Abstract
A novel pentameric structure which differs from the previously reported tetrameric form of the diarrhea-inducing region of the rotavirus enterotoxin NSP4 is reported here. A significant feature of this pentameric form is the absence of the calcium ion located in the core region of the tetrameric structures. The lysis of cells, the crystallization of the region spanning residues 95 to 146 of NSP4 (NSP495-146) of strain ST3 (ST3:NSP495-146) at acidic pH, and comparative studies of the recombinant purified peptide under different conditions by size-exclusion chromatography (SEC) and of the crystal structures suggested pH-, Ca2+-, and protein concentration-dependent oligomeric transitions in the peptide. Since the NSP495-146 mutant lacks the N-terminal amphipathic domain (AD) and most of the C-terminal flexible region (FR), to demonstrate that the pentameric transition is not a consequence of the lack of the N- and C-terminal regions, glutaraldehyde cross-linking of the ΔN72 and ΔN94 mutant proteins, which contain or lack the AD, respectively, but possess the complete C-terminal FR, was carried out. The results indicate the presence of pentamers in preparations of these longer mutants. Detailed SEC analyses of ΔN94 prepared under different conditions, however, revealed protein concentration-dependent but metal ion- and pH-independent pentamer accumulation at high concentrations which dissociated into tetramers and lower oligomers at low protein concentrations. While calcium appeared to stabilize the tetramer, magnesium in particular stabilized the dimer. ΔN72 existed primarily in the multimeric form under all conditions. These findings of a calcium-free NSP4 pentamer and its concentration-dependent and largely calcium-independent oligomeric transitions open up a new dimension in an understanding of the structural basis of its multitude of functions.
INTRODUCTION
Rotaviruses are the major cause of acute gastroenteritis in infants and young children (42). Nonstructural protein 4 (NSP4), an endoplasmic reticulum (ER)-resident glycoprotein of 175 amino acids, has been identified as the viral enterotoxin (3). It exhibits diverse functions in relation to viral morphogenesis and pathogenesis (18), which could be attributed to its localization to different cellular compartments in the infected host cell. During different stages of rotaviral infection, NSP4 has been proposed to be associated with diverse functions, such as the viroporin-associated alteration of Ca2+ homeostasis by the release of Ca2+ from the ER (16, 24, 47), the modification of membrane permeability (39), the triggering of an extracellular Ca2+ signaling pathway (15, 16), and interactions with and the transport of double-layered particles (DLPs) into the lumen of the ER for maturation into triple-layered particles (TLPs) (2, 37). Although it was originally proposed to be an ER-anchored intracellular receptor for DLPs (2, 5, 11, 37, 46), several studies showed its presence in several other cellular sites, including the ER-Golgi intermediate compartment (ERGIC) (6, 50), LC3-positive autophagosomes, and a novel membrane compartment colocalizing with viroplasms (6), membrane rafts (45), microtubules (50), the exofacial surface of the plasma membrane (20), and the extracellular matrix (7). While the N-terminal hydrophobic region anchors the protein to the ER or plasma membrane, the C-terminal part of the protein (residues 45 to 175) is oriented toward the cytoplasm or the extracellular environment (5, 11, 18, 20). It is also known to interact with other rotaviral proteins (2, 23, 33, 37) and cellular proteins, such as tubulin, caveolin, laminin-β3, fibronectin, and integrin (7, 38, 43, 45, 50). Besides its association with several cellular compartments, it is also secreted in both cleaved (51) and native (10, 20) forms from infected and NSP4 gene-transfected cells, which was proposed previously to bind to receptors on neighboring cells and induce diarrhea (43).
With a view toward obtaining the structure-activity relationships of the C-terminal domain of the protein, we have been studying the biochemical, biophysical, and biological properties of a large number of mutants and structural aspects of the segment of NSP4 encompassing the diarrhea-inducing and interspecies variable regions (residues 114 to 135 and 135 to 141, respectively) from different rotavirus strains (13, 14, 26) (Fig. 1). The region spanning residues 95 to 146 of NSP4 (NSP495-146) is the longest one that could be crystallized from among the mutants generated in our laboratory. The structures of this region from two of the strains, simian strain SA11 and human asymptomatic strain I321 (13) (SA11:NSP495-146 and I321:NSP495-146, respectively), showed a coiled-coil domain made up of four α-helices (13) similar to that of the synthetic peptide (8) of SA11 [SA11:NSP495-137(synthetic)]. A calcium ion located in the core of the helical bundle interacts with all four individual chains and stabilizes the homotetramer. The tetramer of SA11:NSP495-146 is formed by the association of a dimer of dimers generated by a crystallographic dyad, whereas that of I321:NSP495-146 exists as a tetramer in the asymmetric unit (13). In contrast to these observations, the structure of the corresponding peptide from asymptomatic human rotavirus strain ST3 (ST3:NSP495-146) reported here reveals a pentameric coiled-coil structure without the bound calcium ion. The presence of the pentameric forms in preparations of ST3:NSP495-146 and SA11:NSP495-146 as well as in preparations of SA11ΔN72 and SA11ΔN94 peptides that contained the complete C-terminal flexible region (FR) but either contained or lacked the N-terminal viroporin domain was further demonstrated by size-exclusion chromatography (SEC) and chemical cross-linking analyses. By SEC, we further demonstrate that while the NSP495-146 peptide undergoes pH- and calcium-dependent transitions easily, ΔN94 exists primarily in the pentameric form at high protein concentrations but dissociates into tetramers and other lower oligomeric forms at low protein concentrations. The tetrameric forms are stabilized by calcium and the dimeric forms are stabilized by magnesium at low protein concentrations.
Fig. 1.
Sequence alignment of residues 95 to 146 of NSP4 peptides from different rotaviral strains whose crystal structures are available, showing the a and d positions of the heptad. Highlighted in red are the residues which differ from the sequence of SA11:NSP495–146. GenBank accession numbers for the NSP4 peptides of strains SA11, I321, and ST3 are JF791804, AF165066, and U59110, respectively. Note the C129R and M135T amino acid substitutions in the I321 clone used for protein expression compared to the previously reported sequence (25).
The present finding of a calcium-free pentamer provides a new direction in an understanding of the structure-function relationships of the viral enterotoxin in light of the newly characterized viroporin function and the proposed alternate topology model for the ER-resident protein (24).
MATERIALS AND METHODS
Cloning, expression, and purification of ST3:NSP495–146.
The DNA fragment of NSP4 from the G4P[6] asymptomatic human strain ST3 (GenBank accession number U59110), corresponding to residues 95 to 146 from the NSP4ΔN72 cDNA clone (13, 26), was amplified by PCR using position-specific primers. The forward primer contained sites for the EcoRI and NdeI restriction endonucleases, and the reverse primer contained the termination codon and a site for HindIII. The EcoRI-HindIII PCR fragment was first cloned into the pBluescript KS(+) (pBS) vector. The NdeI-HindIII fragment liberated from pBS was then cloned into pET 22(b+) between the same sites. The protein was expressed without a histidine tag but contained an additional methionine at the N terminus. The gene sequence was verified by nucleotide sequence analysis (Macrogen, South Korea). N-terminally His-tagged SA11ΔN72 and SA11ΔN94, which contained or lacked the N-terminal amphipathic domain (AD), respectively, but contained the C-terminal FR, were described previously (13, 26). The deletion mutant proteins expressed in Escherichia coli BL21(DE3) cells were highly soluble and purified as described previously (13, 26). Briefly, protein expression was induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.4 for 3 h at 37°C. The bacterial cells expressing the NSP4 mutants were lysed by sonication and purified under the following conditions: cell lysis in 10 mM sodium acetate (NaAc) buffer (pH 5.6) but subsequent steps of purification at pH 7.5 in the absence of CaCl2 (condition A); lysis and purification in the absence of CaCl2 (condition B); lysis in the presence of 0.1, 1.0, or 10 mM CaCl2 but purification in the absence of calcium (condition C); lysis and purification in the presence of 0.1 mM CaCl2 (condition D); lysis in the presence of 10 mM MgCl2 (condition E); and lysis and purification in the presence of 0.1 mM MgCl2 (condition F). Except for condition A, the buffers under all other conditions contained 10 mM Tris-HCl (pH 7.5) and 100 mM NaCl, and the lysis buffer contained phenylmethylsulfonyl fluoride (PMSF) at 3 mM. The cell lysate was incubated on ice for a few minutes with (for ΔN72) or without (for the other mutants) 1% NP-40 and was centrifuged at 17,000 rpm for 40 min. The ST3 protein from the lysate was precipitated in 40% ammonium sulfate and partially purified by anion-exchange chromatography followed by size fractionation.
