Abstract
Adeno-associated virus type 2 (AAV2) capsid assembly requires the expression of a virally encoded assembly-activating protein (AAP). By providing AAP together with the capsid protein VP3, capsids are formed that are composed of VP3 only. Electron cryomicroscopy analysis of assembled VP3-only capsids revealed all characteristics of the wild-type AAV2 capsids. However, in contrast to capsids assembled from VP1, VP2, and VP3, the pores of VP3-only capsids were more restricted at the inside of the 5-fold symmetry axes, and globules could not be detected below the 2-fold symmetry axes. By comparing the capsid assembly of several AAV serotypes with AAP protein from AAV2 (AAP-2), we show that AAP-2 is able to efficiently stimulate capsid formation of VP3 derived from several serotypes, as demonstrated for AAV1, AAV2, AAV8, and AAV9. Capsid formation, by coexpressing AAV1-, AAV2-, or AAV5-VP3 with AAP-1, AAP-2, or AAP-5 revealed the ability of AAP-1 and AAP-2 to complement each other in AAV1 and AAV2 assembly, whereas for AAV5 assembly more specific conditions are required. Sequence alignment of predicted AAP proteins from the known AAV serotypes indicates a high degree of homology of all serotypes to AAP-2 with some divergence for AAP-4, AAP-5, AAP-11, and AAP-12. Immunolocalization of assembled capsids from different serotypes confirmed the preferred nucleolar localization of capsids, as observed for AAV2; however, AAV8 and AAV9 capsids could also be detected throughout the nucleus. Taken together, the data show that AAV capsid assembly of different AAV serotypes also requires the assistance of AAP proteins.
INTRODUCTION
The adeno-associated virus type (AAV) assembly pathway proposed by Myers and Carter (17) suggests that empty capsids form rapidly and then, in a slow reaction, the replicated single-stranded genome is inserted into the capsid. While the process of genome replication has been elucidated in great detail (16, 32), molecular events underlying capsid formation and genome encapsidation are largely unknown. Capsid assembly occurs in the nucleus of infected cells, and the capsids are first detectable in the nucleoli (33). Expression of the cap gene is sufficient for capsid formation (33). In addition to the VP proteins VP1, VP2, and VP3, known to be expressed from open reading frame 1 (ORF1) of the AAV2 cap gene, ORF2 of the cap gene encodes an assembly factor, called assembly-activating protein (AAP), which is essential for the capsid assembly process (29). This protein targets newly synthesized VP proteins to the nucleolus and promotes capsid formation in a still unknown way. The strict localization of the assembly reaction to nucleoli suggests that cellular proteins contribute to the assembly process, too.
In past years a large number of different AAV serotypes have been described (1, 5–7, 15, 26, 27). They are of human or nonhuman primate origin and represent promising tools for establishing optimized vectors for gene transfer. They expand the range of tissue tropism and may help a virus to escape gene transfer neutralization by preexisting antibodies. A striking feature is that vector DNA flanked by inverted terminal repeats of AAV2 can be packaged into capsids of different serotypes if the appropriate Rep proteins (in this case from AAV2) were used for genome replication (8, 25). However, vector production of certain AAV serotypes, is less efficient than for other serotypes. The role of the AAP proteins in capsid assembly for different serotypes has not been studied yet.
We describe here the potential of the newly identified AAP of AAV2 (AAP-2) in promoting capsid formation of serotypes AAV1, AAV2, AAV5, AAV8, and AAV9. Our study shows that AAP-2 can stimulate capsid assembly of VP3 capsids derived from AAV1, AAV2, AAV8, and AAV9, while the respective VP3 proteins alone are assembly incompetent. Furthermore, homologous AAPs from AAV1 and AAV5 have been isolated and tested for their ability to promote capsid formation of VP3 derived from AAV1, AAV2, and AAV5. Although they can also promote capsid formation of heterologous serotypes, they do so with different efficiencies. In particular, AAV5 assembly required AAP-5 for capsid formation and was difficult to detect. The data indicate that capsid assembly of these AAV serotypes depends on the same type of AAP as AAV2. AAP protein sequences reflect the evolutionary relationship of the AAV serotypes described thus far.
MATERIALS AND METHODS
Plasmids and cloning.
Plasmids pBS (Stratagene, Amsterdam, Netherlands), pVP2N-gfp, pCMV-VP3/2809 (29), and pDP1, pDP2, pDP3, pDP4, pDP5, and pDP6 (8) have been described previously. Plasmids pDP8 and pDP9 were constructed by inserting of the cap ORFs of AAV8 and AAV9 obtained from p5E18 VD2/8 and p5E18 VD2/9 (5) (kindly provided by G. P. Gao, University of Pennsylvania School of Medicine, Philadelphia) into pDP2 while replacing the cap gene of AAV2.
Constructs pCMV-AAV5-VP3, pCMV-AAV8-VP3, and pCMV-AAV9-VP3 designed for the expression of VP3 were generated by PCR amplification of the VP3 coding sequences of the respective AAV serotypes (Table 1). The HindIII-SnaBI-digested amplification products were cloned into the HindIII-HincII fragment of pBS-CMV (29). Plasmid pCMV-AAV1-VP3 was cloned as follows. By mutagenesis, a HindIII restriction site was introduced directly before the VP3 ATG start codon of plasmid pUCrep/fs/cap-AAV1 (described within PCT/EP2008/004366) using the primers 5′-CGCTGCTGTGGGACCTAAGCTTATGGCTTCAGGCGGTGGCG-3′ and 5′-CGCCACCGCCTGAAGCCATAAGCTTAGGTCCCACAGCAGCG-3′. The resulting plasmid was digested with AgeI. The AgeI site was blunt ended with Klenow polymerase, and the construct was subsequently digested with HindIII. The generated fragment was cloned into the HindIII/HincII-digested pBS-CMV backbone.
Table 1.
Constructs used in this study
| Construct | Template (reference) | Primer |
|
|---|---|---|---|
| Orientationa | Sequence (5′–3′) | ||
| pCMV-AAV5-VP3 | pRC5 (8) | F | CCAAGTTTGGGAGCTAAGCTTATGTCTGCGGGAGG |
| R | CGAGCGAACGCGACATACGTAAGAGTGCCACACTCTC | ||
| pCMV-AAV8-VP3 | p5E18 VD2/8 (5) | F | GGTGTGGGACCTAAGCTTATGGCTGCAGGC |
| R | GCAGGTTTAAACGAATTCTACGTACGCAGAGACC | ||
| pCMV-AAV9-VP3 | p5E18 VD2/9 (5) | F | CAGGTGTGGGATCTAAGCTTATGGCTTCAGGTGG |
| R | GGTTTAAACGAATTCTACGTACGCAGAGACC | ||
| pAAP-1-AU1 | pDP1 (8) | F | CGGGTTCTCGAACCTCTCAAGCTTGTTGAGGAAGGC |
| R | GCGGTGTCTCGAGTTATATATAGCGATAGGTGTCTGAACACGTCCGCCGGGAACGGAGGGAGGCAGCC | ||
| pAAP-2-AU1 | pTAV2.0 (10) | F | GAGGGTTCTTGAACCTCTAAGCTTGGTTGAGGAACCTG |
| R | GCGGTGTCTCGAGTTATATATAGCGATAGGTGTCGGGTGAGGTATCCATACTGTGGCACCATGAAGAC | ||
| pAAP-5-AU1 | pDP5 (8) | F | CACATCCTTCGGGGGAAGCTTCGGAAAGGCAGTCTTTC |
| R | GCGGTGTCTCGAGTTATATATAGCGATAGGTGTCGCGTCGCGTAACCGTACTGCGGCAGCGTAAAGAC | ||
F, forward; R, reverse.
