virol. 80:1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binley J. M., et al. 1997. Differential regulation of the antibody responses to gag and env proteins of human immunodeficiency virus type 1. J. Virol. 71:2799–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borrow P., et al. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211 [DOI] [PubMed] [Google Scholar]
- 9. Burns D., Collignon C., Desrosiers R. C. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burton D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. U. S. A. 94:10018–10023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casimiro D. R., et al. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 79:15547–15555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cline A. N., Bess J. W., Piatak M., Jr., Lifson J. D. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303–312 [DOI] [PubMed] [Google Scholar]
- 13. Collins K. L., Chen B. K., Kalams S. A., Walker B. D., Baltimore D. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401 [DOI] [PubMed] [Google Scholar]
- 14. Daniel M. D., Kirchhoff F., Czajak S. C., Sehgal P. K., Desrosiers R. C. 1992. Protective effects of a live-attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938–1941 [DOI] [PubMed] [Google Scholar]
- 15. Desrosiers R., et al. 1998. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 72:1431–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desrosiers R. C. 1999. Strategies used by human immunodeficiency virus that allow persistent viral replication. Nat. Med. 5:723–725 [DOI] [PubMed] [Google Scholar]
- 17. Desrosiers R. C., et al. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeWire S. M., Money E. S., Krall S. P., Damania B. 2003. Rhesus monkey rhadinovirus (RRV): construction of a RRV-GFP recombinant virus and development of assays to assess viral replication. Virology 312:122–134 [DOI] [PubMed] [Google Scholar]
- 19. Evans D. T., Bricker J. E., Desrosiers R. C. 2004. A novel approach for producing lentiviruses that are limited to a single cycle of infection. J. Virol. 78:11715–11725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans D. T., et al. 2005. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J. Virol. 79:7707–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans D. T., et al. 2003. Mucosal priming of simian immunodeficiency virus-specific cytotoxic T-lymphocyte responses in rhesus macaques by the Salmonella type III secretion antigen delivery system. J. Virol. 77:2400–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fouts T. R., Binley J. M., Trkola A., Robinson J. E., Moore J. P. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gauduin M. C., et al. 2006. Induction of a virus-specific effector-memory CD4+ T cell response by attenuated SIV infection. J. Exp. Med. 203:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goulder P. J. R., et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217 [DOI] [PubMed] [Google Scholar]
- 25. Hansen S. G., et al. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen S. G., et al. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hel Z., et al. 2001. Potentiation of simian immunodeficiency virus (SIV)-specific CD4(+) and CD8(+) T cell responses by a DNA-SIV and NYVAC-SIV prime/boost regimen. J. Immunol. 167:7180–7191 [DOI] [PubMed] [Google Scholar]
- 28. Hendricks E. E., Lin K. C., Boisvert K., Pauley D., Mansfield K. G. 2004. Alterations in expression of monocyte chemotactic protein-1 in the simian immunodeficiency virus model of disseminated Mycobacterium avium complex. J. Infect. Dis. 189:1714–1720 [DOI] [PubMed] [Google Scholar]
- 29. Horton H., et al. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia B., et al. 2009. Immunization with single-cycle SIV significantly reduces viral loads after an intravenous challenge with SIV(mac)239. PLoS Pathog. 5:e1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson R. P., Desrosiers R. C. 1998. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr. Opin. Immunol. 10:436–443 [DOI] [PubMed] [Google Scholar]
- 32. Johnson R. P., et al. 1997. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. J. Virol. 71:7711–7718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson W., Desrosiers R. C. 2002. Viral persistance: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499–518 [DOI] [PubMed] [Google Scholar]
- 34. Johnson W. E., Lifson J. D., Lang S. M., Johnson R. P., Desrosiers R. C. 2003. Importance of B-cell responses for immunological control of variant strains of simian immunodeficiency virus. J. Virol. 77:375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaur A., et al. 2007. Ability of herpes simplex virus vectors to boost immune responses to DNA vectors and to protect against challenge by simian immunodeficiency virus. Virology 357:199–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim J. J., et al. 2000. Modulation of antigen-specific humoral responses in rhesus macaques by using cytokine cDNAs as DNA vaccine adjuvants. J. Virol. 74:3427–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klasse P. J., Sanders R. W., Cerutti A., Moore J. P. How can HIV-1 Env immunogenicity be improved to facilitate antibody-based vaccine development? AIDS Res. Hum. Retroviruses, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kuroda M. J., et al. 1998. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J. Exp. Med. 187:1373–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lanzavecchia A., Sallusto F. 2005. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17:326–332 [DOI] [PubMed] [Google Scholar]