Anion-exchange chromatography.
The untagged NSP495–146 protein in a 40% ammonium sulfate pellet was dissolved in Tris-HCl (pH 7.5) buffer and dialyzed against 10 mM Tris-HCl (pH 7.5) using dialysis tubing (3,000-molecular-weight [MW] [3K] cutoff), filtered through a 0.2-μm-pore-size filter, and fractionated by anion-exchange chromatography using a HiPrep Mono Q (10/100) column (GE Healthcare) equilibrated with 10 mM Tris-HCl (pH 7.5) buffer at a flow rate of 1 ml/min at room temperature using the Akta Basic fast protein liquid chromatography (FPLC) system (GE Healthcare). The proteins bound to the column were eluted by using a linear gradient of 0 to 1.0 M NaCl in 10 mM Tris-HCl (pH 7.5) buffer. The level of protein in each fraction was further analyzed by SDS-PAGE.
Preparative gel filtration.
The fractions containing the NSP495–146 peptide from the ion-exchange column were pooled and concentrated by using Centricon-3 columns, and the protein was further purified by gel filtration chromatography using the GE Healthcare FPLC system equipped with a Superdex G75 prep-grade column (HiLoad 26/60) using Tris-buffered saline (10 mM Tris [pH 7.5], 100 mM NaCl). The protein fractions were identified spectrophotometrically at 215 nm. The purity and molecular mass (6.495 kDa) of the protein were determined by SDS-PAGE and electrospray ionization (ESI) mass spectrometry. To determine the influence of pH, calcium, and magnesium on the oligomerization of the mutant proteins, the bacterial cells were lysed under different conditions (conditions A to F) as described above. Subsequent steps of purification following cell lysis were identical, unless indicated otherwise.
Analytical gel filtration.
The molecular masses of the native proteins were determined by SEC using a Superdex 75 10/300 analytical column that was calibrated with the protein standards conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), RNase A (13.7 kDa), and aprotinin (6.5 kDa) (GE Healthcare Bio-Sciences AB). The void volume of the column was determined by using blue dextran (2,000 kDa). The purified protein (100 μl) in Tris-HCl (pH 7.5) buffer containing 100 mM NaCl at a concentration of 20 mg/ml or at the desired concentration was used for molecular mass determinations. The molecular weight of the protein was calculated by plotting the Ve/Vo versus the log of the molecular mass, where Ve is the elution volume and Vo is the void volume of the column.
Cross-linking of NSP4 mutant proteins using glutaraldehyde.
The cells expressing ΔN72 or ΔN94 were lysed by sonication in a buffer containing 10 mM Tris-HCl (pH 7.5) and 150 mM NaCl that either contained or lacked 10 mM CaCl2, and debris was removed by centrifugation at 18,000 rpm. The NSP4 mutant proteins were bound to Ni+2-nitrilotriacetic acid (NTA)-agarose resin, and the nonspecific contaminating proteins were removed by washing with a buffer containing 10 mM Tris (pH 7.5), 300 mM NaCl, and 40 mM imidazole. The cross-linking reaction was carried out in a 20-μl reaction mixture containing an equimolar ratio of protein to cross-linker (0.0125% glutaraldehyde) using eluted or resin-bound protein (26). At the end of the reaction, the unreacted cross-linker was quenched by the addition of 10 μl of 1 M Tris (pH 8.0) to the mixture. The cross-linked proteins were resolved by 16% SDS-PAGE, and the bands were visualized by staining with either silver or Coomassie brilliant blue.
Crystallization.
ST3:NSP495–146 purified from cells lysed in pH 5.6 buffer was crystallized at 293K by the hanging-drop vapor diffusion method by mixing 2 μl of approximately 30 mg/ml protein and 2 μl of the reservoir solution. Initial screening was done by using Crystal Screens I and II and Index Screen kits from Hampton Research (27). Small crystals were seen under three different conditions, all at pH 6.5. The best crystals appeared in Hampton Crystal Screen II, condition 23, with a solution containing 10% (vol/vol) dioxane, 0.1 M morpholineethanesulfonic acid (MES) (pH 6.5), and 1.6 M ammonium sulfate. In further trials, the protein concentration was increased to 40 mg/ml, and the drop ratio was modified. Larger crystals with dimensions of 0.3 by 0.25 by 0.1 mm were obtained within 2 weeks.
Data collection.
Crystals of ST3:NSP495–146 were transferred into a cryoprotectant solution containing 10% (vol/vol) ethylene glycol, kept sealed over the reservoir solution containing mother liquor and 10% ethylene glycol for 10 min, and then flash-cooled in liquid nitrogen. A complete data set was collected at the home source to a resolution of 3.2 Å at 100K using a MAR300 imaging-plate system. The Cu Kα radiation of a 1.54-Å wavelength, generated by a Rigaku RU-200 X-ray generator operating at 44 kV and 70 mA and focused with an Osmic mirror system, was used in this experiment. The crystal-to-detector distance was 250 mm. A total of 180 frames were collected, with a 1° oscillation range and a 1° frame width at 100K. Subsequently, data were collected at the synchrotron at SPring-8 BL26B2, Japan, to a resolution of 2.5 Å. One hundred eighty frames with an oscillation range of 1° were collected by using X rays at a 1-Å wavelength at 100K. The crystal-to-detector distance was 184 mm. The data were indexed, integrated, and scaled by using DENZO and SCALEPACK of the HKL2000 suite (40). A close examination of the synchrotron data revealed serious overloads and a missing zone of reflections corresponding to the ice ring, resulting in a sharp dip in the Wilson plot. These reflections were taken from the home source data and appended to the synchrotron data set after shell-wise scaling. The merged data with 277 more reflections were used for refinement. Analysis of the data using the anisotropy correction server (http://www.doe-mbi.ucla.edu/∼sawaya/anisoscale/) revealed that the data had severe diffraction anisotropy.
Structure solution and refinement.
The molecular replacement solution was obtained by using the dimer of SA11:NSP495–137(synthetic) (Protein Data Bank [PDB] accession number 1G1I) as the search model in the space group P42212 using PHASER (35). Refinement was carried out initially in P42212 and later in P42 by using CNS 1.2 (9) and PHENIX (1). Analysis of the data using phenix.xtriage indicated merohedral twinning with the twin operator h,−k,−l and twin fractions in the range of 0.45 to 0.49. Twinning correction and anisotropic B factors were used in the final cycles of refinement.
COOT (17) was used for visualization and model building, the CCP4 package (Collaborative Computational Project Number 4, 1994) was used for other calculations, and figures were generated by using PyMOL (http://www.pymol.org). The final model has a working R factor (Rwork) value of 0.27 and an Rfree value of 0.31 (Table 1). Ramachandran map analysis using PROCHECK (29) indicated that 94.2% of all residues fell into the most favored region and that the remaining 5.8% fell into the additionally allowed region. Structure alignments were carried out with the program ALIGN (12) using all main-chain atoms. The structural pKa values were calculated by using PROPKA (4). HOLLOW (22) and PoreWalker (41) were used to create a graphical representation of the pore. Energy calculations were carried out by using MSDPISA (28).
Table 1.