Plasmid pCMV-AAV9-VP3stop was cloned by site-directed mutagenesis with template pCMV-AAV9-VP3 and the complementary primers 5′- GGTGCCGATGGAGTAGGTAGTTCCTCGGG-3′ and 5′- CCCGAGGAACTACCTACTCCATCGGCACC-3′. Then, the HindIII-XhoI fragment was subcloned from the amplification product into the HindIII-XhoI backbone of pCMV-AAV9-VP3.
Constructs pAAP-1-AU1, pAAP-2-AU1, and pAAP-5-AU1 designed for expressing AAP fused to an AU1-tag were generated by PCR amplification of the AAP coding sequences of the respective AAV serotypes (Table 1). The HindIII-XhoI-digested amplification products were cloned into the HindIII-XhoI fragment of pBS-CMV.
Cell culture.
HeLa and 293T cells were maintained at 37°C and 5% CO2 in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM l-glutamine. X63/Ag8 B lymphoma cells (kindly provided by Peter Krammer, DKFZ, Heidelberg, Germany) were cultivated at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine, and 20 mM HEPES (pH 7.2).
Transfection and preparation of cell lysates.
The 293T cells (5 × 105 per 6-cm dish or 1 × 106 per 10-cm dish) were cultivated for 24 h prior to transfection by calcium phosphate precipitation (18). Afterward, the cells were incubated for another 48 h. To quantify AAV capsid titers, the cells were harvested in the medium (6-cm dish) and lysed by three freeze-thaw cycles (−80°C and 37°C). Cell debris was removed by centrifugation at 10,000 × g for 5 min. To prepare the cell extracts used in immuno-dot blot analysis, cells were harvested (in two 10-cm dishes), washed once in ice-cold phosphate-buffered saline (PBS; 18.4 mM Na2HPO4, 10.9 mM KH2PO4, 125 mM NaCl), resuspended in PBS-MK (PBS containing 1 mM MgCl2 and 2.5 mM KCl), and supplemented with protease inhibitor mix (Complete Mini EDTA-free; Roche, Mannheim, Germany). Three sonication cycles (60% power) were performed on ice to lyse the cells. Lysates were treated with DNase I (Roche) at 500 μg/ml for 2 h at 37°C. After centrifugation at 10,000 × g for 10 min at 4°C, the supernatants were collected.
Analysis of protein expression.
Identical portions of harvested cells were processed for SDS-PAGE. Protein expression was analyzed by Western blot assay with monoclonal antibodies B1 or anti-AU1 (Covance, Emeryville, CA) or polyclonal antibody anti-AAP (29) as described previously (34).
Generation of monoclonal antibodies ADK6, ADK8, and ADK9.
Female C57BL/6 (H-2b) mice were immunized according to the protocol of Kuck et al. (12) using recombinant AAV6, AAV8, or AAV9 vectors that carried a green fluorescent protein (GFP) gene inserted between the AAV2-inverted terminal repeats. Extracted spleen cells and X63/Ag8 B lymphoma cells were fused as described previously (35). Hybridoma supernatants were screened by using an immunofluorescence assay (12).
Determination of antibody isotypes.
Subclasses of antibodies were determined using a mouse monoclonal antibody isotyping kit (RPN29; Amersham, Braunschweig, Germany) according to the manufacturer's guidelines. Both ADK6 and ADK8 were classified as IgG2a/κ, whereas ADK9 was classified as IgA/κ.
Quantification of AAV capsids.
Capsid titers of AAV1, AAV2, and AAV5 were determined using the respective AAV titration enzyme-linked immunosorbent assay (ELISA) kit (Progen, Heidelberg, Germany) according to the manufacturer's manual. The amount of AAV8 or AAV9 capsids was quantified by ELISA based on monoclonal antibodies ADK8 or ADK9, respectively. Flexible Polysorb 96-well microtiter plates (Nunc, Schwerte, Germany) were coated with 50 ng of ADK8 or ADK9 per well (total volume per well was 100 μl) and stored overnight at 4°C. The microtiter plate was washed twice with PBS before adding 100 μl of blocking buffer (0.2% casein in PBS-T [PBS, 0.05% Tween 20]) to each well. After 1 h at 37°C and three PBS-T washes, an AAV8 or AAV9 standard (capsid titers were determined by counting after negative staining and electron microscopy as described by Grimm et al. [9]) and samples were added to the wells in serial dilutions. Viral particles were incubated for 1 h at 37°C. The wells were washed three times with PBS-T and then 100 μl of horseradish peroxidase-conjugated ADK8 or ADK9 (1 μg/ml, diluted in PBS-T) was added. The microtiter plate was kept for another hour at 37°C. After a repeated washing step, 100 μl/well of a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (1 mg of TMB/ml, 0.1 M sodium acetate [pH 6.0], 0.003% H2O2) was applied, producing a color reaction that was stopped with 50 μl/well of 1 M H2SO4. The color intensity was measured at 450 nm in an ELISA reader.
Sucrose density gradient analysis.
Samples were loaded onto a linear 10 to 30% sucrose gradient (sucrose in PBS-MK, 10 mM EDTA, supplemented with protease inhibitor mix) in Beckman tubes (14 by 89 mm). After centrifugation at 160,000 × g for 2 h at 4°C (SW41 rotor; Beckman), 500-μl fractions were collected and further processed in an immuno-dot blot assay.
Immuno-dot blot analysis.
Cell extracts of transfected 293 T cells or fractions obtained after sucrose density gradient centrifugation were analyzed under native conditions by transferring 125 μl of each cell extract or sucrose gradient fraction onto a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany) using a vacuum blotter. Cell extracts of transfected 293 T cells were also analyzed under denaturing conditions by mixing 125 μl of each extract with 125 μl of 2× SDS-PAGE sample buffer, followed by heating for 5 min at 95°C and transfer onto a nitrocellulose membrane. In both cases immune detection was performed as described previously (12).
Capsid assembly in the presence of proteasome inhibitor.
