Abstract
Classical scrapie is a prion disease in sheep and goats. In sheep, susceptibility to disease is genetically influenced by single amino acid substitutions. Genetic breeding programs aimed at enrichment of arginine-171 (171R) prion protein (PrP), the so-called ARR allele, in the sheep population have been demonstrated to be effective in reducing the occurrence of classical scrapie in the field. Understanding the molecular basis for this reduced prevalence would serve the assessment of ARR adaptation. The prion formation mechanism and conversion of PrP from the normal form (PrPC) to the scrapie-associated form (PrPSc) could play a key role in this process. Therefore, we investigated whether the ARR allele substantially contributes to scrapie prion formation in naturally infected heterozygous 171Q/R animals. Two methods were applied to brain tissue of 171Q/R heterozygous sheep with natural scrapie to determine the relative amount of the 171R PrP fraction in PrPres, the proteinase K-resistant PrPSc core. An antibody test differentiating between 171Q and 171R PrP fragments showed that PrPres was mostly composed of the 171Q allelotype. Furthermore, using a novel tool for prion research, endoproteinase Lys-C-digested PrPres yielded substantial amounts of a nonglycosylated and a monoglycosylated PrP fragment comprising codons 114 to 188. Following two-dimensional gel electrophoresis, only marginal amounts (<9%) of 171R PrPres were detected. Enhanced 171Rres proteolytic susceptibility could be excluded. Thus, these data support a nearly zero contribution of 171R PrP in PrPres of 171R/Q field scrapie-infected animals. This is suggestive of a poor adaptation of classical scrapie to this resistance allele under these natural conditions.
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs) or prion diseases are infectious neurological diseases for which susceptibility and transmissibility are at least dependent on the strain of the agent and the prion protein (PrP) genotype of the host, while other host factors also play a role (3, 6, 13, 18). The archetypal example is natural scrapie in sheep, for which the infectious nature was first shown by Cuillé and Chelle following experimental infection of goat and sheep (15). In humans, various forms of TSEs exist, such as Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträussler-Scheinker syndrome (GSS), and kuru (11). The precise nature of the infectious agent is still uncertain, but it is characterized by the presence of PrP in misfolded and aggregated forms and named the scrapie form of PrP (PrPSc) (47). The normal form of the protein is termed PrPC because of its natural occurrence in cell membranes of eukaryotic species. Characteristic for PrPSc is its partial resistance to digestion with potent serine endoproteinases such as proteinase K (PK). While PrPC is fully hydrolyzed by PK, PrPSc is recovered as PrPres, which consists of prion protein core fragments that are usually N-terminally cleaved by approximately 6 kDa. The exact extent of N-terminal cleavage is dependent on strain type-associated conformational conditions of PrPSc (7, 27, 42, 44, 48).
One of the major features of prion disease susceptibility and transmissibility is the PrP-related genetic variability of both host and donor, which, e.g., is evident in sheep (4). The amino acid sequence of PrP is considered to be conserved between mammalian species, yet within species it can be polymorphic, as seen in humans, sheep, and goats, though not typically in cattle (29, 53, 63, 68). Susceptibility for TSE infection is highly influenced by single amino acid polymorphisms. In humans, this has become evident in individuals from Papua New Guinea who developed genetic resistance for kuru by the evolution of a unique resistance PrP allelotype (codon 127, glycine to valine [V]) (38). In sheep, variable levels of resistance to TSEs have been identified and found to be dependent on both prion strain and PrP polymorphisms. For classical scrapie and bovine spongiform encephalopathy (BSE) in sheep, three important amino acid polymorphisms that influence susceptibility and transmission have been described, i.e., alanine (A) to V at codon 136, arginine (R) to histidine (H) at codon 154, and glutamine (Q) to R at codon 171 (3, 28, 29, 57). In atypical/Nor98 scrapie, a form of scrapie that has poor transmission properties, susceptibility mainly correlates to a substitution of R to H at codon 154 or leucine (L) to phenylalanine (F) at codon 141 (19, 43, 53). Taking the major TSE transmission-related polymorphisms of sheep into account, a 3-amino-acid nomenclature for codons 136, 154, and 171 is used, and A136R154Q171 (usually indicated ARQ) is considered to be the wild-type allele. For classical scrapie forms in sheep, the levels of susceptibility in the context of amino acid substitutions have been ranked in the following order: VRQ, ARQ, AHQ, and ARR. Such information has led to successful scrapie eradication programs in different European countries by use of a genetic breeding strategy targeted to the enrichment of the 171R allele (23, 40, 62).
A concern of such breeding strategies is whether this type of genetic selection might lead to the emergence or adaptation of a new TSE strain that would replicate more efficiently using the R171 allele. However, for classical scrapie, such a condition has hardly been reported. It is known that the 171R allele historically occurs in many breeds at relatively high frequencies, though there is little evidence of scrapie in sheep carrying this allele. For example, only a small number of scrapie cases have been associated with ARR/VRQ heterozygous sheep, while scrapie outbreaks in ARQ/ARR sheep with scrapie are very rare, and only three natural cases in sheep that are 171R homozygous have been reported (14, 22, 30, 33). Of significant importance is whether or not scrapie-positive heterozygous ARR/VRQ sheep carry equimolar amounts of both PrP alleles in the PrPSc fraction and if this might be indicative for a tendency that such cases will lead to enhanced scrapie transmissibility within the 171R-carrying sheep population. Thus far, in the one single ARR/VRQ scrapie field case studied, no PK-resistant ARR material was detected (41). In vitro studies already indicate that the binding behaviors of the different PrP allelic forms to PrPSc are comparable on the molecular level of PrP (51), while conversion studies have shown that the 171R allele has a relatively low tendency to become PK resistant (9, 10). After oral BSE challenge of 1- to 2-week-old ARR/ARR lambs, appreciable levels of ARR PrPSc and PrPres were detected only in the spleen of one out of three asymptomatic animals euthanized at 10 months after infection (2).
In this study, we determined the involvement of ARR PrP (further referred to as 171R) in PrPres relative to wild-type 171Q PrP (further referred to as 171Q) in a group of 8 naturally infected sheep with ARR/VRQ scrapie. We used both an immunochemical discrimination method and a biochemical separation technique. For the first method, the binding of monoclonal antibody (MAb) SAF84 to PrP was highly dependent on 171Q-containing allelotypes. The second approach was a novel technique for the quantitative generation of a 6-kDa PrP fragment from codons 114 to 188 (114-188PrP) using endoproteinase Lys-C digestion of PrPres. Generating this PrP polypeptide fragment led to detection of differently charged allelotype polypeptide fragments predicted to be present in scrapie sheep heterozygous at PrP codon 171. Furthermore, to compare the resistance of eventual existing 171R PrPres toward the proteolytic conditions used for these studies, we examined tissues from 171R/R sheep experimentally infected with classical BSE and scrapie.
MATERIALS AND METHODS
Chemicals and reagents.
Chemicals were purchased from Merck (Darmstadt, Germany) unless otherwise specified (and were at least analytical reagent grade). All procedures were performed at room temperature unless otherwise stated. Purified recombinant ovine PrPs (recPrP) with allelic variants 171R and 171Q were prepared by a method similar to the method described previously (45, 50). Endoproteinase Lys-C from Lysobacter enzymogenes was purchased from Sigma (P2289-3UN). PK was obtained from Merck (30 U/mg; 124568; Merck, Germany). Synthetic peptides with and without an acetylated N terminus and with amino acid sequences related to the epitope of antibody 6C2 were purchased from Pepscan Presto (Lelystad, The Netherlands). Gels and reagents for one-dimensional (1D) and two-dimensional (2D) electrophoresis were purchased from Invitrogen (Breda, The Netherlands). ImageQuant (version 5.2) Molecular Dynamics software was from GE Healthcare Life Sciences, Belgium.
Antibodies and epitope mapping.
MAbs used were 12B2, 9A2, 1E4, 6C2, L42, SAF84, and FH6 (16, 25, 26, 33, 52). The epitope specificities of these antibodies assessed by solid-phase peptide epitope mapping (Pepscan analysis) are 93WGQGG97, 102WNK104, 102WNKP105, 114HVAGAAA120, 148YEDRYY153, 166YRPVDQY172, and 225SQAYYQ230, respectively. Antibody 1E4 was from Sanquin BV (Amsterdam, The Netherlands), L42 was from R-Biopharm (Darmstadt, Germany), and SAF84 was from Spi-Bio (Massy, France). FH6 is a newly developed MAb raised by immunization with recombinant ovine 94-233PrP prepared in Escherichia coli as described previously (67). Immunizations in PRNP−/− 129/Ola mice and monoclonal antibody preparation techniques were carried out as described previously (24, 37).