Data collection and refinement statisticsa
| Parameter | Value determined from: |
|
|---|---|---|
| Home source data | Synchrotron data | |
| Data collection | ||
| Space group | P42 | P42 |
| Cell dimensions (Å) | ||
| a | 64.17 | 63.46 |
| c | 123.02 | 123.21 |
| Resolution (Å) | 64.12–3.2 (3.37–3.2) | 50–2.5 (2.59–2.5) |
| Rmerge (%) | 9.9 (48.7) | 5.5 (18.7) |
| I/σI | 13.7 (4.1) | 22.2 (2.2) |
| Completeness (%) | 99.7 (99.3) | 95.1 (74.6) |
| Redundancy | 6.3 (6.3) | 6.6 (4.7) |
| Refinement | ||
| Resolution (Å) | 28-2.5 | |
| No. of reflections | 16,272 | |
| Rwork/Rfree | 0.27/0.31 | |
| No. of atoms | ||
| Protein | 3,328 | |
| Ligand/ion | 108 | |
| Water | 162 | |
| B factors (Å2) | ||
| Protein | 72.8 | |
| Ligand | 72.7 | |
| Water | 65.0 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.53 | |
Values in parentheses are for the highest-resolution shell. RMS, root mean square.
Protein structure accession number.
The atomic coordinates and structure factors reported here have been deposited in the Protein Data Bank under accession number 3MIW.
RESULTS
Data collection and refinement.
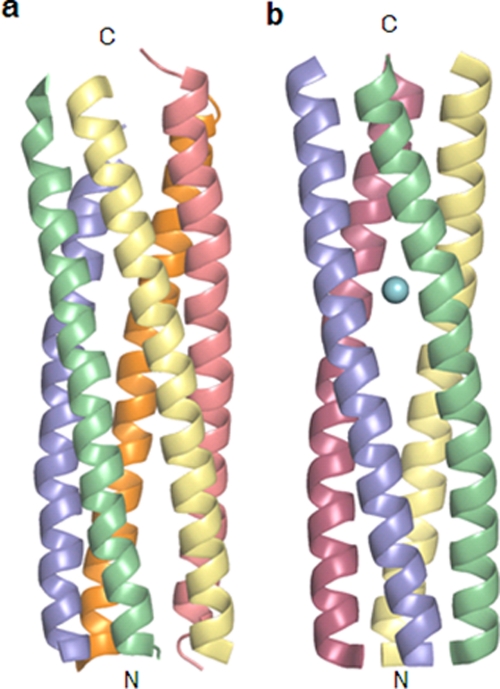
The crystal structure of ST3:NSP495–146 spanning the diarrhea-inducing, Ca2+- and VP4-binding, and interspecies variable regions (Fig. 1) of an asymptomatic human rotavirus strain, ST3, has been determined at 2.5-Å resolution. As in the two previously described structures (13) (PDB accession numbers 2O1K and 2O1J) of the recombinant NSP4 proteins from strains SA11 and I321 reported from our laboratory, no clear electron density was seen for the C-terminal 9 residues corresponding to the interspecies variable region due to its highly flexible nature (8, 13, 26). The rest of the structure folds into a hitherto unobserved pentameric form, in contrast to the known tetrameric coiled-coil stretches found in the two recombinant proteins mentioned above as well as the synthetic peptide (8) (PDB accession number 1G1I) corresponding to residues 95 to 137 (Fig. 2) of NSP4 from rotavirus strain SA11. There are two dissimilar pentamers in the asymmetric unit of the P42 space group. A noncrystallographic axis, 1.5° away from the a axis, relates the two pentamers by a rotation of 179.5°, whereas the 5-fold axes are approximately parallel to the crystallographic c axis. This type of packing of long helical structures gives rise to highly intense diffracted rays from the planes passing through the helices, resulting in overloads. Diffraction anisotropy also arises for the same reason, giving stronger data along the c* axis than in the other two directions. Data collection and refinement details are given in Table 1.
Fig. 2.
Two oligomeric structures of the coiled-coil domain of NSP4. Shown are the pentameric structure of ST3:NSP495–146 presented in this paper (a) and the tetrameric structure of SA11:NSP495–146 reported previously (PDB accession number 2O1K) (b). The bound calcium ion is shown as a sphere.
The pentamer.
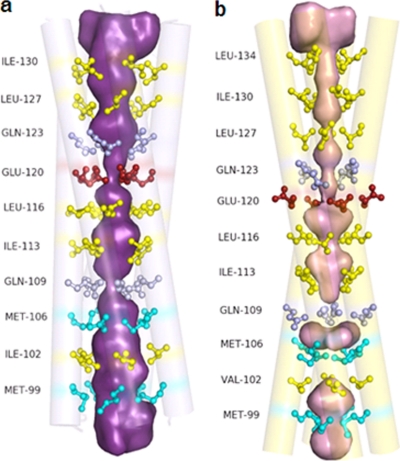
The homopentamers are formed by the parallel association of the α-helices of the NSP4 fragments. The right-handed α-helices of each pentamer form coiled coils of superhelices with a left-handed twist. The five helical bundle can be considered a cylinder with a length of ∼62 Å and a width of ∼28 Å. The angles of rotation relating successive helices in the pentamers vary between 67° and 77°. The core of the pentamers is predominantly hydrophobic, with three polar layers, as discussed below. The outer surface of the pentamers has several charged residues. A major difference in the tetrameric and pentameric structures is that while all the reported tetrameric structures contain Ca2+ at the core, no metal ion could be detected in the pentamers (Fig. 2 and 3).
Fig. 3.
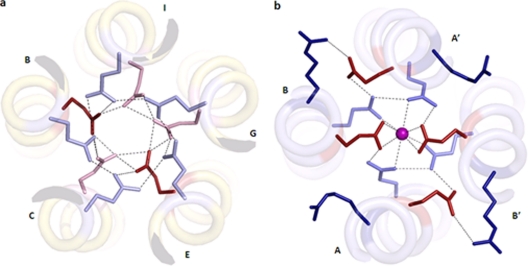
(a) Two polar layers at the helical core of the ST3:NSP495–146 pentamer. Interactions between Glu120 (red, normal pKa; pink, high pKa) and Gln123 (light blue) are marked as dotted lines. (b) SA11:NSP495–137 tetramer. The calcium ion is shown as a sphere. The ionic interactions between Arg119 (dark blue) and Glu120 (red) and hydrogen bonds between Glu120 (red) and Gln123 (light blue) and the calcium ion are marked as dotted lines.
An examination of the free energies of both forms revealed that while the pentameric form is more stable (ΔG of ∼−100 kcal/mol) than the tetrameric structure without any metal ion (ΔG of ∼−87 kcal/mol), the presence of the Ca2+ ion at the core of the tetramer makes it more stable (ΔG of ∼−110 kcal/mol) than the pentamer. The buried surface areas are 8,620 Å2 for the tetramer and 10,640 and 10,390 Å2 for the two pentamers in the asymmetric unit.
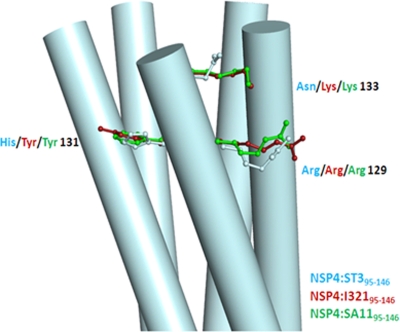
The majority of the sequence differences in NSP495–146 from various strains occurs at the C-terminal end region (Fig. 1 and 4). A key difference in the sequences of the previously determined tetrameric proteins and the pentamer of ST3:NSP495–146 is the residue at position 131, which is a histidine in the pentamer instead of a tyrosine. The presence of tyrosine appears to be crucial for diarrhea induction and DLP binding (2, 3, 14). The amino-aromatic interaction between the Tyr131 and Arg129 residues observed for the tetrameric structures is lost in the pentamers. In the present structure, histidine was observed to have a conformation similar to that of the tyrosine without causing any change in the α-helical nature of the chains. It contributes to the increase in the net charge on the surface of the molecule. The implications of this change are unknown at present, but it is likely to affect the way in which NSP4 interacts with other proteins. Of the 10 independent histidines present in the asymmetric unit, only two interact with the next residue, Asp132, through the side-chain atoms.