For studying capsid assembly in the presence of proteasome inhibitor MG132, transfected 293T cells were incubated 24 h after transfection via a medium exchange with 2 μM MG132 in the culture medium for another 24 h. Thereafter, MG132-containing medium was withdrawn and replaced by fresh medium without MG132. All other conditions of the capsid transcomplementation assay were identical to the experiments without MG132 as described here and in Results.
Immunofluorescence analysis.
HeLa cells were cultivated for 24 h on coverslips. Then, 20 h after transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), the cells were fixed with 100% methanol (10 min, −20°C) and washed with PBS. They were subsequently incubated with primary antibodies for 1 h at room temperature or overnight at 4°C. Coverslips were washed three times with PBS and thereafter incubated with appropriate secondary antibodies (purchased from Dianova [Hamburg, Germany] or Molecular Probes [Leiden, Netherlands]) for 1 h at room temperature. The coverslips were then washed again, dipped into 100% ethanol, and embedded in Permafluor mounting medium (Beckman Coulter, Marseille, France). Confocal images (0.3-μm sections) were obtained with a Leica TCS SP2 laser scanning microscope and further processed using Adobe Photoshop CS software.
Preparation and electron cryomicroscopy of empty AAV2 capsids.
Empty AAV2 capsids were purified using two consecutive double sucrose cushions as described previously (30). For electron microscopy, the samples were vitrified as previously described (11) and imaged under low-dose conditions in a Philips CM200 FEG, which was operated at an accelerating voltage of 200 kV. Micrographs were recorded on a 2kx2k charge-coupled device camera (TVIPS GmbH) at a nominal magnification of 50,000 corresponding to a calibrated pixel size of 2.68 Å. A total of 3,345 particles were manually selected from 162 micrographs using Boxer (13). The orientation and origin of the boxed particles (box size, 150 × 150 pixels) were calculated by cross-common lines (4) against ten projections of a reference map. The map of empty AAV2 capsids was taken as the first reference map (11). Afterward, the last calculated map of VP3 VLPs was used as a reference. Determining orientations followed by calculating a new map was repeated three times. After the third round, the resolution did not improve any further. The resolution of the map was estimated by Fourier-Shell correlation using the spatial frequency at which the correlation dropped to 0.5 (3). The estimated resolution was 15.5 Å. Differences between empty AAV2 capsids (11) and VP3 VLPs were calculated according to the Spider DR DIFF procedure (28). This procedure scales the gray values of the maps to minimize the differences between them. The surface representations were generated with Chimera (19).
RESULTS
Structure of VP3 capsids.
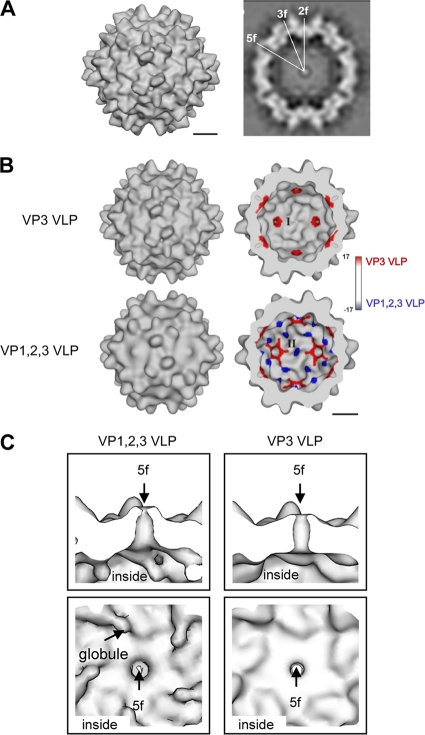
The AAP induces capsid formation of VP3 in the absence of VP1 and VP2 (29). VP3 of AAV2 expressed without AAP cannot form capsids, whereas coexpression of AAP restores capsid assembly to a level comparable to that obtained by expression of all three capsid proteins. To determine whether capsids assembled from VP3 alone (VP3-VLPs) have the basic characteristics of AAV capsids, which are assembled of VP1, VP2, and VP3, we analyzed VP3-VLPs by electron cryomicroscopy and image processing. Image reconstruction of VP3-VLPs showed all features of AAV capsids, including the 3-fold spikes and the pores at the 5-fold and the dimple at the 2-fold symmetry axes (Fig. 1A). However, the form of the pores at the 5-fold symmetry axes was slightly different and globules were lacking at the 2-fold axes compared to empty VP1-, VP2-, and VP3-containing capsids (11). These areas at the inner capsid surface show the largest differences between VP3-VLPs and VP1,2,3-VLPs, as visualized by the color-coded difference map (Fig. 1B). The globules at the 2-fold symmetry axes of VP1,2,3-VLPs (colored in blue, negative difference), which are characteristic for empty AAV2 capsids (11), were absent on VP3-VLPs, and VP3-VLPs showed an increased density around the pores of the 5-fold symmetry axes (indicated in red, positive difference) restricting the inner pore diameter and forming a more cylinder-shaped channel compared to the funnel-like channel formed in the presence of all three VP proteins (Fig. 1C). At the inner surface, the VP3-VLPs resemble the conformation of the crystal structure of AAV2 (36) much closer than VP1,2,3-VLPs. The data indicate that the assembly assay using only VP3 capsid proteins together with AAP validly reflects AAV capsid formation, but they also show that the VP1/VP2 N termini affect the morphology of the inner AAV capsid surface.
Fig. 1.
Image reconstruction of AAV2 VP3 VLPs in comparison to VLPs consisting of VP1, VP2, and VP3. (A) Surface representation of the capsid composed of VP3 only (VP3 VLP) (left) and equatorial slice through the capsid (right). For representation, the map was sharpened by applying an inverse B-factor of 300 Å2. (B) Surface representations of the outer (left) and inner capsid surfaces (right) of the maps of VP3 VLPs (top) and VP1,2,3 VLPs (bottom) (11). The surfaces are color coded to show the difference between the map of VP3 VLPs and the map of VP1,2,3 VLPs. Red represents extra densities in the map of the VP3 VLPs, and blue represents extra densities in the map of the VP1,2,3 VLPs. The major differences are located at the 5- and 2-fold symmetry axes at the inner surface, indicated by I and II. Bars, 5 nm. (C) Cross-section through a pore at a 5-fold symmetry axis (upper panel) and view onto the inner surface at the 5-fold symmetry axis (lower panel) showing the map of VP1,2,3 VLPs (left) and the map of VP3 VLPs (right). No B-factor sharpening was applied in panels B and C.
Capsid assembly of different AAV serotypes.