Epitope mapping by Pepscan analysis using solid-phase single-amino-acid position-shifted overlapping 15-mer N-acetylated peptides was performed in miniaturized 3-microliter-well cards (20, 56). Further epitope specificity determinations were performed by a blocking enzyme-linked immunosorbent assay (ELISA). In brief, recPrP was coated on 96-well polystyrene microtiter plates at 0.1 μg/ml in 6 M guanidine-HCl in phosphate-buffered saline, pH 7.3. Antibody 6C2 at 6 ng/ml was premixed with epitope 114-120PrP-related peptides in various concentrations from 10 to 0.0001 μg/ml, and the mixture was applied to the coated wells for 1 h at ambient temperature, washed with 0.05% (wt/vol) Tween 20 in water, and further subjected to 1 h of incubation with rabbit anti-mouse IgG conjugated to horseradish peroxidase (Dako). Antibody binding was visualized with 3,3′,5,5′-tetramethylbenzidine substrate development, and the reaction was terminated by the addition of 0.7 M H2SO4, before analysis at 450 nm.
Animals and tissues.
Tissue samples from sheep with classical scrapie were obtained from naturally infected flocks following passive and active monitoring of slaughtered and fallen stock. Some sheep were from the naturally scrapie-infected Central Veterinary Institute of Wageningen UR (CVI) institutional flock. Scrapie was confirmed using immunohistochemical (IHC) procedures, as described for both brain and tonsil tissues (61, 64), and also Western blotting (WB) techniques (31). PrP genotyping was performed using TaqMan analysis and pyrosequencing as described before (9, 60).
For evaluating the PK resistance of 171R PrP in PrPres brain stem or thalamus homogenates, samples of sheep experimentally infected with either BSE or scrapie were obtained from the Institut National de la Recherche Agronomique (INRA), Tours and Toulouse, France. All cases showed clinical signs at tissue collection. The tissues were generated by intracerebral inoculation after different passages: first-passage BSE from three Poll Dorset ARR/ARR sheep (sheep PD337, PD368, and PD369, inoculated with bovine BSE), second-passage BSE from three Suffolk ARQ/ARQ sheep (sheep 1252, 486, and 136, inoculated with Poll Dorset first-passage ARQ/ARQ BSE), third-passage BSE from two Suffolk ARQ/ARR sheep (sheep P30 and P130, inoculated with Suffolk second-passage ARQ/ARQ BSE; thus, it is not an adaptation to the 171R allele during the first two passages), and classical scrapie from an ARR/ARR sheep from the Langlade flock. Methods for these studies have been partly published (2).
Tissue treatments.
Ten percent (wt/vol) brain stem homogenates were prepared in lysis buffer and digested with PK at 37°C, and PrPres was partially purified by precipitation with 1-propanol as described previously (31). For PrPres analyses, pellets were directly subjected to SDS-PAGE and Western blotting.
For further biochemical identification of 171Q and 171R allele fragments, pellets containing 10 mg tissue equivalents (TEs) were denatured by suspension in 50 μl of 8 M guanidine hydrochloride containing 40 mM dithiothreitol, and after 5 min at ambient temperature, the protein was precipitated by mixing with 10 volumes of ice-cold (−20°C) methanol, kept for 30 min at −20°C, and then centrifuged at 18,000 × g for 20 min at 4°C (5417R; Eppendorf centrifuge). The final pellet was resuspended in 150 μl of 1% (wt/vol) N-lauroylsarcosine sodium salt in 0.1 M Tris-HCl, pH 8.5, containing 40 mM dithiothreitol, heated for 5 min at 96°C, and cooled down to ambient temperature. Cleavage of protein at carboxy-terminal sites of lysines by Lys-C was performed by adding 15 μl of a stock solution of 0.015 U/μl of enzyme and incubating at 37°C overnight.
1D electrophoresis (SDS-PAGE).
Three volumes of Lys-C-digested sample were mixed with 1 volume of NuPAGE 4× lithium dodecyl sulfate (LDS) sample buffer containing 0.1 M dithiothreitol and denatured by heating at 96°C for 5 min. After cooling to room temperature, the sample was centrifuged at 16,000 × g for 5 min in a Biofuge Pico microcentrifuge. Sample supernatants (10 μl, 0.5 mg TE per lane) were then subjected to 1D electrophoresis in 12% bis-Tris NuPAGE gels using morpholineethanesulfonic acid (MES) with antioxidant as running buffer.
2D electrophoresis.
The protein in the Lys-C-digested sample (165 μl) was precipitated with 10 volumes of ice-cold (−20°C) methanol (without detectable PrP loss). After consecutive mixing and incubating at −20°C for 30 min, the sample was centrifuged at 18,000 × g (5417R; Eppendorf centrifuge) at 4°C for 20 min. The supernatant was discarded. Further procedures used the supplier's (Invitrogen) instructions for 2D electrophoresis. The pellet was resuspended and diluted into 165 μl of sample rehydration buffer (SRB2; 90% [vol/vol] ZOOM 2D protein solubilizer 2, 0.6% [vol/vol] ZOOM carrier ampholytes, pH 3 to 10, 0.002% [wt/vol] bromophenol blue, 0.5% [vol/vol] 2 M dithiothreitol in water). After centrifugation at 16,000 × g for 5 min in a Biofuge Pico microcentrifuge for debris removal, the supernatant was diluted in SRB2 to obtain 2.5 mg TEs in 140 μl per immobilized pH gradient (IPG) strip. ZOOM strips, pH 3 to 10, were rehydrated with the sample for 1 h in an IPG runner cassette. First-dimension isoelectric focusing occurred in the IPG runner system with a stepwise voltage program of from 200 V to 2,000 V for a total run time of 80 min until the dye front reached the anode. The IPG strips were then equilibrated for the second-dimension separation by incubation in reducing and denaturing solution for 15 min at ambient temperature according to the supplier's protocol, followed by alkylation with iodoacetamide for 15 min. The equilibrated IPG strip was applied to the NuPAGE Novex bis-Tris ZOOM gel. The reference lane was loaded with a mixture of 5 μl SeeBlue Plus2 marker and 5 μl of the 1D SDS-PAGE sample prepared in the same way described above for the IPG strip to obtain 0.25 mg TE. Second-dimension electrophoresis was performed for 35 min at 200 V with MES running buffer and antioxidant.
WB, detection, and quantitation.
The subsequent WB procedures—electrotransfer, immunostaining, and the development of luminescence (using CDP-Star as the substrate for alkaline phosphatase)—were performed as described previously (33). Antibodies were used at the concentrations indicated. To obtain digital images, individual films were exposed to the blots for different time periods (i.e., 30 s, 1 min, 3 min, and 6 min) and digitalized using a scanner. Calculations on these digital readings were performed using ImageQuant (version 5.2) software. Additional WB studies on PK resistance of PrPres were performed using a triplex WB system, which uses a recently published fluorimetric method which allows immunochemical quantification of protein without enzymatic enhancement (31). Using both Western blot systems, the fractions of R and Q PrPres present in a typical digest were obtained from the ratios of the signals obtained with MAbs L42 and SAF84 with L42 binding to both PrP allelotypes and SAF84 binding only to the 171Q PrP allelotype. These fractions were calculated as follows: the fraction of the PrP 171Q product in scrapie or BSE was obtained by applying the formula ratiox/ratioQ/Q where ratiox is the SAF84/L42 ratio of an unknown sample and ratioQ/Q is the SAF84/L42 ratio determined for Q/Q homozygous material, which was an average of measurements of at least three different scrapie or BSE 171Q/Q samples; likewise, the fraction of PrP 171R product could be deduced from the formula (ratioQ/Q − ratiox)/ratioQ/Q.
RESULTS
Discrimination of R and Q at codon 171 in synthetic PrP peptides and recombinant PrP using SAF84 binding.