Fig. 4.
The C-terminal end of the coiled-coil domain of the pentameric structure of NSP4:ST395–146. Residues at positions 129, 131, and 133 of one of the chains of the pentamer are shown, and the same residues from two tetrameric structures are superposed on these residues.
The core.
The core of the coiled-coil structures of the tetrameric NSP4 is distinct from those of other tetrameric coiled coils due to the presence of a negatively charged core neutralized by a metal ion. The predominantly hydrophobic core is interrupted by three polar layers at residues Gln109, Glu120, and Gln123. Calcium binding is facilitated by both Glu120 and Gln123 (Fig. 3a). Although a 4-fold symmetry exists for the backbone atoms at the metal binding site, it does not extend to the side-chain level. Only two Glu120 residues coordinate the calcium ion, whereas the other two point away from the core and are stabilized by ionic interactions with the Arg119 of the neighboring chain. All four Gln123 residues, however, are oriented toward the core. In contrast, in the pentameric structure, all five Glu120 and Gln123 residues point toward the core, and the calcium ion is absent (Fig. 3b). Glu120 and Gln123 residues are involved in the formation of an extensive hydrogen-bonding network. All Gln123 side chains interact with the side chains of the neighboring Gln123 residues. Similarly, Glu120 residues interact with those on the neighboring helices, except that chains A and D are not close enough to form a hydrogen bond. Glu120 side chains also interact with Gln123 side chains of the neighboring helices, with a few exceptions.
It is also interesting that Bowman et al. (8) observed previously that the region between residues 95 and 120 of the tetrameric form is less 4-fold symmetric than the rest of the structure. To obtain the exact relation, we superimposed the main-chain atoms of this region of the two chains present in the asymmetric unit of the tetrameric structure of SA11:NSP495–146 (PDB accession number 2O1K) and found that the chains are in fact related by a 5-fold rotation. This observation reveals the structural plasticity within a single oligomeric state and implies that Glu120 could act as a switch between the two oligomeric states.
The structural pKa values of the charged residues of the protein were calculated to find the correlation between their ionization/protonation states and the structural environment. The only residue which showed significant deviations from the theoretical value and variations in the pKa values between the pentameric and tetrameric structures is Glu120, which resides inside the core. For the two Glu120 residues interacting with the Ca2+ ion in the tetramers, the pKa values were between 1 and 2, which clearly indicates the negatively charged state of the glutamates. Out of the five glutamates in the pentamers, two had pKa values close to the theoretical value, whereas the remaining three had very high values (>10). These elevated values suggest that a majority of the five glutamates are protonated, enabling them to interact with one another and also reduce the net charge at this location without the requirement for charge compensation by metal binding. A similar case of Ca2+-dependent protonation of an aspartate residue was observed previously for bacteriorhodopsin as well (49).
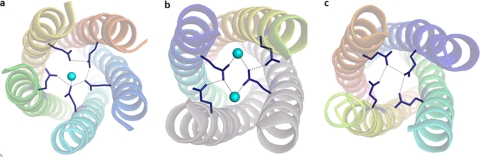
The Gln109 residues form another polar layer away from the metal binding site. The 4-fold symmetry of the backbone near this residue is not maintained at the core of the tetramers, as two of them point inwards and the other two point outwards in SA11:NSP495–146 and SA11:NSP495–137 (synthetic), as shown in Fig. 5. Two water molecules fill the space in the core left by the two Gln109 residues that turn outside. In contrast, in the tetramer of I321:NSP495–146, the side chains of all four Gln109 residues are located in the interior of the core following the 4-fold relation. Thus, there is a clear difference between the two tetramers at this particular polar layer, probably due to the deviation of SA11:NSP495–146 from 4-fold symmetry in this region, as discussed above. In the pentamer of ST3:NSP495–146, however, all five Gln109 residues face the core and coordinate a water molecule.
Fig. 5.
Interactions in the Gln109 polar layer. The water molecules are shown as cyan spheres. Interactions between Gln109 (blue) and the water molecule are marked as dotted lines. (a) ST3:NSP495–146 (pentamer); (b) SA11:NSP495–146 (dimer of dimers); (c) I321:NSP495–146 (tetramer).
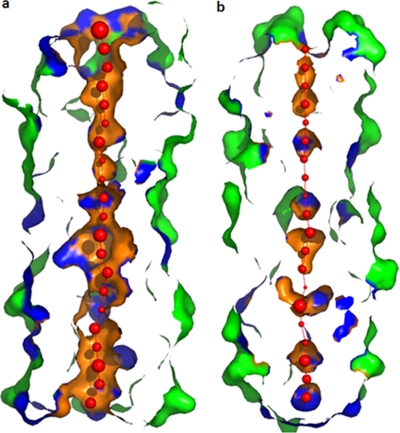
Two water molecules were found inside the core of pentamer 1. One is located on the helix axis close to the plane containing Glu120 residues within 2.60 to 3.25 Å from the glutamates. The other water molecule resides between the layers formed by Met106 and Gln109. In pentamer 2, four water molecules were located in the core, three of which were between the layers of Met106 and Ile113, and the fourth water molecule was close to the Ile130 layer. Except in the case of the water molecule in pentamer 1, no interactions involving the other four water molecules were observed. The interior of the tetrameric structures is severely constricted at the Met106, Gln109, Glu120, Gln123, and Leu127 residues and is clearly inaccessible for the passage of water or other ions (Fig. 6). The pentameric structure, however, has an open channel between the N- and the C-terminal-end regions, which can allow the flow of ions or water molecules through the interior of the helical bundle (Fig. 7).
Fig. 6.
The pore in the ST3:NSP495–146 pentamer (a) and the SA11:NSP495–146 tetramer (b). Shown in a ball-and-stick representation are the residues at the a and d positions of the heptad, as indicated in Fig. 1.
Fig. 7.
Surface representation of the pore in the pentameric (a) and tetrameric (b) structures of NSP495–146. Red spheres represent pore centers at 3-Å steps, with their diameters proportional to the measured diameters.
Oligomeric transitions in NSP495–146.
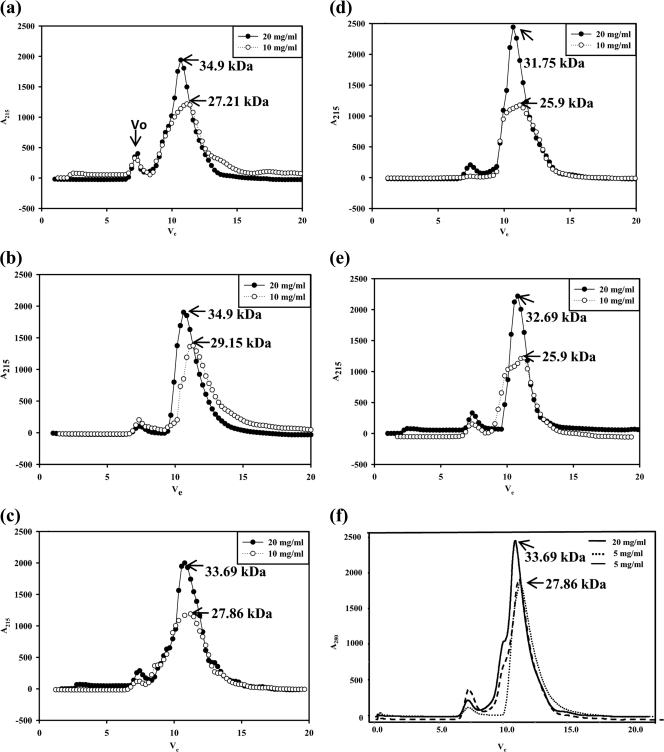
Since the crystals were obtained under acidic buffer conditions using the peptide prepared from cells lysed in pH 5.6 buffer, SEC analyses were performed to establish the conditions that determine the oligomeric state of the ST3:NSP495–146 recombinant peptide. This study suggested that the peptide exists primarily as pentamers under all conditions (Fig. 8a to e) at high concentrations (20 mg/ml). However, when analyzed at low concentrations (10 mg/ml), the peptide prepared from cells lysed under normal conditions (condition B) existed primarily in the tetrameric form (Fig. 8b), whereas that prepared under other conditions appeared to exist in both pentamers and tetramers at this concentration, as seen from the broader peaks (Fig. 8a and c to e). However, at 5 mg/ml, the peptide existed primarily as a tetramer (Fig. 8f). The oligomeric state of NSP495–146 at low concentrations thus appeared to depend on both the pH of the buffer in which the cells were lysed and presence or absence of Ca2+ during purification, although at high concentrations, only pentamers and, at 5 mg/ml, primarily tetramers are seen. Similar results were observed with SA11:NSP495–146 (data not shown).