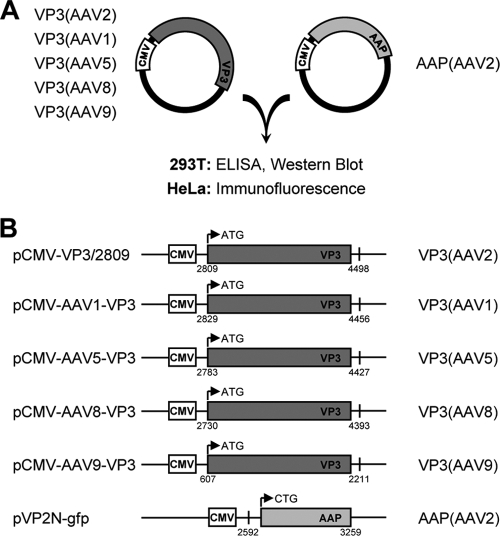
In order to analyze whether AAP from AAV2 promotes capsid formation of other serotypes as well, we chose VP3 of AAV1, AAV5, AAV8, and AAV9 to investigate the capacity of AAP-2 to direct the assembly process. To address this question, we used our previously established assembly transcomplementation assay (Fig. 2A) (29). The VP3 sequences of the cap genes of AAV1, AAV5, AAV8, and AAV9 were isolated by PCR from corresponding genomic AAV plasmids and inserted into expression vectors under the control of the human cytomegalovirus immediate-early promoter (CMV) (Fig. 2B). The assembly reaction was monitored by capsid-specific ELISAs and by indirect immunofluorescence. While for AAV1, AAV2, and AAV5 capsid-specific ELISAs were already available (9, 12), we had to generate new antibodies to monitor AAV8 and AAV9 capsid formation.
Fig. 2.
Transcomplementation assay used to compare capsid assembly of different AAV serotypes. (A) VP3 of the indicated serotypes was coexpressed with AAP derived from AAV2 in 293T and HeLa cells. Capsid assembly was analyzed by ELISA and immunofluorescence. (B) VP3 and AAP expression constructs, including denotations of the constructs (left) and the expressed proteins (right). Small numbers indicate nucleotide positions relative to the corresponding AAV serotype genome sequences (see Fig. 7 for the accession numbers). Arrows represent the translation start sites of the VP3 and AAP proteins. CMV, human cytomegalovirus immediate-early promoter.
Generation of new capsid-specific monoclonal antibodies.
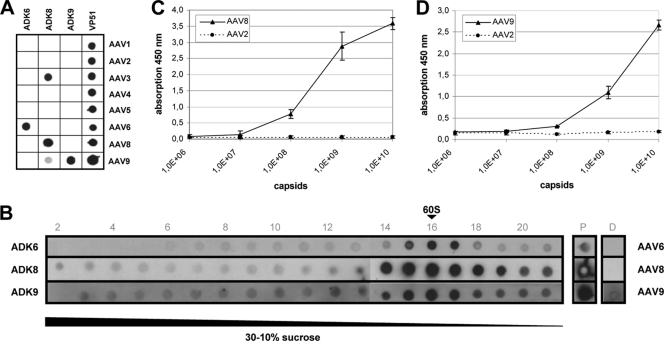
Fig. 3A shows the reaction of newly generated monoclonal antibodies ADK6, ADK8, and ADK9 with capsids of a representative panel of different AAV serotypes in a dot blot assay under nondenaturing conditions. The cross-reaction of ADK8 antibody with AAV3 in the dot blot assay is overrepresented (Fig. 3A) and was not confirmed by an indirect immunofluorescence assay (data not shown). ADK9 reacted only with AAV9 capsids. In the same series of experiments, we isolated an antibody, ADK6, which distinguished between the closely related serotypes AAV1 and AAV6. Sucrose density gradient analysis of cell extracts containing assembled and nonassembled capsid proteins by immune dot blots under nondenaturing conditions showed that the new antibodies preferentially reacted with assembled capsids (Fig. 3B). None of the antibodies reacted with denatured capsid proteins (lane D in Fig. 3B). Capsid ELISAs with ADK8 and ADK9 antibodies showed that the detection limit of ADK8 is in the range of 5 × 107 capsids and 1 × 108 capsids for ADK9 (Fig. 3C and D). Antibody ADK6 was not used in the following experiments.
Fig. 3.
Characterization of newly generated AAV capsid antibodies. (A) Monoclonal antibodies ADK6, ADK8, and ADK9 were reacted in a native dot blot assay with 293T cell extracts, each containing empty capsids of one of the indicated AAV serotypes. Reaction with a polyclonal AAV2 antiserum (VP51) indicated the presence of respective capsid proteins and capsids. (B) Cell extracts containing AAV6, AAV8, or AAV9 capsids were fractionated on sucrose gradients and analyzed by the native dot blot assay. Sedimentation position of 60S was determined in a parallel gradient using empty AAV2 capsids detected by antibody A20. Lane P shows the reaction with the loaded cell extract under native conditions and lane D after denaturation of proteins with SDS and heating. (C) Reaction of ADK8 antibody in an ELISA measuring absorption at 450 nm with increasing amounts of AAV8 capsids. (D) Reaction of ADK9 antibody in an ELISA measuring the absorption at 450 nm with increasing amounts of AAV9 capsids. The dotted lines in panels C and D show the reaction with equivalent amounts of AAV2 capsids.
AAP-2 stimulates capsid assembly of several other AAV serotypes.
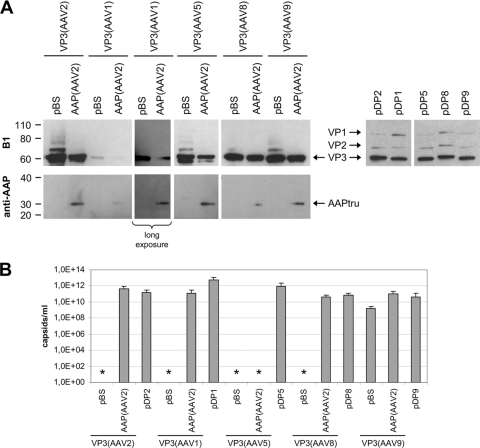
To analyze the effect of AAP-2 on the capsid assembly of different AAV serotypes, the respective VP3 expression plasmids were either cotransfected with a plasmid expressing the assembly helper protein AAP-2 in a truncated variant (pVP2N-gfp, Fig. 2B) (29) or with the empty vector plasmid (pBS) into 293T cells. To control the efficiency of capsid assembly, plasmids pDP1, pDP2, pDP5, pDP8, and pDP9 were transfected, which express the corresponding complete cap genes—including the respective AAP proteins—in combination with the rep gene of AAV2 and the adenovirus genes E2A and E4orf6 and VA-RNAs (8).