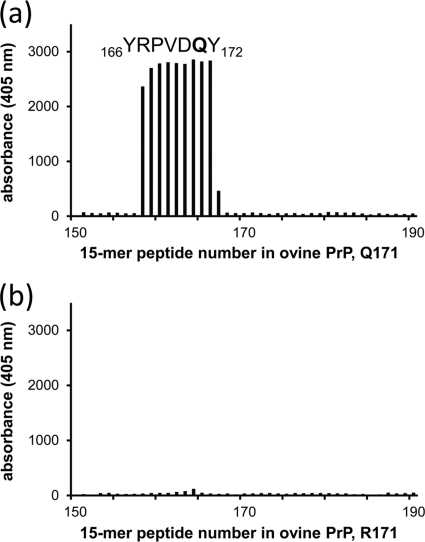
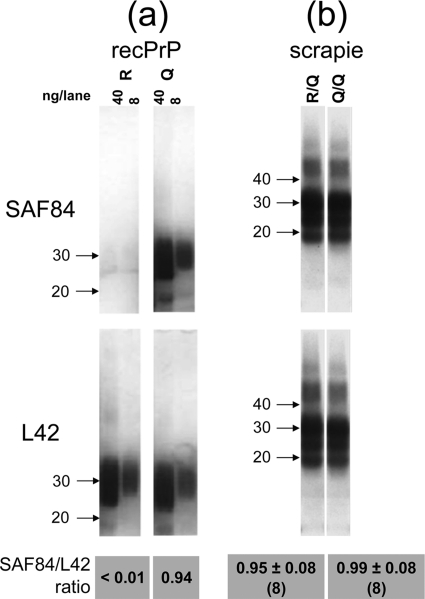
The presence or absence of the allele 171R expression product in PrPres material from heterozygous (171Q/R) sheep was first investigated by an antibody discrimination assay. To this end, the PrP-specific antibody SAF84 was employed since, using epitope mapping with Pepscan analysis, it appeared to bind the 166YRPVDQY172 sequence containing the 171Q codon (underlined) but not the polymorphic substitution 171R (Fig. 1). The PrP codon with 171Q specificity for SAF84 binding was further assessed using an ELISA in which three different recombinant PrP variants (ovine VRQ, ARQ, and ARR) were coated to microtiter plates. The ARR variant (carrying R at codon 171) exhibited approximately 250 to 1,000 times less binding to SAF84 than either the VRQ or ARQ allelotype, whereas antibodies that bound other PrP sites (e.g., MAbs 6C2 and L42) bound all PrP variants equally well (data not shown). Western blotting, after 1D SDS-PAGE, confirmed the specificity of SAF84 to 171Q-containing recombinant PrP variants. RecPrP with the 171R polymorphism (Fig. 2a) showed little or no binding of SAF84 but showed obvious binding to the 171Q recPrP variant. The SAF84/L42 binding ratio was less than 0.01 for 171R, while for the 171Q PrP variant a SAF84/L42 ratio of 0.94 was determined. These data confirmed that MAb SAF84 reactivity is specific for the sequence of ovine PrP from codons 166 to 172 with Q at position 171 and that the 171R polymorphism does not permit binding. Hence, we concluded that SAF84 is a suitable antibody to discriminate between PrP carrying either an R or a Q at codon 171.
Fig. 1.
Pepscan analysis of antibody SAF84 on overlapping solid-phase 15-mer peptides of two ovine PrP sequences differing only at codon 171. (a) Analysis of 171Q PrP; (b) analysis of 171R allelic variants. With wild-type ovine PrP, this gives 166YRPVDQY172 as the SAF84 core epitope sequence with 171Q, while there is no binding observed when the polymorphic amino acid R171 is present. SAF84 was applied at 0.05 μg/ml. Of all 220 15-mers, only peptides 151 to 190 are displayed. All other overlapping peptides (data not shown) of the 256 residues of the ovine PrP sequence (21) exhibited background signals similar to those shown in panel b.
Fig. 2.
Comparing affinity of PrP-specific antibodies between 171Q and 171R variants of ovine PrP. (a) Western blot of 171R and 171Q recPrPs. The 171Q-dependent MAb SAF84 binds with the 171Q allele but not with the 171R variant, while reference MAb L42 binds equally well to both 171Q and 171R recombinant PrP variants. (b) PrPres material from a scrapie-positive VRQ/VRQ sheep (Q/Q lane) reacts equally strongly with both MAbs L42 and SAF84. Likewise, PrPres of scrapie-positive ARR/VRQ sheep (R/Q lane) remained equally strongly reactive with both L42 and SAF84, yielding nearly equal SAF84/L42 ratios that were statistically significantly equal by the two-sided Student t test (P = 0.18). For sample loading, 40 and 8 ng recombinant PrP was applied in the left and right lanes, respectively (a), and 0.5 mg TE was applied for the scrapie samples (b). Concentrations used for L42 and SAF84 antibodies were 0.1 and 0.5 μg/ml, respectively. Arrows indicate molecular mass markers (in kDa). The SAF84/L42 ratios below the blots are calculated from digitalized film images of blotting results for eight different 171R/Q cases and eight different 171Q/Q (2 ARQ/ARQ, 6 ARQ/VRQ) cases.
Estimation of 171R/Q-containing allelotype in PrPres from sheep brain tissue using SAF84.
PrPres was prepared from brain homogenates of scrapie-positive sheep with the ARR/VRQ, VRQ/VRQ, or ARQ/VRQ genotype. From results with recPrP, it was assumed that 171Q-dependent SAF84 binding to PrPres would be less strong if 171R PrPres was present in the brain homogenates than if only 171Q PrPres was present (compared to a non-polymorphism-dependent MAb like L42). Using 1D SDS-PAGE followed by Western blotting with MAbs SAF84 and L42 in parallel, the SAF84/L42 ratios were only slightly lower in the 171R/Q heterozygotes than the 171Q/Q homozygotes (0.95 ± 0.08 and 0.99 ± 0.08, respectively), whereby the differences were not statistically significant (Fig. 2b). If there had been more than 10% of the 171R product present, the ratio would have been ≤0.89 (using the 0.99 ratio in the 171Q/Q sheep as a 100% 171Q reference). Although the observed ratios were considered equal, if there was any 171R PrPres present in the brain of 171R/Q heterozygote sheep, it was calculated that the 171R PrP relative level constituted ≤4% of the total PrPres material on the basis of these ratio differences.
Design of a biochemical approach for identification of 171R and 171Q PrPres in scrapie material.
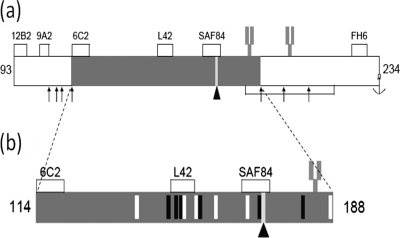
Since the Q-to-R amino acid change at position 171 involves an obvious charge inversion, we attempted to find potential cleavage fragments of PrPres that could be measured by 2D analysis. We rationalized, in principle, that following treatment with the enzyme endoproteinase Lys-C (which cleaves after lysine [K] residues), a 75-amino-acid-residue PrP fragment spanning codons 114 to 188 of ovine PrP would be produced. Furthermore, this putative polypeptide would be of a size sufficiently large to be studied by gel electrophoresis and Western blotting. The resultant PrP fragment was calculated to have an approximate molecular mass of 8 kDa and an isoelectric point (pI) of between 7 and 8 (34). It was also postulated that the PrP fragment, when produced from 171Q- or 171R-containg PrP variants, would yield a net difference in charge, whereby when derived from wild-type 171Q PrP, it would have a net ionic charge of 0, whereas when derived from 171R PrP, it would carry a net ionic positive charge of 1 (Fig. 3). Thus, the 171R-containing fragment would have a calculated pI of between 8 and 9 and the two allelic products would differ by about 1 pI unit and could be readily distinguished using 2D electrophoresis. Figure 3b suggests also that the 114-188PrP fragment could potentially carry one asparagine-linked glycosylation site at codon 184. Following these hypotheses, we optimized experimental conditions for the preparation, digestion, denaturation, and collection of PrPres fragments from sheep scrapie brain.
Fig. 3.