Fig. 8.
Analytical size-exclusion chromatogram of ST3:NSP495–146 under different conditions. (a) Lysis of cells in NaAc buffer (pH 5.6) (condition A). (b) Lysis in the absence of calcium (condition B). (c) Lysis in the presence of 10 mM calcium (condition C). (d) Lysis and purification in the presence of 0.1 mM calcium (condition D). (e) Lysis and purification in the presence of 0.1 mM magnesium (condition F). (f) SEC of the peptide purified under condition B at concentrations of 20 mg/ml and 5 mg/ml and under condition D at a concentration of 5 mg/ml. The NSP4 peptide in the bacterial cell lysate was precipitated in 40% ammonium sulfate. All subsequent purification steps, irrespective of cell lysis conditions, were carried out under physiological buffer conditions in the absence or presence of added calcium or magnesium. The protein was purified by ion-exchange chromatography followed by size-exclusion chromatography, and the molecular masses were determined as described in Materials and Methods. Vo, void volume.
Cross-linking of SA11:NSP4ΔN72 and SA11:NSP4ΔN94 mutants demonstrates that the protein forms pentamers.
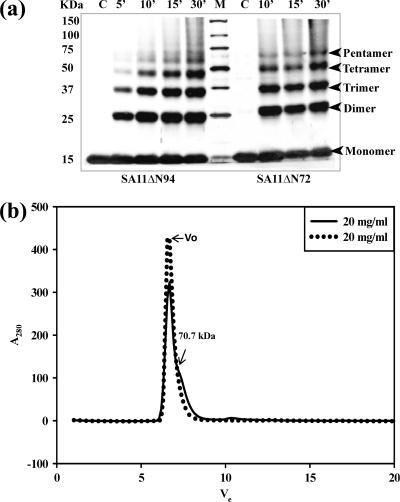
We have previously shown that the AD from residues 73 to 85 in the ΔN72 mutant in cooperation with the C-terminal FR promotes ordered multimerization and diarrhea-inducing and DLP binding properties of the protein (26). Furthermore, the ΔN72 mutant was more efficient in its biological functions than the full-length protein. Deletions or mutations in either the AD or the FR resulted in a severe loss of multimerization and significantly affected biological functions (14, 26). To demonstrate that the pentameric structure reported in this study is not specific just to the NSP495–146 mutant due to the lack of both the AD and FR but that the NSP4 protein in general exists as pentamers as well as the previously demonstrated tetramers, we subjected purified ΔN72 (containing the AD) and ΔN94 (lacking the AD) mutant proteins, both of which contained the complete C-terminal FR, to chemical cross-linking using glutaraldehyde. As shown in Fig. 9a, both mutant proteins showed bands corresponding to dimers, trimers, tetramers, and pentamers. These multiple oligomeric forms were observed for both mutants independent of the presence or absence of calcium, suggesting that NSP4 in general could exist in different oligomeric forms.
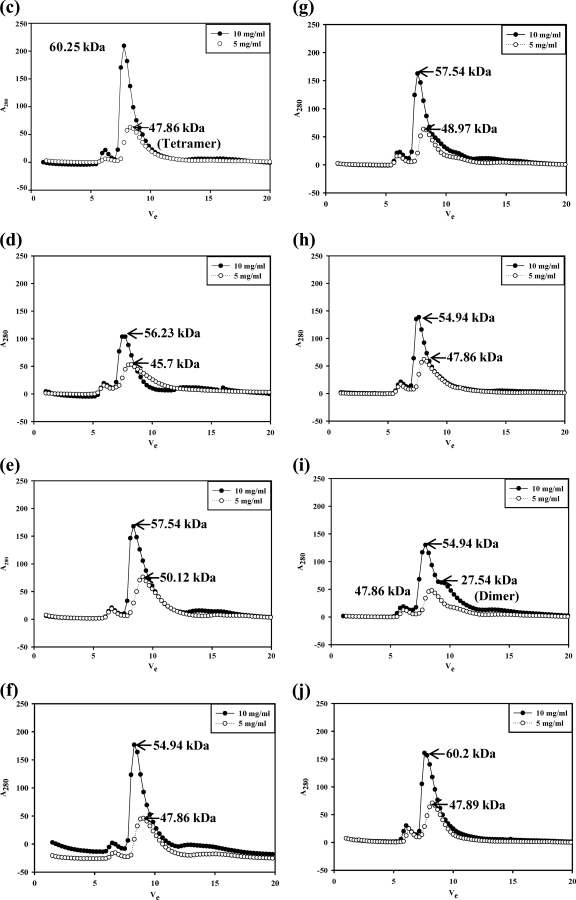
Fig. 9.
NSP4 exists in multiple oligomeric states, as demonstrated by glutaraldehyde cross-linking and analytical gel filtration of the SA11ΔN72 and SA11ΔN94 proteins by use of a Superdex G75 column. (a) Glutaraldehyde cross-linking. Bacterial cells expressing ΔN94 were lysed in a buffer containing 10 mM Tris-HCl (pH 7.5) and 100 mM NaCl in the absence of calcium, and bacterial cells expressing ΔN72 were lysed in the presence of 10 mM CaCl2, but both were purified further under identical conditions in the absence of added calcium. The proteins were cross-linked for the indicated time periods directly on the beads using an equimolar ratio of protein to cross-linker. The proteins were resolved by SDS-PAGE, and the bands were visualized by silver staining. Note the pentamers and other oligomeric forms of both mutant proteins purified after the lysis of cells in the presence or absence of calcium. M, Precision Plus protein standards from Bio-Rad. (b) Analytical size-exclusion chromatography of ΔN72. The protein was analyzed after purification from cells lysed in Tris-HCl (pH 7.5) buffer in the presence (−) or absence (.....) of 10 mM CaCl2. Vo, void volume. (c to j) Analytical size-exclusion chromatography of ΔN94 purified under different conditions. (c) Cells lysed in NaAc buffer (pH 5.6) (condition A); (d) lysis and purification in the absence of calcium (condition B); (e) lysis in the presence of 0.1 mM CaCl2 (condition C0.1); (f) lysis in the presence of 1.0 mM CaCl2 (condition C1); (g) lysis in the presence of 10 mM CaCl2 (condition C10); (h) lysis and purification in the presence of 0.1 mM CaCl2 (condition D); (i) lysis in the presence of 10 mM MgCl2 (condition E); (j) lysis and purification in the presence of 0.1 mM MgCl2 (condition F).
In the cross-linking experiments, we did not find significant differences in the intensities of bands corresponding to different oligomers in the two mutants prepared under different conditions of pH and the presence or absence of calcium. Furthermore, it is difficult to correlate the intensities of the bands corresponding to different oligomeric species to their relative abundance in solution, as the probability of a cross-linking of all five chains in a pentamer is much lower than that for 2, 3, or 4 chains. Thus, under the experimental conditions of the substrate-to-cross-linker ratio (1:1) and the time and dynamics of the reaction, the intensity of the band corresponding to the pentamer is always lower than those of the dimer, trimer, and tetramer. However, the results of cross-linking experiments clearly demonstrate the existence of pentameric forms of NSP4.