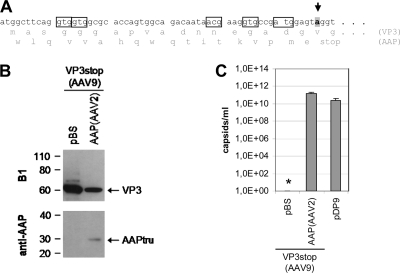
Western blot analysis showed that the different constructs expressed the expected VP proteins at comparable levels (Fig. 4A). Only expression of AAV1 VP3 was relatively low; however, it could be readily detected upon longer exposure of the Western blot. The AAP-2 protein was detected by an AAP peptide-directed antiserum (Fig. 4A). While in combination with VP3 of AAV2, AAV5, AAV8, and AAV9 expression could be traced, it was poor in combination with AAV1 VP3. By quantifying capsid formation, we showed that the VP3 of AAV1, AAV2, AAV5, and AAV8 alone was not able to form capsids. Coexpression of AAP-2 induced capsid assembly of AAV1, AAV2, and AAV8 VP3 to levels close to those of the control or only slightly lower (Fig. 4B). AAV5 VP3 capsid assembly could not be stimulated by AAP-2 under these conditions. Unexpectedly, expression of VP3 of AAV9 showed capsid formation at a low level in the absence of coexpressed AAP. DNA sequence analysis of the AAV9-VP3 expression plasmid revealed that it contains potential translation initiation codons in ORF2, which could induce the expression of an N-terminally truncated AAP-9 and stimulate VP3 capsid formation (Fig. 5A). In order to exclude this possibility, we introduced a stop codon in the corresponding ORF2 sequence and analyzed capsid assembly with the mutated VP3 expression plasmid (Fig. 5B and C). In this case, AAV9 VP3 alone was not able to form capsids, while coexpression of AAP-2 efficiently reconstituted the assembly reaction. Thus far, the data indicate that an AAP is required for VP3 capsid assembly of all tested AAV serotypes. In addition, the assembly helper protein of AAV2 was able to provide this function in all cases except for AAV5-VP3.
Fig. 4.
Comparison of capsid assembly of different AAV serotypes. (A) Western blot analysis of VP protein expression was performed with monoclonal antibody (MAb) B1, which detects all three capsid proteins of the used serotypes. Samples also analyzed for AAP expression were processed with a polyclonal anti-AAP serum. VP3 was complemented with a truncated, but functional variant of AAP-2 (AAPtru) resulting from expression by pVP2N-gfp (29). (B) Capsid formation was quantified by ELISAs based on MAb A20 (AAV2), ADK1 (AAV1), ADK5 (AAV5), ADK8 (AAV8), or ADK9 (AAV9). The means and standard deviations (SDs) of at least three independent experiments are shown; asterisks indicate samples for which no capsids could be detected. Detection limits: A20 ELISA, 5 × 107 capsids/ml; ADK1 ELISA, 5 × 108 capsids/ml; ADK5 ELISA, 2 × 109 capsids/ml; ADK8 ELISA, 5 × 107 capsids/ml; ADK9 ELISA, 1 × 108 capsids/ml.
Fig. 5.
Prevention of AAP expression from the AAV9 VP3 coding sequence. (A) In order to prevent AAP expression from the AA9 VP3 coding sequence, a stop codon was introduced into the corresponding reading frame (arrow) downstream of potential translation initiation codons (boxes). (B) Western blot analysis of VP3 protein expression was performed using MAb B1. Samples were also analyzed for AAP expression with a polyclonal anti-AAP serum detecting AAPtru (a truncated variant of AAP-2) resulting from construct pVP2N-gfp. (C) Capsid formation was quantified by ELISA based on MAb ADK9 (AAV9). The means and SDs of at least three independent experiments are shown; an asterisk indicates a sample for which no capsids could be detected.
Subcellular localization of capsid proteins and assembled capsids of different AAV serotypes.
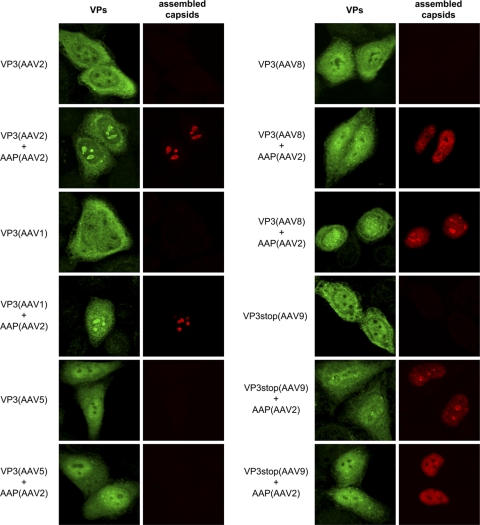
Previous studies have shown that capsid assembly of AAV2 starts in the nucleoli of cells coinfected with AAV2 and adenovirus type 2 (33). By characterizing AAP-2 activity, we also demonstrated that it targets VP3 proteins of AAV2 to the nucleoli for capsid assembly (29). It was now of interest to determine whether these characteristics also apply to the VP3 capsid proteins of the different AAV serotypes and whether coexpression of AAP also affects the cellular distribution of these capsid proteins. The subcellular localization of nonassembled and assembled capsid proteins of AAV1, AAV2, AAV5, AAV8, and AAV9 was determined by indirect immunofluorescence 20 h after transfecting HeLa cells. A polyclonal antiserum was used to visualize the localization of total—nonassembled and assembled—capsid proteins. Assembled capsids were detected by the monoclonal antibodies also used for the capsid-specific ELISAs.
In the absence of AAP, VP3 of all serotypes was evenly distributed over the nucleus and cytoplasm (Fig. 6). The nucleoli were negative for VP3 in all cases. When AAP-2 was coexpressed, a strong signal appeared in the nucleoli of cells expressing VP3 of AAV1 and AAV2 that coincided with the signal for assembled capsids. This was also observed for VP3 of AAV8 and AAV9, although the nucleolar localization was less pronounced. In addition, assembled capsids of these serotypes were also distributed over the nucleus, and the nucleoli of some cells were negative for capsid proteins and capsids. VP3 of AAV5 was slightly enriched in the nucleus but showed no nucleolar localization and no capsid assembly even in the presence of AAP-2, confirming the ELISA data.
Fig. 6.
Intracellular localization and capsid assembly of VP3 of different AAV serotypes. Indirect double immunofluorescence of HeLa cells expressing VP3 in the absence or presence of AAP derived from AAV2. Total expressed capsid proteins were localized using a polyclonal VP antiserum VP51 (VPs, green). Assembled capsids (red) were detected using MAb A20 (AAV2), ADK1 (AAV1), ADK5 (AAV5), ADK8 (AAV8), or ADK9 (AAV9).
Activation of AAV capsid assembly with AAP-1 and AAP-5.