Postulated PrPres polypeptide fragment generated by specific Lys-C cleavage. (a) Mature ovine PrP extending from amino acids 25 to 234. The lysine residues which are cleaved following treatment with Lys-C are indicated by arrows. (b) Magnified region of the polypeptide fragment from codons 114 to 188 obtained after Lys-C cleavage of recombinant PrP and scrapie brain-derived PrPres. White vertical bars, 6 basic arginine (R) or lysine (K) residues; black vertical bars, the 6 acidic glutamic acid (E) and aspartic acid (D) amino acid residues; bar marked by an arrowhead, the polymorphic 171 codon that is either a neutral glutamine (Q) or a basic arginine (R). Asparagine at position 184 and 200 can be glycosylated (fork-like structure). The accumulated charge differences between the Lys-C-generated fragments are 0 for the 171Q allele and +1 for the 171R allele. The antibody binding sites for MAbs 12B2, 9A2, 6C2, L42, SAF84, and FH6 are displayed.
Generation of a 144-188PrP fragment from scrapie brains.
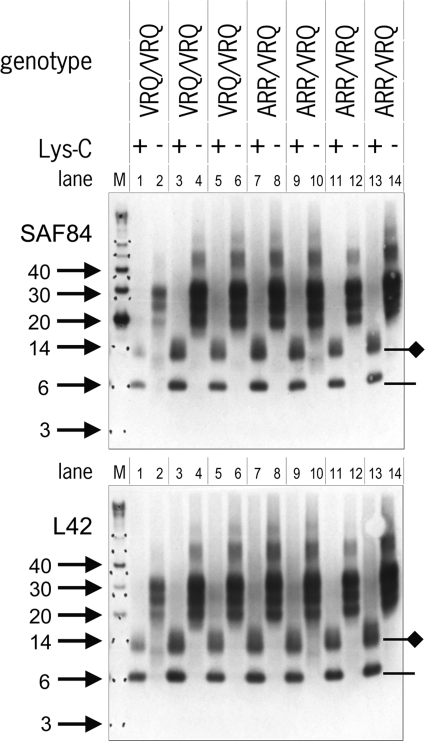
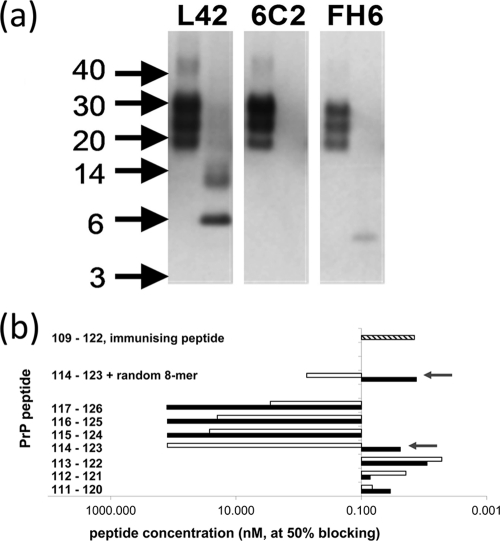
PrPres from scrapie tissue samples was digested with Lys-C and first resolved by 1D electrophoresis. Subsequent analysis using Western blotting with antibodies L42 and SAF84 showed the disappearance of the typical three protein bands corresponding to PrPres and the emergence of smaller polypeptide species migrating at 6 kDa and at 14 kDa (Fig. 4). These were presumed to be the nonglycosylated and monoglycosylated forms, respectively, of the generated 114-188PrP fragment containing the epitopes of MAbs L42 and SAF84. The epitopes of antibodies 12B2, 9A2, and FH6 were removed, as expected (Fig. 3 to 5). The 2-kDa discrepancy between a nonglycosylated 6-kDa product (shown in Fig. 4) and the 8-kDa product that we theorized would result from this approach is likely a consequence of the imprecision of the technique for estimating the molecular masses of proteins with molecular masses below 12 kDa and where the charges of amino acid side chains influence migration behavior. Unexpectedly, however, the N-terminal domain of the PrP fragment, which we anticipated to contain the epitope of the MAb 6C2 (amino acids 114 to 120), was not present, as illustrated by failure of this antibody to bind the PrP fragment (Fig. 5a). Since Lys-C cleavage leads to a free amino terminus, it is conceivable that antibody binding is dependent on an intact peptide bond at residue 114, the first amino acid of the 6C2 epitope. Indeed, in a blocking ELISA, the binding of 6C2 to coated ovine PrP could be prevented by peptides only if the entire peptide sequence from codons 114 to 120 was present with an intact peptide bond but not when the 114 residue was terminally present with a free amino terminus (Fig. 5b). This observation, in conjunction with the binding of MAbs L42 and SAF84 but not 12B2, 9A2, and FH6 and the resultant 6-kDa fragment, collectively indicated that the postulated cleavage by Lys-C at residues 113K and 188K had occurred to completion (as opposed to the variable levels of cleavage that could have occurred at lysines at positions 25, 26, 29, 104, 107, 109, 197, and 207 within PrP).
Fig. 4.
Result of Lys-C cleavage displayed in 1D Western blot. Shown are PK digests of PrPres in seven scrapie isolates from sheep with either the Q/Q or R/Q genotype at codon 171 before and after cleavage by Lys-C. Lanes 1 to 6, sample sets from three individual VRQ/VRQ sheep; lanes 7 to 14, sample sets from four ARR/VRQ sheep. In each lane, 0.5 mg TE was applied. Lane M, molecular mass markers (in kDa, arrows). At the right, the positions of the 114-188PrP fragment with (diamond line) or without (line) a glycosyl group are indicated. Again, blotting with either SAF84 or L42 yielded nearly equal staining for all seven sheep, indicative of the minimal presence of the 171R allele product. Antibody concentrations are as described in the legend to Fig. 2.
Fig. 5.
Evidence for cleavage at lysine residue 113 by Lys-C in PrPres of ovine scrapie brain material. (a) Electrophoretic pattern of PrP fragments after cleavage of ovine scrapie material with PK (left lanes) or PK followed by Lys-C (right lanes) on Western blots immunostained with antibodies L42, 6C2, and FH6. Surprisingly, the band reactive with L42 at 6 kDa was not reactive with 6C2 (Fig. 3). The absence of reactivity with C-terminal MAb FH6 at the 6-kDa position was as expected, and the reactive band below the 6-kDa position could well be an intermediate digestion product from codons 189 to 234 carrying a glycophosphatidyl anchor. In each lane, 0.5 mg TE was applied. Concentrations used for L42, 6C2, and FH6 antibodies were 0.1, 0.5, and 0.5 μg/ml, respectively. (b) Blocking ELISA to show the requirement of an N-terminal peptidyl link at lysine 114 of PrP for MAb 6C2 binding. Peptides with (shaded bars) and without (open bars) an amino-terminal N-acetyl group differed profoundly in their affinity only when residue 114 of ovine PrP is the N-terminal residue (arrows). A high blocking effect corresponds to peptide concentrations of about or below 0.1 μM at 50% blocking. Thus, the presence of the acetyl group appeared to be essential for 6C2 binding and was considered representative of an intact peptide bond. For comparison, other peptide sequences were tested, i.e., the peptide from codons 109 to 122 used for immunization as a positive control (hatched bar) and a peptide set containing the sequence from codons 114 to 123 at the C terminus, followed by an 8-amino-acid randomized sequence with or without an N-terminal N-acetyl group (the randomized sequence used as a control to exclude the influence of C-terminal variations in proximity to the epitope).
Identification of 171Q- and 171R-containing PrPres fragments after 2D electrophoretic separation.
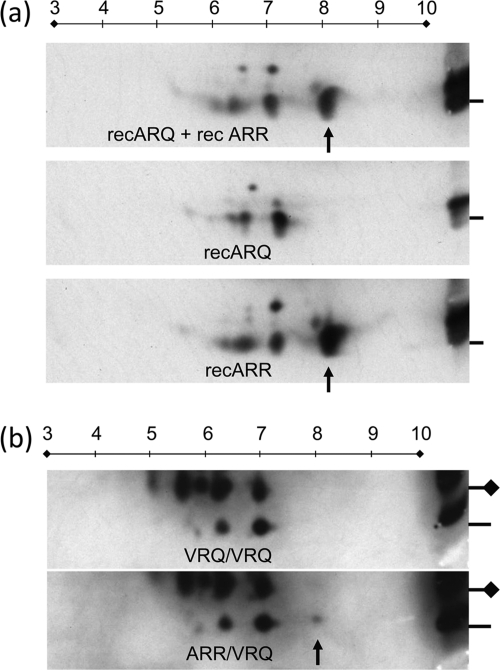
The 114-188PrP fragment generated by Lys-C treatment of recombinant ovine 171R- and 171Q-containing PrP variants was further characterized using 2D electrophoresis and Western blotting (with MAb L42). Figure 6a shows that the major cleavage products from the Lys-C digest migrated as expected, whereby pIs of approximately 8 for the 171R recPrP-derived polypeptide and 7 for the 171Q-containing recPrP polypeptide were observed. Also, in 171R recPrP, the pI 8 protein spot showed a positive reactivity against MAb L42, which was absent when the 171Q-specific MAb SAF84 was used. This confirmed that the pI 8 spot consisted of 171R protein.