Note that the ST3:NSP495–146 peptide in the present study as well as the equivalent recombinant peptides of SA11 and I321 (13) were purified under physiological conditions in the absence of calcium, the only difference being that the cells expressing the former were lysed in pH 5.6 buffer, as the yield of this peptide was observed to be higher under this condition. All three peptides were also crystallized in buffer that lacked added calcium. However, while SA11 and I321 peptides yielded crystals of calcium-bound tetramers at pH 7.5, the ST3 peptide crystallized at pH 6.5 as calcium-free pentamers. Furthermore, the concentration of the peptide preparations used for setting up crystallization differed, with that of the ST3 peptide being 40 mg/ml and those of the SA11 and I321 peptides being 30 mg/ml.
Oligomeric transitions in SA11ΔN94 and SA11ΔN72 peptides.
To determine if there exists an equilibrium among different oligomeric forms and to examine in detail the influence of the N-terminal AD and the C-terminal flexible region on pH-, calcium-, or protein concentration-dependent oligomeric transitions and the relative equilibrium between pentamers and tetramers, the SA11ΔN94 and SA11ΔN72 mutant proteins were purified under different conditions (conditions A to F), as described in Materials and Methods, and analyzed by analytical SEC. As seen from the elution profiles and the calculated apparent molecular weights (MWs) corresponding to the major peaks (Fig. 9c to j and Table 2), when analyzed at a 10-mg/ml concentration, the ΔN94 protein consistently eluted as a relatively sharp peak corresponding to an apparent molecular mass in the range of 54.94 to 57.54 kDa independent of the conditions of purification. However, at 5 mg/ml, the major peak shifted to an apparent molecular mass in the range of 45.70 to 50.12 kDa (Fig. 9c to j). The protein purified under condition B (Fig. 9d) eluted as a broad peak corresponding to tetramers and other oligomeric forms at 5 mg/ml, suggesting an extensive dissociation of the pentamer. Although calcium and, to a lesser extent, magnesium appear to stabilize the tetramer, the latter significantly stabilized the dimeric form, which eluted at an apparent molecular mass in the range of 21.3 to 29.5 kDa, in contrast to the former. Note that the observed molecular masses corresponding to different major and minor peaks are slightly higher than those expected for a pentamer (54.75 kDa), a tetramer (43.80 kDa), or a dimer (21.90 kDa) of His-tagged SA11ΔN94 (Table 2), which could be attributed to the extended conformation of the C-terminal region and the flexible extreme C terminus rather than the globular nature of the standard proteins. The present results clearly demonstrate that NSP4 exists as pentamers at high concentrations and as tetramers and lower forms at low protein concentrations independent of the conditions of preparation. ΔN72 existed primarily in multimeric forms independent of the conditions of preparation. Interestingly, a minor shoulder peak corresponding to a pentameric form of an apparent molecular mass of 70.7 kDa (with the predicted molecular mass for this pentamer being 66.75 kDa) was consistently observed when the protein was prepared from cells lysed in the presence of calcium. Only at very low concentrations (1.0 mg/ml) did ΔN72 show detectable lower oligomeric forms (data not shown).
Table 2.
SEC analysis at different concentrations of NSP495-146 and ΔΝ94 purified under different conditionsa
| Purification process | Apparent molecular mass (no. of subunits) |
|||
|---|---|---|---|---|
| SA11ΔΝ94 at concn (mg/ml) of: |
ST3:NSP494-29 at concn (mg/ml) of: |
|||
| 10 | 5 | 20 | 10 | |
| Lysis in acidic pH 5.0 buffer (condition A) | 60.25 (5.51) | 47.86 (4.36) | 34.9 (5.41) | 27.21 (4.2) |
| Lysis in the absence of CaCl2 (condition B) | 56.23 (5.14) | 45.70 (4.08) | 34.9 (5.41) | 29.15 (4.5) |
| Lysis in the presence of CaCl2 (condition C) at concn (mM) of: | ||||
| 0.1 | 57.54 (5.26) | 50.12 (4.58) | ND | ND |
| 1.0 | 54.94 (5.02) | 47.86 (4.38) | ND | ND |
| 10 | 57.54 (5.26) | 48.97 (4.48) | 33.69 (5.21) | 27.86 (4.3) |
| Lysis and purification in the presence of 0.1 mM CaCl2 (condition D) | 54.94 (5.02) | 47.86 (4.38) | 31.75 (4.95) | 25.9 (4.0) |
| Lysis in the presence of 10 mM MgCl2 (condition E) | 54.94 (5.02) | 47.86 (4.36) | ND | ND |
| 27.54 (2.52) | ||||
| Lysis and purification in the presence of 0.1 mM MgCl2 (condition F) | 60.2 (5.51) | 47.89 (4.38) | 32.69 (5.0) | 25.9 (4.0) |
Shown are the apparent molecular masses (numbers of subunits) corresponding to the major peaks of the SA11ΔN94 and ST3:NSP495-146 peptides purified under different conditions. Conditions A to F represent different conditions of purification. ND, not determined.
Note that in our previous report (26), the pentameric peak could not be clearly distinguished, as the analyses were carried out with relatively low protein concentrations (2 mg/ml) using Sephacryl S-200 resin. Furthermore, the existence of bands corresponding to pentameric forms in previous cross-linking experiments has been overlooked in view of the accepted tetrameric structure of the diarrhea-inducing region (8, 13) and the membrane-anchored tetrameric topology models of NSP4 (5, 11).
DISCUSSION
Prior to the present study, NSP4 was considered primarily a calcium-bound tetramer. We have previously reported that NSP4 forms ordered multimeric structures mediated by the AD located between residues 73 and 85 (14, 26), which has been also observed in other studies (23, 24). Here we report a novel pentameric structure for the NSP495–146 region from strain ST3, which was previously determined to be a calcium-bound tetramer in strains SA11 and I321, with implications for its biological functions. One of the distinct features of the pentameric structure is the absence of the Ca2+ ion present in the core of the tetrameric form coordinating with the side chains of four Gln123 and two Glu120 residues (Fig. 3). At pH 7.5, it is likely that the glutamates in the tetramer are charged, as indicated by their pKa values, and as the proximity of four negative charges is highly unfavorable in the interior of the protein, the side chains of only two of them are accommodated inside. The Ca2+ ion coordinates with these two glutamates and neutralizes the charge. The charges of the other two glutamates are neutralized as they interact with Arg119 residues of the neighboring helices by forming salt bridges. In the pentamers, all five glutamates remain protonated and are involved in forming a hydrogen-bonding network inside the helical core in a ringlike manner. Ca2+ does not bind in the pentamers, as there is no requirement for charge compensation, and the coordinating distances will not be favorable, as the pore diameter of the pentamer is more than that of the tetramer.
The angle of rotation between the two chains in the asymmetric unit of the two tetrameric structures of NSP4 (PDB accession numbers 1G1I and 2O1K) is about 81°, and that between successive chains in the third structure (PDB accession number 2O1J), which is a tetramer in the asymmetric unit, varies between 88° and 95°. The fact that the relation between the individual chains of the tetrameric NSP4 is significantly different from the exact 4-fold rotational symmetry might be an indication that the chains are poised to take up different types of associations. The N-terminal part (residues 95 to 120) of the chains of the tetramer being related by a 5-fold relation indicates flexibility within single chains. Similarly, in the present structure, the angles of rotation between successive chains do not correspond to a strict 5-fold rotation but vary between 67° and 77°. Transitions between tetrameric and pentameric structures were observed previously for other proteins with parallel helical arrangements similar to that observed for the present structure. The assembly domain of a cartilage oligomeric matrix protein (34) and an E. coli major outer membrane lipoprotein (31, 32) were reported previously to undergo such transitions. Note that these are also membrane proteins. In the case of the E. coli protein, mutations at the a and d positions changed the oligomeric state, whereas in the cartilage matrix protein, the mutation of an Asn residue involved in interhelical H bonds to a Leu residue changed the pentameric structure to a tetramer. In the structure of the peptide reported here, a brief exposure to acidic pH during cell lysis, the presence or absence of metal ions, or high protein concentrations appeared to be responsible for the observed transition. Furthermore, many bacterial enterotoxins, such as the heat-labile enterotoxin of E. coli, cholera toxin, and Shiga toxin, are also known to form pentamers (30, 36, 44).