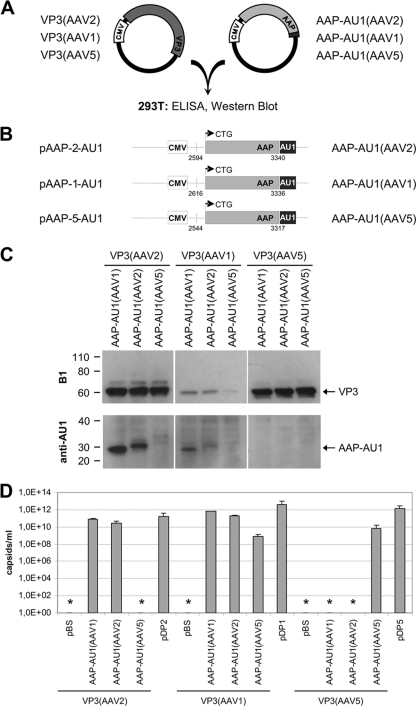
Thus far, capsid assembly analysis of several AAV serotypes revealed that an assembly helper activity is required to induce capsid formation and that AAP-2 was able to activate capsid assembly activity for AAV1-, AAV2-, AAV8-, and AAV9-VP3. However, assembly of AAV5-VP3 was not restored by AAP-2 and assembly of AAV1-VP3 was 10- to 50-fold less efficient in the presence of AAP-2 than in the control. Sequence alignment of the proposed AAP proteins of serotypes 1 to 13 shows that homologous AAPs are encoded by the cap genes of all serotypes (Fig. 7). To test whether the AAPs of AAV1 and AAV5 are able to support VP3 capsid assembly of homologous or heterologous VP3 proteins, we prepared expression constructs of AAP-1, AAP-2, and AAP-5 and tested them for stimulation of capsid assembly of VP3 from AAV1, AAV2, and AAV5 in the transcomplementation assay (Fig. 8A and B). As shown by Western blotting, VP3 of AAV1 was again poorly expressed, whereas AAV5-VP3 expression approached the level of AAV2-VP3 (Fig. 8C). AAP-1 and AAP-2, which could be readily detected when coexpressed with AAV2-VP3, could not be detected after coexpression with AAV5-VP3. AAP-5 was below the detection level in all combinations. In addition, coexpression of AAP-1 and AAP-2 with AAV1-VP3 resulted in a reduced AAP protein level. These results suggest that the nature of capsid protein VP3 affects the steady-state level of AAP proteins.
Fig. 7.
AAP sequences of different AAV serotypes. Alignment of predicted AAP protein sequences derived from ORF2 of the corresponding cap genes (multiway protein alignment; scoring matrix, linear). A conserved ctg codon in ORF2 of the cap gene was presumed as translation initiation codon. A shaded background indicates 100% identity and at least 60% conserved. The GenBank accession numbers of AAV serotype complete or partial genome sequences are as follows: AF043303 (AAV2), AF063497 (AAV1), AF028705 (AAV3B), NC_001829 (AAV4), AF085716 (AAV5), AF028704 (AAV6), NC_006260 (AAV7), AF513852 (AAV8), AY530579 (AAV9), AY631965 (AAV10), AY631966 (AAV11), DQ813647 (AAV12), and EU285562 (AAV13; AAV VR-942).
Fig. 8.
Comparison of VP3 capsid assembly activated by AAP derived from AAV1, AAV2, or AAV5. (A) Transcomplementation assay used for analysis of capsid assembly. VP3 and AAP of AAV serotypes 1, 2, and 5 were coexpressed in different combinations in 293T cells. (B) AAP expression constructs including denotations of the constructs (left) and the expressed proteins (right). AAPs were fused to an AU1 tag at the C terminus. Small numbers indicate nucleotide positions relative to the corresponding AAV serotype genome sequences (see legend to Fig. 7 for accession numbers). Arrows represent the presumed translation start sites of the AAP proteins. CMV, human cytomegalovirus immediate-early promoter. (C) Western blot analysis of VP3 protein expression was performed with MAb B1. AAP-AU1 expression was monitored by using MAb anti-AU1. (D) Capsid formation was quantified by ELISAs based on MAb A20 (AAV2), ADK1 (AAV1), or ADK5 (AAV5). The means and SDs of at least three independent experiments are shown; asterisks indicate samples for which no capsids could be detected. Detection limit: A20 ELISA, 5 × 107 capsids/ml; ADK1 ELISA, 5 × 108 capsids/ml; ADK5 ELISA, 2 × 109 capsids/ml.
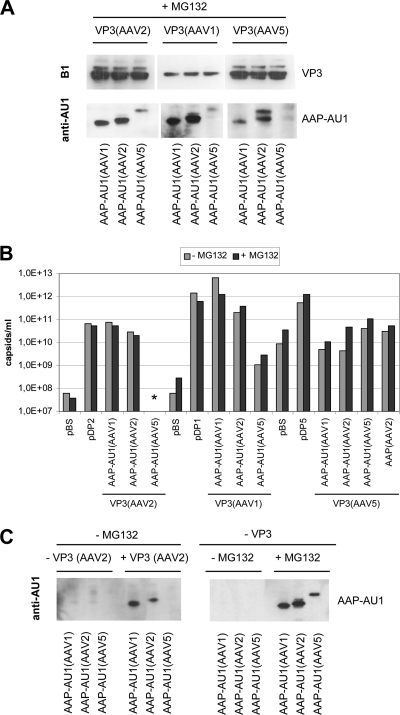
Coexpression of AAV2-VP3 with the helper protein of AAV1 or AAV2 resulted in capsid formation (Fig. 8D). AAP-5 did not stimulate assembly of AAV2-VP3 to detectable levels. The highest titers were obtained with the AAP-1 assembly protein. AAV1-VP3 proteins showed capsid assembly in the presence of AAP-1 and AAP-2 and weak but detectable assembly with AAP-5. Assembly of AAV5-VP3 was not stimulated by AAP-1 or AAP-2. However, AAP-5 was able to restore capsid formation, although at a level ∼100-fold lower than capsid assembly observed after transfection of the pDP5 plasmid. Because the AAP proteins could only be weakly detected in these assays—and not at all when coexpressed with VP3 of AAV5—we performed the assembly experiments in the presence of proteasome inhibitor MG132 (Fig. 9A and B). While AAP protein detection improved in the presence of MG132, capsid assembly did not change significantly. Assembly of AAV1-VP3 and AAV2-VP3 was the same as without MG132 treatment. The average of two independent experiments showed a weak stimulation of AAV5 capsid assembly by two distinct AAP-2 expression constructs in addition to AAP-5. However, these results should be judged cautiously due to the high background noise and low sensitivity of the AAV5 capsid ELISA.
Fig. 9.
Comparison of VP3 capsid assembly activated by AAP derived from AAV1, AAV2, or AAV5 after incubating cells with MG 132. (A) Western blot analysis of VP3 protein expression using MAb B1. AAP-AU1 expression was monitored by using MAb anti-AU1. Transfected cells were incubated with MG132. Arrowheads indicate AAP-specific signals. (B) Capsid formation was quantified by ELISAs based on MAb A20 (AAV2), ADK1 (AAV1), or ADK5 (AAV5). ELISA results are shown as the averages of two independent experiments in the presence of MG132 in comparison to a single experiment without MG132 incubation. Detection limit: A20 ELISA, 5 × 107 capsids/ml; ADK1 ELISA, 5 × 108 capsids/ml; ADK5 ELISA, 2 × 109 capsids/ml. (C) Western blot analysis of AAP-AU1 expression using MAb anti-AU1. The left panel shows AAP expression in the absence or presence of coexpressed VP3 of AAV2, without MG132. The right panel shows AAP expression with or without MG132 incubation of transfected cells in the absence of coexpressed VP3 protein.