Fig. 6.
Separation based on charge and molecular mass by 2D electrophoresis. The 114-188PrP polypeptide fragments generated by Lys-C cleavage are shown. (a) Result obtained with 171Q and 171R recPrP applied as a mixture or separately (only Lys-C digestion was applied); (b) result obtained with VRQ/VRQ and ARR/VRQ scrapie PrPres (PK and subsequent Lys-C digestion were applied). In panel a, only the 171R fragment shows a dominant L42-positive protein spot at pI 8 (arrow) which was not reactive with the 171Q-dependent antibody SAF84 (data not shown). The reference samples in panels a and b, shown on the right, contain the respective recombinant PrP and sheep samples mixed with molecular mass markers and were run only in the vertical 1D dimension. In panel b, the lines on the right refer to the 6-kDa markers (coinciding with the monoglycosylated 114-188PrPres fragment), and the lines with diamonds indicate the migration positions of the glycosylated 114-188PrPres fragment at 14 kDa. Applied amounts for 1D and 2D analyses were 165 and 330 ng of recombinant PrP, respectively, in panel a and 0.5 and 2.5 mg TEs of brain stem tissue, respectively, in panel b. The MAb L42 concentration was 0.1 μg/ml. In five out of eight cases, only the ARR/VRQ isolates exhibited a faint spot at pI 8 (arrow), which was probably also present at the glycosylated position but too faint for detection due to microheterogeneity of sugar chains. Numbers on lines above the panels indicate isoeletric points.
This assay was repeated using PrPres prepared from homozygous 171Q/Q (VRQ/VRQ or ARQ/ARQ) sheep. The digestion products resolved at pIs of 7 and lower for both the nonglycosylated (6 kDa) and the glycosylated 114-188PrP fragment (14 kDa), but none did so above this pI value (Fig. 6b). Yet, in five out of eight codon 171 heterozygous (all ARR/VRQ) cases, 171R 114-188PrP fragment material was marginally evident at pI 8 only at the nonglycosylated 6-kDa migration position (Fig. 6b, spot above the arrow). This suggests that 171R-containing polypeptide material is of a lower concentration at pI 8, while nonglycosylated and glycosylated 171Q materials appeared to be major spots at pI 7 and lower. The pI 8 spot was not detectable in the remainder of the scrapie-infected 171R/Q animals investigated, nor was it detectable in any 171Q/Q animals (Table 1). In the five cases where a 171R protein spot was observed, the amount of ARR material in the pI 8 spot approximated, on average, 2.6% (range, 0 to 8.1%) of the total protein spots at the glycosylated and nonglycosylated positions (ARR plus VRQ spots, following densitometric analysis of digitalized films). These five 171R/Q heterozygous cases having the pI 8 spot were by 1D analysis among the strongest positive PrPres cases. In summary, the 171R/Q heterozygous scrapie-infected sheep investigated did contain the 171R PrP in their PK-resistant PrPSc material, though at variable concentrations and levels below 9% of total PrPres.
Table 1.
Details of sheep scrapie cases analyzed by 2D electrophoresisa
| Case code | Condition at death | Age (mo) | Genotype | IHC result |
ARR spotb (%) | |
|---|---|---|---|---|---|---|
| Tonsil | Brain IHC | |||||
| 2004–31 | Fallen stock | Unk | VRQ/VRQ | Pos | Pos | ND |
| 2006–38 | Fallen stock | Unk | VRQ/VRQ | Pos | Pos | ND |
| 621553–4035 | Clinical | 32.8 | ARQ/VRQ | Pos | Pos | ND |
| 573862 | Clinical | Unk | ARR/VRQ | Neg | Pos | ND |
| 2006–25 | Fallen stock | Unk | ARR/VRQ | Neg | Pos | 1.5 |
| 2007–19 | Fallen stock | Unk | ARR/VRQ | Pos | Pos | 1 |
| 2006–42 | Fallen stock | Unk | ARR/VRQ | Neg | Pos | ND |
| 601677–2861 | Found dead | 67.2 | ARR/VRQ | Neg | Pos | ND |
| 547189–4333 | Healthy | 81.9 | ARR/VRQ | Neg | Pos | 8.1 |
| 2004–01 | Slaughter | Unk | ARR/VRQ | Neg | Pos | 6 |
| 2007–20 | Slaughter | Unk | ARR/VRQ | Wk pos | Pos | 6 |
Cases were either from active surveillance, i.e., those starting with a year number; passive surveillance, i.e., those with a single case number (case 573862); or CVI institutional flock with circulating natural scrapie (i.e., those with a double case number). Unk, unknown; Pos, positive; Neg, negative; Wk pos, weakly positive; ND, not determined.
Polypeptide material in spots at pI 8 was calculated as the percentage of all spots at the 6-kDa and 14-kDa positions, as explained in the Materials and Methods section. ND, not detected.
Estimation of proteinase K susceptibility of 171R PrPres.
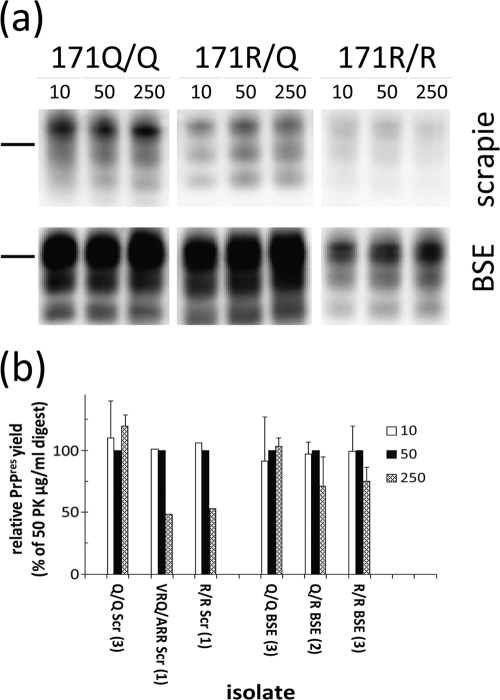
The differential susceptibility of 171R PrPres to PK treatment was investigated by applying enzyme concentrations of from 10 to 250 μg/ml. In this experiment, not only were field scrapie-derived materials used, but also materials derived from experimental animals inoculated intracerebrally with scrapie and BSE. It appeared that in all cases the 171R material was equally resistant to the protease at concentrations between 10 and 50 μg PK/ml and by use of typical experimental conditions (Fig. 7), notwithstanding 1- to 100-fold variations in PrPres level between individual samples (see Fig. S1 in the supplemental material). For PK concentrations of from 50 to 250 μg PK/ml, there was a 25 to 50% reduction in PrPres signal in the R allele-containing samples but not in the homozygous 171Q/Q cases of scrapie and BSE. Interestingly, when brain material from 171R/Q heterozygous sheep experimentally infected with BSE was digested under standard PK conditions of 50 μg PK/ml, the amount of 171R-containing PrPres (on the basis of antibody binding ratios between SAF84 and L42) was about 64% (standard deviation [SD], 10%). This was much higher than that for ARR/VRQ field case material from sheep with classical scrapie, where the 171R-containing fraction represented less than 9% (Fig. S2 in the supplemental material).
Fig. 7.