Although the lysis of cells in acidic buffer as well as its crystallization at pH 6.5 suggested that exposure to acidic pH might have facilitated ST3:NSP495-146, which lacks the N- and C-terminal modulatory regions, to form pentamers, detailed analyses of the peptide from strains ST3 and SA11 purified under different conditions indicated that it forms pentamers at relatively high concentrations (∼20 mg/ml) and that it forms tetramers at or below a ∼10-mg/ml concentration irrespective of the conditions of preparation. In contrast, ΔN94 existed as pentamers at 10 mg/ml and as tetramers and other lower forms at a 5-mg/ml concentration, suggesting that the longer mutant is more efficient in pentamer formation or that the pentamers are more stable at relatively low concentrations than those of the NSP495-146 peptide. Similarly, while the pentamer of the NSP495-146 peptide dissociated into the tetrameric form at a relatively high concentration (∼10 mg/ml), that of ΔN94 dissociated into tetramers at ∼5 mg/ml. Thus, NSP495-146 appears to be easily susceptible to the oligomeric transition from a pentamer to a tetramer at relatively high concentrations (∼10 mg/ml), whereas the ΔN94 pentamer appears to be more stable, and calcium has little influence on its oligomeric transitions. The tetrameric forms in both the shorter and the longer mutants are stabilized by Ca2+ at a low concentration. These results clearly demonstrated the concentration-dependent formation of pentamers by both mutants independent of the conditions of preparation. Cross-linking experiments with ΔN94 and ΔN72 mutant proteins further confirmed the presence of pentameric, tetrameric, and other oligomeric forms in solutions of both proteins.
It is of interest that no equilibrium between the pentameric and tetrameric forms was observed for the longer mutants at the concentrations at which the experiments were performed. At high concentrations, only pentamers were detected, and at low concentrations, only tetramers and lower oligomers were detected. The level of pentamers appears to be dependent primarily on the concentration of the protein, but not calcium, although calcium and, to some extent, magnesium appear to stabilize or prevent the further dissociation of the tetrameric forms at low protein concentrations. ΔN72 existed primarily in a high-MW multimeric form, but in the presence of calcium (condition C), a minor shoulder peak corresponding to that of a pentamer was consistently seen, even at high concentrations, suggesting calcium-independent pentamer formation (Fig. 9b), and low levels of oligomers of ΔN72 were observed only at very low (1 mg/ml) concentrations (data not shown). These data and our previous results (14, 26) suggest that the N-terminal AD and the C-terminal flexible region are important for the concentration-dependent formation and stability, as well as for equilibrium maintenance among the different oligomeric states, of the protein.
Very recently, the viroporin activity of NSP4 was shown to be associated with the region at residues 54 to 81, in which lysines 62, 66, and 69 within the pentalysine domain (PD) were shown to be important for membrane insertion and the AD from residues 71 to 81 of the viroporin domain (VD) were shown to be important for its viroporin activity (24). An alternate two-helix transmembrane hairpin model was proposed, since the previously accepted membrane-associated topology model of NSP4 could not account for the viroporin function of this domain (24). Although viroporin functions of several viral proteins have been reported (19, 21, 48), the precise oligomeric organization in the membrane of any of the viroporins is yet to be understood. Since Ca2+ is tightly bound to the core in the previously reported tetrameric structures of NSP4 (8, 13), this model fails to support the viroporin activity of the protein (24), due to the narrow pore size. In contrast, the larger pore size of the present pentameric form would allow the free passage of mono- and divalent ions. In this context, recent studies indicated that the amphipathic coiled-coil helical structure extends further N terminal to residue 94 (24, 26), thus likely extending the internal pore into the lumen of the ER. The viroporin activity of NSP4 correlates with ER and plasma membrane permeability, the depletion of ER Ca2+ stores, and an increase in cytoplasmic Ca2+ levels. The increase in cytoplasmic calcium levels was proposed previously to mobilize the ER-associated NSP4 into the punctate structures surrounding the viroplasms (24), which are the cytoplasmic inclusions in which genome replication and the assembly of the immature DLPs occur (18). NSP4 was proposed previously to function as a sensor for changes in cytoplasmic Ca2+ levels (24), leading to the stabilization of tetramers at relatively low protein concentrations in different cellular locations. The present studies clearly demonstrate that the oligomeric state of the protein is driven primarily by its effective concentration and that calcium has little role in the formation or stability of higher oligomers, and divalent metal ions, including calcium, appear to stabilize the tetrameric forms at low concentrations of the protein. Since the protein is synthesized on the ER membrane, its effective concentration is likely to be higher at the site of synthesis than at other sites in the cell. It is possible that while the ER-anchored NSP4 exists predominantly in pentamers, the lower oligomeric forms, including tetramers, could be present in other cellular compartments, where its effective concentration would be low.
Note that NSP4 binding to integrin α1β1 and α2β1 receptors was shown to correlate with diarrhea induction in neonatal mice and that a mutation of Glu120 abolished the receptor binding and diarrhea-inducing properties of the protein. However, in the presence of Ca2+, but not Mg2+ or Mn2+, NSP4 failed to bind to the receptor (43). This suggests a Ca2+-independent role for Glu120 in NSP4 function. In this context, unpublished studies from our laboratory on the secreted and diarrheagenic form of the ΔN112 protein reveal that it fails to form tetramers and it is likely stabilized by magnesium.
NSP4 plays an essential role in the transport of DLPs into the lumen of the ER, where they mature into TLPs, the mechanism of which is not yet understood. Our studies as well as others reveal that NSP4 exists in oligomeric as well as ordered multimeric states (14, 23, 24). Although it is difficult to predict a correlation between different structural states and the varied activities of NSP4, it is tempting to speculate a role for the pentameric form in its function as a viroporin that supports the passage of Ca2+. Furthermore, unlike the tetrameric model, the pentameric model possesses the 5-fold symmetry required for the ER-resident receptor to interact with the VP6 molecules related by a 5-fold axis on the surface of the DLPs. It is apparent that NSP4, which carries out multiple tasks in the life cycle of rotavirus and which has been reported to exist in tetrameric, multimeric, cleaved, and secreted forms, has a high degree of conformational flexibility and most likely exists in different oligomeric states in different compartments in the infected cell. Prior to this study, our knowledge of the structure and function of rotavirus NSP4 was limited to that based on the calcium-bound tetrameric model. The exact implications of the existence of multiple oligomeric forms of NSP4 are yet to be established, but this structural diversity, demonstrated for the first time in this report, provides new directions for future experiments and interpretations of results in rotavirus research involving this multifunctional protein.
ACKNOWLEDGMENTS
Preliminary X-ray diffraction data were collected at the X-Ray Facility for Structural Biology, supported by the Department of Science and Technology, Government of India. We acknowledge financial support from the Indian Council of Medical Research and the Department of Biotechnology Infrastructure Program to the Indian Institute of Science. A.R.C. thanks the University Grants Commission, India, for a research fellowship.
We acknowledge the technical assistance of D. Dhivya, P. Aishwarya, Vembuli Amman, Sudheendra Kumar, and A. Raghavendra.