The data show that the helper proteins from different AAV serotype cap genes are able to induce capsid assembly activity in other serotype capsid proteins, although with differing efficiency.
DISCUSSION
We establish here the principle that AAV capsid assembly generally requires an AAP. This has experimentally been proven for five serotypes, and protein sequence alignments predict that also all other AAV serotype cap genes encode a homologous protein. In accordance with the high degree of amino acid sequence homology, we could show that capsid assembly of several serotypes is stimulated by the AAP protein derived from AAV2. However, this was not the case for all serotypes analyzed. Considerable differences were observed with VP proteins and AAP proteins of AAV5.
Attempts to cross-complement the capsid assembly of VP3 proteins from AAV1, AAV2, or AAV5 with AAP-1, AAP-2, or AAP-5 showed that AAP-1 and AAP-2 were able to support capsid assembly of AAV1 and AAV2 but not (or insufficiently) of AAV5 capsid proteins. On the other hand, AAP-5 also only stimulated the assembly of AAV5 capsids weakly, and only to a low level of AAV1, but not of AAV2 capsids. The functional difference between AAP-5 and AAP-1 and AAP-2 is reflected in a lower degree of sequence homology. AAP-5—together with AAP-4, AAP-11, and AAP-12—differs in the N-terminal part of the protein, including a highly conserved box between amino acid (aa) positions 30 and 40 of the other AAV serotypes. There are also differences in the C terminus of AAP-4, AAP-11, and AAP-12 versus the other serotypes which are not present in AAP-5, illustrating that AAP-5 has a special role in the AAP protein family. The assembly data indicate that the sequence differences between the various serotype AAPs likely also have functional consequences and do not only reflect differences in the overlapping ORF1 coding for the VP proteins. Assuming that the AAP proteins interact with the capsid proteins during the assembly process, the differences in AAP protein sequences may correspond to specific interaction sites at the capsid proteins. Differences in the VP protein sequence of AAV5 versus the other serotypes cluster in the C-terminal portion of the molecule (aa 612 to 636 and aa 665 to 692) and in the N-terminal part that overlaps with the AAP protein. In addition, there is another region of sequence differences between aa 406 and 421, which may constitute a potential interaction site for AAPs. Thus far, it is not known which capsid protein sequences are recognized by the AAP proteins and which AAP protein sequences are essential for the assembly function.
Some unexpected variations of AAP protein expression levels were observed. First, the AAP-5 protein level was generally lower than the AAP-1 and AAP-2 levels, and second, the protein steady-state levels of all three AAP proteins were influenced by the coexpressed capsid proteins and the proteasome. The generally lower steady-state level of AAP-5 might be due to higher instability or lower synthesis of AAP-5, but it is unlikely that it is the result of a mutant AAP-5 protein obtained by PCR amplification. Two AAP-5 expression-constructs with different 5′-untranslated extensions (data not shown) were completely sequenced excluding point mutations or deletions leading to reduced protein levels. Furthermore, AAP-5 supported the assembly of AAV1 and AAV5, also arguing against a complete lack of expression. Finally, when cells were incubated with proteasome inhibitor MG132, AAP-5 could be detected by Western blotting. This is most strikingly seen in the absence of coexpressed capsid proteins (Fig. 9C) and indicates that proteasomal turnover is one factor involved in maintaining the steady-state levels of AAP proteins. Current studies indicate that the 5′ untranslated region of AAP-2 has an influence on translation initiation at the nonconventional initiation codon of ORF2. It could well be that the unusual mRNA processing of AAV5 (22–24) compared to AAV2 affects translation of the AAP-5 mRNA. Thus, we cannot exclude that our constructs are inefficiently spliced and do not provide the authentic AAV5 upstream sequences for translation initiation and may therefore be less efficiently expressed. The influence of the coexpressed VP proteins on the AAP protein levels is most clearly seen in the Western blots of AAP-1 and AAP-2. Both proteins were well detectable in the presence of AAV2 VP3, while AAP-2 could barely be detected in the presence of AAV1 VP3, and neither could be detected in the presence of AAV5 VP3 (in the absence of proteasome inhibitor) (Fig. 8). Similarly, the AAP-5 protein level was also influenced by the VP proteins as seen in the presence of MG 132 (Fig. 9A). All three AAP proteins were completely below detection level in the absence of coexpressed VP3 and without proteasome inhibitor (Fig. 9C). Interestingly, MG132 incubation did not abrogate the influence of the capsid proteins on AAPs, although it increased the steady-state levels of all three AAPs. The mechanism by which VP proteins influence AAP protein levels is unknown. AAP proteins may become stabilized by interaction with the VP proteins, whereas AAP proteins interacting weakly or not at all with VP proteins might be prone to proteolytic degradation. However, a differential destabilizing activity of VP proteins from different serotypes toward AAP also has to be considered based on the AAP steady-state levels in the presence of MG 132. In this context, it should be noted that the high capsid titers of AAV1 VP3 were obtained despite low VP3 and AAP-1 or AAP-2 expression. This result confirms our previous observation that substoichiometric amounts of AAP proteins are sufficient for activating the assembly process and suggests that only a portion of the expressed VP3 proteins is used for capsid formation, as also indicated by the immunofluorescence data.
Localization of assembled capsids to the nucleoli is a hallmark of AAV capsid assembly. It is clear from earlier studies analyzing a productive AAV2 infection that capsids can first be detected in the nucleoli of infected cells (33). Moreover, nucleolar proteins have been shown to associate with AAV2 capsids (2, 21). However, nucleolar localization of assembled capsids is transient (33). With progression of a productive infection, capsids are spread throughout the nucleus and are finally released from the cell. Several factors are known to influence nucleolar structure and the nucleolar localization of capsids. Coinfection with adenovirus induces nucleolar fragmentation and may contribute to the release of AAV capsids from the nucleoli (14, 20, 31). Furthermore, expression of Rep proteins—in the absence of adenovirus—induces the relocalization of assembled capsids from the nucleoli to the nucleoplasm (33), whereas capsids assembled from VP proteins expressed alone accumulate and remain in the nucleoli. It has recently been shown that AAP translocates a portion of the VP proteins to the nucleolus, where the assembly process takes place (29). However, nucleolar localization of VP proteins is not sufficient for capsid formation. A role of the nucleolar capsid-interacting proteins in the assembly process has not been analyzed thus far. The cellular localization of capsids assembled from VP proteins of different serotypes showed some variation compared to the original observations made for the AAP-2-mediated capsid assembly. Capsids of AAV8 and AAV9 were not only detected in nucleoli but also in the nucleoplasm, although no Rep proteins or adenovirus proteins were coexpressed. Since these capsids were assembled with the help of AAP-2—which is localized to the nucleolus—the association of these capsids with AAP or nucleolar components might be less stringent. The observations in the present study are all made with VP3-only capsids. The contribution of VP1/VP2 to nuclear or nucleolar localization of AAV capsids is still unknown. Further investigations are needed, however, to elucidate the physiological role of the transient nucleolar localization of capsids during assembly and infection.