Resistance to digestion with PK of 171Q and 171R PrPres-containing ovine brain samples from either scrapie- or BSE-infected animals shown after Western blotting. Blots were developed using a triplex WB system. Samples were subjected to three PK concentrations. (a) (Top) Results of classical scrapie of genotypes ARQ/VRQ 532644, ARR/VRQ 2006-42, and ARR/ARR G361 from left to right (amounts applied, 0.5, 0.5, and 2.5 mg TEs, respectively); (bottom) results with BSE of genotypes and identity codes ARQ/ARQ 486, ARQ/ARR P130, and ARR/ARR PD368 from left to right (amounts applied, 0.5, 0.5, and 1.0 mg TE, respectively). Antibody used was L42 (0.2 μg/ml). Bars, 28-kDa molecular mass markers. (b) Overall results of digestion conditions varying only in applied PK concentrations. The bars indicate the relative signal to the 50-μg PK/ml treatment, presented where possible as averages ± SDs per number (between parentheses) of sheep analyzed. Sheep genotypes with their identity codes were ARQ/VRQ 532644, ARQ/ARQ 537272, VRQ/VRQ 553237, ARR/VRQ 2006-42, and experimental ARR/ARR case Langlade G361 for scrapie cases and ARQ/ARQ cases 1252, 486, and 136, ARQ/ARR cases P30 and P130, and ARR/ARR cases PD337, PD368, and PD369 for experimental BSE.
DISCUSSION
An altered form of prion protein is believed to be the major, if not sole, protein component of the infectious agent in TSE diseases, and expression of the normal isoform in the host is essential for disease transmission. Being so critically involved in disease development, PrP polymorphic sequence variants genetically determine transmissibility properties, such as the species barrier, lymphotropism, and within-species susceptibility, by modulating the interconvertibility of PrP itself. In the study described in this paper, evidence was found for the presence of the 171R protein in PrPres from heterozygous ARR/VRQ scrapie sheep, which usually incubate the disease over 6 years under natural conditions and which carry PrPC of both the 136V allele, associated with high susceptibility, and the 171R allele, associated with resistance. However, the level of the 171R protein in PrPres of heterozygous sheep is very low (if present at all), and in this study, it was quantified to constitute a maximum of 8.1% of the total PrPres material in such sheep, contrasting with an expected value of 50% if the expression level and conversion of both allelic products had been equal in the tissue of origin. These data were deduced using two independent methods: a discriminatory antibody tool and a novel procedure to generate and separate 114-188PrP fragments from PrPres containing either the 171Q or 171R codon.
First, the use of MAb SAF84 was chosen because of its selective affinity to bind ovine PrP containing 171Q, in contrast to a lack of reactivity for PrP containing the polymorphic amino acid, 171R. The estimation of the relative amount of the 171R allele in scrapie material obtained using SAF84 was possible by comparison to that obtained using the nondiscriminative antibody L42, which binds both allelic expression products. The approach of using a polymorphism-dependent PrP-specific antibody has proven to be a useful way of determining the genotype status at the codon 171 position in serum of sheep and for confirming the presence of the 171R allele in PrPres in spleen and brain of 171R/R homozygous sheep experimentally challenged with bovine BSE (8). These kinds of estimations can be safely carried out since the PRNP gene that encodes PrP is likely expressed equally at the mRNA level for different genotypes and different cells, even though PrP levels do vary depending on the tissue (8, 17, 36, 41, 54). Thus, it is assumed that the different alleles of PrPC studied are present in equal amounts in brains of healthy heterozygous sheep, yet in heterozygous ARR/VRQ scrapie-infected individuals, the relative contributions of the various levels of allele expression products in the PrPres material are not equal.
The second procedure that we employed is a new tool to generate a polypeptide core fragment from codons 114 to 188 of ovine PrP from PrPres by using endoproteinase Lys-C digestion. This allowed a clear-cut discrimination of both codon 171 allotypic PrP forms (R/Q) in 2D electrophoresis on the basis of charge differences. This enzyme has been used before in a different context for studying the C-terminal fragment carrying a glycosylphosphatidylinositol anchor (58). In principle, Lys-C would release the 114-188PrP fragment from PrPs of nearly all animal species, including humans, the exceptions being some examples where the C-terminal lysine for cleavage would be expected to occur at PrP codon 167K (mink) or 197K (felines) (1, 35, 65). In recombinant PrP as well as proteinase K-treated scrapie PrPres, the 114-188PrP polypeptide fragment generation method appeared to be highly selective and sufficiently robust for the discrimination of 171R/Q-containing ovine PrP variants under the experimental conditions described. This was evidenced in the 1D Western blotting analyses, whereby the nonglycosylated and monoglycosylated PrP fragment forms were selectively detected using various PrP-specific antibodies; i.e., the binding of MAb 6C2 appeared to be absent if the epitope N terminus was processed into a free amino terminus (as was seen to occur after lysine 113).
Both the selective antibody approach using MAb SAF84/L42 ratios and the isolation of the 171R fragment by 2D electrophoresis demonstrated that only marginal amounts of the 171R-containing PrP were present (if at all) and constituted less than 9% of total PrPres. The differential migration at pI 7 and pI 8 of the 6-kDa PrP fragment generated after Lys-C digestion when using 171Q and 171R recPrP, respectively, was a consistent indicator that equivalent PrPres fragments were generated from scrapie brain. We concluded that the involvement of the 171R PrP allele product in PrPres formation is limited in classical scrapie field cases in ARR/VRQ sheep. The nature of the additional protein spots at a pI of <7 that were observed in both recPrP samples and sheep PrPres preparations is, as yet, unknown and requires further investigation. Unlike other studies, incomplete processing by PK seems to be an unlikely explanation since the migration differences in the second dimension are very small and the ablation of antibody binding by MAb 6C2 indicates that cleavage at lysine 113 was efficient. Therefore, it is conceivable that posttranslational differences between the additional protein spots are the more likely explanation. For example, methionine oxidation or methylated arginines could result in an increase of more acidic methionine residues or a decrease in the number of basic arginine residues, which would subsequently affect the overall net charge and thus explain the differently charged spots at a pI of <7 (12, 49).
The limited involvement of resistance allele 171R PrP in PrPSc formed during scrapie infection corroborates the findings observed in in vitro conversion studies with cellular and recombinant ovine PrPs when sheep are exposed to scrapie or BSE, in addition to data generated by mass spectrometric study of a single scrapie-infected ARR/VRQ sheep (9, 10, 32, 41). Our findings that 171R in PrPres was detected in limited amounts and in only 5 out of 8 ARR/VRQ sheep naturally infected with classical scrapie indicate that the 171R PrPres is not always present in PK-resistant material. Alternatively, it may be present, but at levels less than our current assay limits will permit detection.
A selective conversion of susceptibility-related 171Q PrPC to PrPSc in classical sheep scrapie might occur when in heterozygotes a highly susceptible allele, such as a codon 136V PrP, and a highly resistant allele, such as 171R PrP, are present, as was suggested before on the basis of in vitro conversion assays with sheep PrP (9). Similarly, in human heterozygous M/V129 carriers with GSS having either the susceptibility mutant codon 198 (Phe → Ser) or codon 217 (Gln → Arg), this mutant PrP appeared to be selectively involved in amyloid formation (59). Other studies on the inherited codon 102L mutant in human GSS patients (P102L heterozygous) revealed a variable but minor presence of wild-type PrP of up to 10% of total PrPres using immunochemical techniques with codon 102-insensitive and -sensitive PrP-specific antibodies, similar to the method used here with MAbs L42 and SAF84, respectively (66). However, in patients either with familial CJD and heterozygous for the inherited prion disease-related PrP codon 210V/I or with sporadic CJD with codon 129 M/V, both alleles were equally present in the proteinase K-resistant prion fraction (55). Our analysis of experimental BSE samples from sheep with the ARR/ARQ allele containing up to 65% 171R PrP in PrPres might indicate either that in this combination both ARQ (wild-type) and the resistant ARR alleles convert equally well or that BSE behaves differently from classical scrapie. A difference in allele composition in PrPres of heterozygotes between infections in the field and experimental infections might also play a role, though such information is lacking for sheep. Furthermore, heterozygosity can be a protective factor by itself, as was shown in cell culture (46).