We declare that we have no financial conflict of interest.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Adams P. D., et al. 2002. Crystallographic structure determination. Acta Crystallogr. D 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 2. Au K. S., Chan W. K., Burns J. W., Estes M. K. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS 28. J. Virol. 63:4553–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ball J. M., Tian P., Zeng C. Q.-Y., Morris A. P., Estes M. K. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104 [DOI] [PubMed] [Google Scholar]
- 4. Bas D. C., Rogers D. M., Jensen J. H. 2008. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins 73:765–783 [DOI] [PubMed] [Google Scholar]
- 5. Bergman C. C., Maass D., Poruchynsky M. S., Atkinson P. H., Bellamy A. R. 1989. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 8:1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berkova Z., et al. 2006. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasm. J. Virol. 80:6061–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boshuizen J. A., et al. 2004. Rotavirus enterotoxin NSP4 binds to the extracellular matrix proteins laminin-β3 and fibronectin. J. Virol. 78:10045–10053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bowman G. D., et al. 2000. Crystal structure of the oligomerization domain of NSP4 from rotavirus reveals a core metal-binding site. J. Mol. Biol. 304:861–871 [DOI] [PubMed] [Google Scholar]
- 9. Brunger A. T., et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905–921 [DOI] [PubMed] [Google Scholar]
- 10. Bugarcic A., Taylor J. A. 2006. Rotavirus nonstructural glycoprotein NSP4 is secreted from the apical surfaces of polarized epithelial cells. J. Virol. 80:12343–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan W.-K., Au K. S., Estes M. K. 1988. Topography of the simian rotavirus nonstructural glycoprotein (NS28) in the endoplasmic reticulum membrane. Virology 164:435–442 [DOI] [PubMed] [Google Scholar]
- 12. Cohen G. H. 1997. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr. 30:1160–1161 [Google Scholar]
- 13. Deepa R., Rao C. D., Suguna K. 2007. Structure of the extended diarrhea-inducing domain of rotavirus enterotoxigenic protein NSP4. Arch. Virol. 152:847–859 [DOI] [PubMed] [Google Scholar]
- 14. Deepa R., et al. 2008. The flexible C terminus of the rotavirus non-structural protein NSP4 is an important determinant of its biological properties. J. Gen. Virol. 89:1485–1496 [DOI] [PubMed] [Google Scholar]
- 15. Diaz Y., et al. 2008. Expression of nonstructural rotavirus protein NSP4 mimics Ca2+ homeostasis changes induced by rotavirus infection in cultured cells. J. Virol. 82:11331–11343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dong Y., Zeng C. Q.-Y., Ball J. M., Estes M. K., Morris A. P. 1997. The rotavirus enterotoxin NSP4 mobilizes intracellular calcium in human intestinal cells by stimulating phospholipase C-mediated inositol 1,4,5-trisphosphate production. Proc. Natl. Acad. Sci. U. S. A. 94:3960–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Estes M. K., Kapikian A. Z. 2007. Rotaviruses, p. 1917–1974 In Knipe D. M., et al. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 19. Fischer W. B., Sansom M. S. P. 2002. Viral ion channels: structure and function. Biochim. Biophys. Acta 1561:27–45 [DOI] [PubMed] [Google Scholar]
- 20. Gibbons T. F., et al. 2011. Rotavirus NSP4: cell type-dependent transport kinetics to the exofacial plasma membrane and release from intact infected cells. Virol. J. 8:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gonzalez M. E., Carrasco L. 2003. Viroporins. FEBS Lett. 552:28–34 [DOI] [PubMed] [Google Scholar]
- 22. Ho B. K., Gruswitz F. 2008. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hyser J. M., Zeng C. D., Beharry Z., Palzkill T., Estes M. K. 2008. Epitope mapping and use of epitope-specific antisera to characterize the VP5* binding site in rotavirus SA11 NSP4. Virology 373:211–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hyser J. M., Collinson-Pautz M. R., Utama B., Estes M. K. 2010. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 1:e00265–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jagannath M. R., Vethanayagam R. R., Reddy B. S. Y., Raman S., Rao C. D. 2000. Characterization of symptomatic rotavirus isolates MP409 and MP480 having ‘long’ RNA electropherotype and subgroup I specificity, highly related to the P[6], G8 type bovine rotavirus A5, from Mysore, India. Arch. Virol. 145:1339–1357 [DOI] [PubMed] [Google Scholar]
- 26. Jagannath M. R., et al. 2006. N- and C-terminal cooperation in rotavirus enterotoxin: novel mechanism of modulation of the properties of a multifunctional protein by a structurally and functionally overlapping conformational domain. J. Virol. 80:412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jancarik J., Kim S. H. 1991. Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 24:409–411 [Google Scholar]
- 28. Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 29. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 30. Ling H., et al. 1998. Structure of the Shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry 37:1777–1788 [DOI] [PubMed] [Google Scholar]
- 31. Liu J., Yong W., Deng Y., Kallenbach N. R., Lu M. 2004. Atomic structure of a tryptophan-zipper pentamer. Proc. Natl. Acad. Sci. U. S. A. 101:16156–16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., Zheng Q., Deng Y., Kallenbach N. R., Lu M. 2006. Conformational transition between four and five-stranded phenylalanine zippers determined by a local packing interaction. J. Mol. Biol. 361:168–179 [DOI] [PubMed] [Google Scholar]
- 33. Maass D. R., Atkinson P. H. 1990. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J. Virol. 64:2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malashkevich V. N., Kammerer R. A., Efimov V. P., Schulthess T., Engel J. 1996. The crystal structure of a five-stranded coiled coil in COMP: a prototype ion channel? Science 274:761–765 [DOI] [PubMed] [Google Scholar]
- 35. McCoy A. J., et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merritt E. A., et al. 1994. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 3:166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer J. C., Bergmann C. C., Bellamy A. R. 1989. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology 171:98–107 [DOI] [PubMed] [Google Scholar]
- 38. Mir K. D., Parr R. D., Schroeder F., Ball J. M. 2007. Rotavirus NSP4 interacts with both the amino- and carboxyl-termini of caveolin-1. Virus Res. 126:106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newton K., Meyer J. C., Bellamy A. R., Taylor J. A. 1997. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J. Virol. 71:9458–9465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Otwinowsky Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 41. Pelligrini-Calace M. T., Maiwald T., Thornton J. M. 2009. PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structures. PLoS Comput. Biol. 5:e100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roberts L. 2004. Vaccines: rotavirus vaccines' second chance. Science 305:1890–1893 [DOI] [PubMed] [Google Scholar]
- 43. Seo N.-S., et al. 2008. Integrins α1β1 and α2β1 are receptors for the rotavirus enterotoxin. Proc. Natl. Acad. Sci. U. S. A. 105:8811–8818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sixma T. K., et al. 1993. Refined structure of Escherichia coli heat-labile enterotoxin, a close relative of cholera toxin. J. Mol. Biol. 230:890–918 [DOI] [PubMed] [Google Scholar]
- 45. Storey S. M., et al. 2007. Full-length, glycosylated NSP4 is localized to plasma membrane caveolae by a novel raft isolation technique. J. Virol. 81:5472–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor J. A., O'Brien J. A., Yeager M. 1996. The cytoplasmic tail of NSP4, the endoplasmic reticulum-localized non-structural glycoprotein of rotavirus, contains distinct virus binding and coiled coil domains. EMBO J. 15:4469–4476 [PMC free article] [PubMed] [Google Scholar]
- 47. Tian P., et al. 1994. The nonstructural glycoprotein of rotavirus affects intracellular calcium levels. J. Virol. 68:251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Kuppeveld F. J. M., Melchers W. J. G., Kirkegaard K., Doedens J. R. 1997. Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227:111–118 [DOI] [PubMed] [Google Scholar]
- 49. Varo G., Brown L. S., Needleman R., Lanyi J. K. 1999. Binding of calcium ions to bacteriorhodopsin. Biophys. J. 76:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu A., Bellamy A. R., Taylor J. A. 2000. Immobilization of the early secretory pathway by a virus glycoprotein that binds to microtubules. EMBO J. 19:6465–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang M., Zeng C. Q.-Y., Morris A. P., Estes M. K. 2000. A functional NSP4 enterotoxin peptide secreted from rotavirus-infected cells. J. Virol. 74:11663–11670 [DOI] [PMC free article] [PubMed] [Google Scholar]