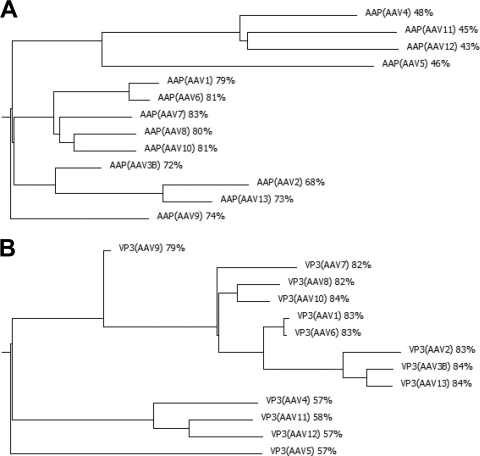
Phylogenetic trees developed from VP protein sequences correlate with the trees deduced from AAP sequences, supporting the notion that the two ORFs have undergone a coevolution process (Fig. 10). It is possible that evolutionary pressure is imposed not only by the overlapping sequences of AAP and VP proteins but also by nonoverlapping sequences of VP proteins involved in the AAP-VP interactions necessary for capsid assembly. The different AAP sequences are highly conserved in three blocks from aa 46 to 89, from aa 100 to 133, and from aa 163 to 175 of the AAP-2 sequence. The conserved regions contain clusters of hydrophobic amino acids, clusters rich in serine and threonine and basic clusters in the C-terminal part of the molecule. The highly conserved blocks are also conserved among the AAPs of the genus dependovirus (29), suggesting that they are essential elements for protein function. A homologue of AAP is not encoded in autonomous parvoviruses. Taking the fundamental similarity of the capsid structure of different parvoviruses into account, it still remains elusive at which step of the assembly reaction the AAP protein is required for capsid formation in the AAV family and how autonomous parvoviruses solved the respective assembly problem without a homologous assembly factor.
Fig. 10.
Evolutionary relationships among AAV serotype gene products. Phylogenetic trees of predicted AAP protein sequences (A) and VP3 protein sequences (B) of the different AAV serotypes were determined on the basis of multiway protein alignments (scoring matrix, linear).
ACKNOWLEDGMENTS
F.S. was supported by grant KL 516/7-1 of the Deutsche Forschungsgemeinschaft.
We thank Andrea Jungmann, University of Heidelberg, for providing AAP-5 expression constructs, including AAV5 (nucleotides 2150 to 3317).
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Bantel-Schaal U., Delius H., Schmidt R., zur Hausen H. 1999. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J. Virol. 73:939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bevington J. M., et al. 2007. Adeno-associated virus interactions with B23/Nucleophosmin: identification of sub-nucleolar virion regions. Virology 357:102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bottcher B., Wynne S. A., Crowther R. A. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386:88–91 [DOI] [PubMed] [Google Scholar]
- 4. Crowther R. A., et al. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943–950 [DOI] [PubMed] [Google Scholar]
- 5. Gao G., et al. 2004. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao G., Vandenberghe L. H., Wilson J. M. 2005. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 5:285–297 [DOI] [PubMed] [Google Scholar]
- 7. Gao G. P., et al. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grimm D., Kay M. A., Kleinschmidt J. A. 2003. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 7:839–850 [DOI] [PubMed] [Google Scholar]
- 9. Grimm D., et al. 1999. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 6:1322–1330 [DOI] [PubMed] [Google Scholar]
- 10. Heilbronn R., Burkle A., Stephan S., zur Hausen H. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kronenberg S., Kleinschmidt J. A., Bottcher B. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuck D., Kern A., Kleinschmidt J. A. 2007. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J. Virol. Methods 140:17–24 [DOI] [PubMed] [Google Scholar]
- 13. Ludtke S. J., Baldwin P. R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 14. Matthews D. A. 2001. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 75:1031–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mori S., Wang L., Takeuchi T., Kanda T. 2004. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330:375–383 [DOI] [PubMed] [Google Scholar]
- 16. Muzyczka N., Berns K. I. 2001. Parvoviridae: the viruses and their replication, 4th ed Lippincott/Williams & Wilkins Co., Philadelphia, PA [Google Scholar]
- 17. Myers M. W., Carter B. J. 1980. Assembly of adeno-associated virus. Virology 102:71–82 [DOI] [PubMed] [Google Scholar]
- 18. Naldini L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 19. Pettersen E. F., et al. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 20. Puvion-Dutilleul F., Christensen M. E. 1993. Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol. 61:168–176 [PubMed] [Google Scholar]
- 21. Qiu J., Brown K. E. 1999. A 110-kDa nuclear shuttle protein, nucleolin, specifically binds to adeno-associated virus type 2 (AAV-2) capsid. Virology 257:373–382 [DOI] [PubMed] [Google Scholar]
- 22. Qiu J., Cheng F., Pintel D. 2007. Distance-dependent processing of adeno-associated virus type 5 RNA is controlled by 5′ exon definition. J. Virol. 81:7974–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qiu J., Nayak R., Pintel D. J. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by both a downstream element within the intron 3′ splice acceptor and an element upstream of the P41 initiation site. J. Virol. 78:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu J., Nayak R., Tullis G. E., Pintel D. J. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rabinowitz J. E., et al. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rutledge E. A., Halbert C. L., Russell D. W. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmidt M., et al. 2008. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J. Virol. 82:1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaikh T. R., et al. 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3:1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sonntag F., Schmidt K., Kleinschmidt J. A. 2010. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. U. S. A. 107:10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steinbach S., Wistuba A., Bock T., Kleinschmidt J. A. 1997. Assembly of adeno-associated virus type 2 capsids in vitro. J. Gen. Virol. 78:1453–1462 [DOI] [PubMed] [Google Scholar]
- 31. Walton T. H., Moen P. T., Jr., Fox E., Bodnar J. W. 1989. Interactions of minute virus of mice and adenovirus with host nucleoli. J. Virol. 63:3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward P. (ed.). 2006. Replication of adeno-associated virus DNA, p. 189–211 In Kerr J., et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom [Google Scholar]
- 33. Wistuba A., Kern A., Weger S., Grimm D., Kleinschmidt J. A. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wistuba A., Weger S., Kern A., Kleinschmidt J. A. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wobus C. E., et al. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xie Q., et al. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]