It seems unlikely that the low/absent levels of the 171R-containing PrP in PrPres from ARR/VRQ sheep with scrapie result from the fact that while the 171R allele is converted to PrPSc, it is more susceptible to proteinase K and therefore would not be detected. The analysis of samples from a single ARR/ARR sheep with classical scrapie or sheep with BSE (3 cases studied) did not show differences in susceptibility of PrPres to proteolytic digestion at the PK concentrations (50 μg PK/ml) used for our studies. At higher concentrations of PK, however, PrPres 171R allele carriers appeared to be more PK susceptible than the 171Q/Q homozygotes. In two 171R/R field cases with classical scrapie, PrPres was shown to be more protease susceptible than that from 171Q/Q homozygous scrapie individuals, where higher PK concentrations, at and above 50 μg PK/ml, were used (22). A difference in the methodology used must also be considered, since we used Western blotting for PrPres detection, which revealed the whole remaining part of the PrP molecule present in PK-digested PrPSc, while the study referred to above used an ELISA with octarepeat (56-88PrP)-specific antibody for PrPres capture. The octarepeat region might be more prone to removal by PK than the large region at the C terminus of the octarepeats. Indeed, under our conditions, such octarepeats can be considered largely removed in both classical scrapie and BSE PrPres from sheep, while the part of PrP at the C terminus of these repeats remains available for detection (60). Finally, the possibility remains that under certain conditions the infectious entity associated with PrP conformations may be protease sensitive.
In the study of Rigter and Bossers, the binding of the 171Q or 171R allele product to PrPSc appeared to be equal (51). Therefore, it will be important to understand the mechanisms by which PrPC undergoes conversion to its disease-associated isoform and whether resistance to disease in heterozygotes is a consequence of poor conversion of a resistance allele (in this case, 171R PrP) or whether resistance-associated alleles somehow inhibit the entire process of conversion (including the conversion of alleles associated with susceptibility to infection). For other TSEs, including those caused by new and emerging strains, it is likely that other polymorphisms will play a more significant role in modulating the conversion process. For example, it is known that in the atypical/Nor98 scrapie TSE isolates, the 171R PrP is not a key polymorphism encoding absolute resistance (5). In our study, the methods used were not appropriate to estimate the level of 171R PrPres in atypical scrapie material of 171Q/R heterozygotes since, when using PK, our digestion conditions destroy most of the PrP region around position 171 eventually present.
Our data, showing the limited presence of 171R PrP in the PrPSc fraction of infected heterozygous sheep, could imply that 171R/R sheep infected with classical scrapie have a low risk of developing a new form of prion disease which has specifically adapted for this resistance-associated allele. If there was a significant risk for such adaptation, scrapie or new forms of TSE in sheep carrying this resistance allele would likely occur at frequencies much higher than those actually observed in 171R carriers, given that this allele historically represents more than 30% of the gene pool in most breeds (39). For other strains of prion disease, i.e., atypical/Nor98 scrapie or BSE, it is likely that the contributions of allelic PrP variants other than the sole contribution of the 171R allele in PrPres formation should be taken into account, though it appears that atypical/Nor98 scrapie does not transmit under field conditions and that experimental infection with BSE in 171R allele carriers is facilitated by intracerebral inoculation and disease manifests only after long incubation periods (5, 19, 28). In conclusion, our results strongly support current genetic breeding programs in sheep aiming at eradication of classical scrapie with the concurrent potential benefit of preventing BSE infection.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Geert-Jan Willems for skilled assistance in MAb 6C2 characterization. We are grateful to B. Jones and G. Hill from the Microbiological Services Department at the Institute for Animal Health, Compton, United Kingdom, for assistance in the generation of MAb FH6. Recombinant 94-233PrP was kindly provided by Andrew C. Gill (The Roslin Institute, University of Edinburgh). The help of H. Leroux and C. Rossignol in processing BSE tissues was greatly appreciated, as was the support of the INRA-PFIE level 3 animal facility. We thank DEFRA for providing “TSE-free” Pol Dorset and Suffolk NZ sheep.
These investigations were largely supported by the Dutch Ministry of Economic Affairs, Agriculture and Innovation, projects WOT-01-002-001.01 and WOT-01-002-001.05. Experimental BSE and scrapie samples were generated within EU project BSE in sheep (QLRT-2001-01309), led by Olivier Andreoletti, INRA-Toulouse-ENVT. Antibody characterization of FH6 was supported by the United Kingdom-Netherlands Partnership Programme in Science.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Agrimi U., et al. 2008. Prion protein amino acid determinants of differential susceptibility and molecular feature of prion strains in mice and voles. PLoS Pathog. 4:e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreoletti O., et al. 2006. Bovine spongiform encephalopathy agent in spleen from an ARR/ARR orally exposed sheep. J. Gen. Virol. 87:1043–1046 [DOI] [PubMed] [Google Scholar]
- 3. Baylis M., Goldmann W. 2004. The genetics of scrapie in sheep and goats. Curr. Mol. Med. 4:385–396 [DOI] [PubMed] [Google Scholar]
- 4. Belt P. B., et al. 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 76(Pt 3):509–517 [DOI] [PubMed] [Google Scholar]
- 5. Benestad S. L., Arsac J. N., Goldmann W., Noremark M. 2008. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet. Res. 39:19. [DOI] [PubMed] [Google Scholar]
- 6. Beringue V., Vilotte J. L., Laude H. 2008. Prion agent diversity and species barrier. Vet. Res. 39:47. [DOI] [PubMed] [Google Scholar]
- 7. Bessen R. A., Marsh R. F. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68:7859–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bilheude J. M., et al. 2007. Discrimination of sheep susceptible and resistant to transmissible spongiform encephalopathies by an haplotype specific monoclonal antibody. J. Virol. Methods 145:169–172 [DOI] [PubMed] [Google Scholar]
- 9. Bossers A., et al. 1997. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. U. S. A. 94:4931–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bossers A., de Vries R., Smits M. A. 2000. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol. 74:1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown P. 1990. Transmissible spongiform encephalopathies in humans: kuru, Creutzfeldt-Jakob disease and Gerstmann-Straussler-Scheinker disease. Can. J. Vet. Res. 54:38–41 [PMC free article] [PubMed] [Google Scholar]
- 12. Canello T., et al. 2008. Methionine sulfoxides on PrPSc: a prion-specific covalent signature. Biochemistry 47:8866–8873 [DOI] [PubMed] [Google Scholar]
- 13. Carp R. I., Moretz R. C., Natelli M., Dickinson A. G. 1987. Genetic control of scrapie: incubation period and plaque formation in I mice. J. Gen. Virol. 68(Pt 2):401–407 [DOI] [PubMed] [Google Scholar]
- 14. Corbiere F., et al. 2007. Advanced survival models for risk-factor analysis in scrapie. J. Gen. Virol. 88:696–705 [DOI] [PubMed] [Google Scholar]
- 15. Cuillé J., Chelle P. L. 1939. Transmission de la tremblante à la chèvre. C. R. Hebd. Seances Acad. Sci. 207:1058–1060 [Google Scholar]
- 16. Demart S., et al. 1999. New insight into abnormal prion protein using monoclonal antibodies. Biochem. Biophys. Res. Commun. 265:652–657 [DOI] [PubMed] [Google Scholar]
- 17. Denman R., Potempska A., Wolfe G., Ramakrishna N., Miller D. L. 1991. Distribution and activity of alternatively spliced Alzheimer amyloid peptide precursor and scrapie PrP mRNAs on rat brain polysomes. Arch. Biochem. Biophys. 288:29–38 [DOI] [PubMed] [Google Scholar]
- 18. Dickinson A. G. 1975. Host-pathogen interactions in scrapie. Genetics 79(Suppl.):387–395 [PubMed] [Google Scholar]