- 40. Lewis M. G., et al. 1994. Titration and characterization of two rhesus derived SIVmac challenge stocks. AIDS Res. Hum. Retroviruses 10:213–220 [DOI] [PubMed] [Google Scholar]
- 41. Lifson J. D., et al. 2001. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187–10199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J., et al. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457:87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu S., et al. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retroviruses 14:151–155 [DOI] [PubMed] [Google Scholar]
- 44. Manrique M., et al. 2011. Long-term control of simian immunodeficiency virusmac251 viremia to undetectable levels in half of infected female rhesus macaques nasally vaccinated with simian immunodeficiency virus DNA/recombinant modified vaccinia virus Ankara. J. Immunol. 186:3581–3593 [DOI] [PubMed] [Google Scholar]
- 45. Mansfield K. G., et al. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Means R. E., Greenough T., Desrosiers R. C. 1997. Neutralization sensitivity of cell culture passaged simian immunodeficiency virus. J. Virol. 71:7895–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murphy C. G., et al. 2000. Vaccine protection against simian immunodeficiency virus by recombinant strains of herpes simplex virus. J. Virol. 74:7745–7754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ourmanov I., et al. 2009. Improved survival in rhesus macaques immunized with modified vaccinia virus Ankara recombinants expressing simian immunodeficiency virus envelope correlates with reduction in memory CD4+ T-cell loss and higher titers of neutralizing antibody. J. Virol. 83:5388–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parren P. W. H. I., et al. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Piantadosi A., Chohan B., Chohan V., McClelland R. S., Overbaugh J. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Regier D. A., Desrosiers R. C. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 6:1221–1231 [DOI] [PubMed] [Google Scholar]
- 52. Reynolds M. R., et al. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J. Virol. 84:9190–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Richman D. D., Wrin T., Little S. J., Petropoulos C. J. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roederer M. 2001. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry 45:194–205 [DOI] [PubMed] [Google Scholar]
- 55. Rosati M., et al. 2009. DNA vaccination in rhesus macaques induces potent immune responses and decreases acute and chronic viremia after SIVmac251 challenge. Proc. Natl. Acad. Sci. U. S. A. 106:15831–15836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Salisch N. C., et al. 2010. Inhibitory TCR coreceptor PD-1 is a sensitive indicator of low-level replication of SIV and HIV-1. J. Immunol. 184:476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sattentau Q. J., Moore J. P. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwartz O., Marechal V., LeGall S., Lemonnier F., Heard J.-M. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 nef protein. Nat. Med. 2:338–342 [DOI] [PubMed] [Google Scholar]
- 59. Speck S. H., Ganem D. 2010. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8:100–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Swigut T., et al. 2004. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J. Virol. 78:13335–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Veazey R. S., et al. 2001. Emergence and kinetics of simian immunodeficiency virus-specific CD8(+) T cells in the intestines of macaques during primary infection. J. Virol. 75:10515–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Walker B. D., Burton D. R. 2008. Toward an AIDS vaccine. Science 320:760–764 [DOI] [PubMed] [Google Scholar]
- 63. Walsh S. R., et al. 2009. Diverse recognition of conserved orthopoxvirus CD8+ T cell epitopes in vaccinated rhesus macaques. Vaccine 27:4990–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Watanabe D., et al. 2007. Properties of a herpes simplex virus multiple immediate-early gene-deleted recombinant as a vaccine vector. Virology 357:186–198 [DOI] [PubMed] [Google Scholar]
- 65. Wilson N. A., et al. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wright P. F., et al. 2004. Comparison of systemic and mucosal delivery of 2 canarypox virus vaccines expressing either HIV-1 genes or the gene for rabies virus G protein. J. Infect. Dis. 189:1221–1231 [DOI] [PubMed] [Google Scholar]
- 67. Wyand M. S., et al. 1999. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 73:8356–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wyand M. S., Manson K. H., Garcia-Moll M., Montefiori D., Desrosiers R. C. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xiao P., et al. 2010. Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J. Virol. 84:7161–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]