- 19. Fediaevsky A., et al. 2008. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Vet. Res. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geysen H. M., Meloen R. H., Barteling S. J. 1984. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. U. S. A. 81:3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldmann W., et al. 1998. Two alleles of a neural protein gene linked to scrapie in sheep. Proc. Natl. Acad. Sci. U. S. A. 87:2476–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groschup M. H., et al. 2007. Classic scrapie in sheep with the ARR/ARR prion genotype in Germany and France. Emerg. Infect. Dis. 13:1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hagenaars T. J., et al. 2010. Scrapie prevalence in sheep of susceptible genotype is declining in a population subject to breeding for resistance. BMC Vet. Res. 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harlow E., Lane D. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 25. Harmeyer S., Pfaff E., Groschup M. H. 1998. Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J. Gen. Virol. 79(Pt 4):937–945 [DOI] [PubMed] [Google Scholar]
- 26. Hoffmann C., et al. 2011. BSE infectivity in jejunum, ileum and ileocaecal junction of incubating cattle. Vet. Res. 42:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hope J., Multhaup G., Reekie L. J., Kimberlin R. H., Beyreuther K. 1988. Molecular pathology of scrapie-associated fibril protein (PrP) in mouse brain affected by the ME7 strain of scrapie. Eur. J. Biochem. 172:271–277 [DOI] [PubMed] [Google Scholar]
- 28. Houston F., et al. 2003. Prion diseases: BSE in sheep bred for resistance to infection. Nature 423:498. [DOI] [PubMed] [Google Scholar]
- 29. Hunter N., Bossers A. 2006. The PrP genotype as a marker for scrapie susceptibility in sheep, p. 640–647 In Hörnlimann B., Riesner D., Kretzschmar H. (ed.), Prions in humans and animals. de Gruyter, Berlin, Germany [Google Scholar]
- 30. Ikeda T., et al. 1995. Amino acid polymorphisms of PrP with reference to onset of scrapie in Suffolk and Corriedale sheep in Japan. J. Gen. Virol. 76(Pt 10):2577–2581 [DOI] [PubMed] [Google Scholar]
- 31. Jacobs J. G., et al. 2011. Differentiation of ruminant transmissible spongiform encephalopathy isolate types, including bovine spongiform encephalopathy and CH1641 scrapie. J. Gen. Virol. 92:222–232 [DOI] [PubMed] [Google Scholar]
- 32. Kirby L., Goldmann W., Houston F., Gill A. C., Manson J. C. 2006. A novel, resistance-linked ovine PrP variant and its equivalent mouse variant modulate the in vitro cell-free conversion of rPrP to PrP(res). J. Gen. Virol. 87:3747–3751 [DOI] [PubMed] [Google Scholar]
- 33. Langeveld J. P., et al. 2006. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet. Res. 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lehninger A. L., Lee Nelson D., Cox M. M. 2005. Lehninger principles of biochemistry, vol. 1 W. H. Freeman & Co., New York, NY [Google Scholar]
- 35. Lysek D. A., Nivon L. G., Wuthrich K. 2004. Amino acid sequence of the Felis catus prion protein. Gene 341:249–253 [DOI] [PubMed] [Google Scholar]
- 36. Manson J., et al. 1992. The prion protein gene: a role in mouse embryogenesis? Development 115:117–122 [DOI] [PubMed] [Google Scholar]
- 37. Manson J. C., et al. 1994. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol. Neurobiol. 8:121–127 [DOI] [PubMed] [Google Scholar]
- 38. Mead S., et al. 2009. A novel protective prion protein variant that colocalizes with kuru exposure. N. Engl. J. Med. 361:2056–2065 [DOI] [PubMed] [Google Scholar]
- 39. Melchior M. B., et al. 2011. Active surveillance for scrapie in the Netherlands: effect of a breeding programme on the prevalence of scrapie in sheep (2002-2010). Tijdschr. Diergeneeskd. 136:84–93 (In Dutch.) [PubMed] [Google Scholar]
- 40. Melchior M. B., et al. 2010. Eradication of scrapie with selective breeding: are we nearly there? BMC Vet. Res. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morel N., Andreoletti O., Grassi J., Clement G. 2007. Absolute and relative quantification of sheep brain prion protein (PrP) allelic variants by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 21:4093–4100 [DOI] [PubMed] [Google Scholar]
- 42. Oesch B., et al. 1985. A cellular gene encodes scrapie PrP 27-30 protein. Cell 40:735–746 [DOI] [PubMed] [Google Scholar]
- 43. Orge L., et al. 2010. Putative emergence of classical scrapie in a background of enzootic atypical scrapie. J. Gen. Virol. 91:1646–1650 [DOI] [PubMed] [Google Scholar]
- 44. Parchi P., Capellari S., Gambetti P. 2000. Intracerebral distribution of the abnormal isoform of the prion protein in sporadic Creutzfeldt-Jakob disease and fatal insomnia. Microsc. Res. Tech. 50:16–25 [DOI] [PubMed] [Google Scholar]
- 45. Paspaltsis I., et al. 2006. Titanium dioxide photocatalytic inactivation of prions. J. Gen. Virol. 87:3125–3130 [DOI] [PubMed] [Google Scholar]
- 46. Priola S. A., Caughey B., Race R. E., Chesebro B. 1994. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68:4873–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prusiner S. B. 1998. Prions. Proc. Natl. Acad. Sci. U. S. A. 95:13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prusiner S. B., Groth D. F., Bolton D. C., Kent S. B., Hood L. E. 1984. Purification and structural studies of a major scrapie prion protein. Cell 38:127–134 [DOI] [PubMed] [Google Scholar]
- 49. Reporter M. 1973. Methylation of basic residues in structural proteins. Mech. Ageing Dev. 1:367–372 [DOI] [PubMed] [Google Scholar]
- 50. Rezaei H., et al. 2000. High yield purification and physico-chemical properties of full-length recombinant allelic variants of sheep prion protein linked to scrapie susceptibility. Eur. J. Biochem. 267:2833–2839 [DOI] [PubMed] [Google Scholar]
- 51. Rigter A., Bossers A. 2005. Sheep scrapie susceptibility-linked polymorphisms do not modulate the initial binding of cellular to disease-associated prion protein prior to conversion. J. Gen. Virol. 86:2627–2634 [DOI] [PubMed] [Google Scholar]
- 52. Rigter A., et al. 2007. Mapping of possible prion protein self-interaction domains using peptide arrays. BMC Biochem. 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saunders G. C., et al. 2007. Polymorphisms of the prion protein gene coding region in born-after-the-reinforced-ban (BARB) bovine spongiform encephalopathy cattle in Great Britain. J. Gen. Virol. 88:1374–1378 [DOI] [PubMed] [Google Scholar]
- 54. Scott M. R., et al. 1988. Prion protein gene expression in cultured cells. Protein Eng. 2:69–76 [DOI] [PubMed] [Google Scholar]
- 55. Silvestrini M. C., et al. 1997. Identification of the prion protein allotypes which accumulate in the brain of sporadic and familial Creutzfeldt-Jakob disease patients. Nat. Med. 3:521–525 [DOI] [PubMed] [Google Scholar]
- 56. Slootstra J. W., Puijk W. C., Ligtvoet G. J., Langeveld J. P., Meloen R. H. 1996. Structural aspects of antibody-antigen interaction revealed through small random peptide libraries. Mol. Divers. 1:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smits M. A., Bossers A., Schreuder B. E. 1997. Prion protein and scrapie susceptibility. Vet. Q. 19:101–105 [DOI] [PubMed] [Google Scholar]
- 58. Stahl N., Baldwin M. A., Burlingame A. L., Prusiner S. B. 1990. Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry 29:8879–8884 [DOI] [PubMed] [Google Scholar]
- 59. Tagliavini F., et al. 1994. Amyloid fibrils in Gerstmann-Straussler-Scheinker disease (Indiana and Swedish kindreds) express only PrP peptides encoded by the mutant allele. Cell 79:695–703 [DOI] [PubMed] [Google Scholar]
- 60. Thuring C. M., et al. 2004. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J. Clin. Microbiol. 42:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thuring C. M., et al. 2005. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J. Comp. Pathol. 132:59–69 [DOI] [PubMed] [Google Scholar]
- 62. Truscott J. E., Ferguson N. M. 2009. Control of scrapie in the UK sheep population. Epidemiol. Infect. 137:775–786 [DOI] [PubMed] [Google Scholar]
- 63. Vaccari G., et al. 2009. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet. Res. 40:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van Keulen L. J., et al. 1996. Immunohistochemical detection of prion protein in lymphoid tissues of sheep with natural scrapie. J. Clin. Microbiol. 34:1228–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Rheede T., Smolenaars M. M., Madsen O., de Jong W. W. 2003. Molecular evolution of the mammalian prion protein. Mol. Biol. Evol. 20:111–121 [DOI] [PubMed] [Google Scholar]
- 66. Wadsworth J. D., et al. 2006. Phenotypic heterogeneity in inherited prion disease (P102L) is associated with differential propagation of protease-resistant wild-type and mutant prion protein. Brain 129:1557–1569 [DOI] [PubMed] [Google Scholar]
- 67. Whyte S. M., et al. 2003. Stability and conformational properties of doppel, a prion-like protein, and its single-disulphide mutant. Biochem. J. 373:485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Windl O., et al. 1996. Genetic basis of Creutzfeldt-Jakob disease in the United Kingdom: a systematic analysis of predisposing mutations and allelic variation in the PRNP gene. Hum. Genet. 98:259–264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.