Abstract
Hepatitis C virus (HCV) infection is a prominent risk factor for the development of hepatocellular carcinoma (HCC). Similar to most solid tumors, HCCs are believed to contain poorly differentiated cancer stem cell-like cells (CSCs) that initiate tumorigenesis and confer resistance to chemotherapy. In these studies, we demonstrate that the expression of an HCV subgenomic replicon in cultured cells results in the acquisition of CSC traits. These traits include enhanced expression of doublecortin and CaM kinase-like-1 (DCAMKL-1), Lgr5, CD133, α-fetoprotein, cytokeratin-19 (CK19), Lin28, and c-Myc. Conversely, curing of the replicon from these cells results in diminished expression of these factors. The putative stem cell marker DCAMKL-1 is also elevated in response to the overexpression of a cassette of pluripotency factors. The DCAMKL-1-positive cells isolated from hepatoma cell lines by fluorescence-activated cell sorting (FACS) form spheroids in Matrigel. The HCV RNA abundance and NS5B levels are significantly reduced by the small interfering RNA (siRNA)-led depletion of DCAMKL-1. We further demonstrate that HCV replicon-expressing cells initiate distinct tumor phenotypes compared to the tumors initiated by parent cells lacking the replicon. This HCV-induced phenotype is characterized by high-level expression/coexpression of DCAMKL-1, CK19, α-fetoprotein, and active c-Src. The results obtained by the analysis of liver tissues from HCV-positive patients and liver tissue microarrays reiterate these observations. In conclusion, chronic HCV infection appears to predispose cells toward the path of acquiring cancer stem cell-like traits by inducing DCAMKL-1 and hepatic progenitor and stem cell-related factors. DCAMKL-1 also represents a novel cellular target for combating HCV-induced hepatocarcinogenesis.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common malignancy worldwide, accounting for approximately 1 million deaths annually (10, 40, 58, 59). The high mortality associated with HCC is attributed to the failure of early-stage diagnosis and lack of effective treatment (10, 55, 56). Chronic infection with hepatitis C virus (HCV) is considered to be a prominent risk factor for the development of HCC (6, 23, 57). More than 170 million people (>4 million in the United States alone) are infected, and HCV-related liver disease is increasing globally. Although a strong relationship between HCV-induced chronic liver diseases and the development of HCC is widely accepted, the molecular mechanism of HCV-induced hepatocarcinogenesis is not clearly understood.
HCV is a positive-strand RNA virus classified as a hepacivirus of the family Flaviviridae (see reviews in references 35, 41, and 45). Among the 6 genotypes, 12 subtypes, and various quasispecies (32), 1a and 1b are the most prevalent strains in the United States and are less responsive to the antiviral treatments (11, 27, 45). The HCV genome (9,600 nucleotides [nt]) encodes a single polyprotein that is processed cotranslationally into three structural (C, E1, and E2) and seven nonstructural (NS) polypeptides (p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B) (25). Similar to other positive-strand RNA viruses, HCV replicates via synthesis of negative-strand RNA using replication complexes (RCs) comprising most of the NS proteins and as-yet-undefined cellular factors (5, 41). During infection, HCV induces weblike membranous structures and uses lipid raft and microtubule filaments (MTFs) for its replication and transport (19, 35, 54). Additionally, the viral NS3/4A protein cleaves the mitochondrial antiviral signaling protein (MAVS, also known as IPS-1 or VISA) and toll-like receptor 3 adaptor protein (TRIF) to suppress innate immunity (13, 42, 43). It also induces endoplasmic reticulum (ER) stress and alters a cascade of signal transduction pathways that control cell cycle and cellular growth (12, 49, 53).
HCV-induced molecular alterations in infected cells contribute significantly to HCC development and progression. These alterations may include (i) loss of tumor suppressor proteins, (ii) activation of oncoproteins, such as c-Myc, (iii) activation and secretion of cytokines, such as transforming growth factor β (TGF-β), and (iv) alterations in the Wnt/β-catenin signaling, leading to nuclear accumulation of β-catenin, which are found in 33 to 67% of HCC cases (6, 38). Activation of β-catenin is essential for liver development; deletion of the protein in mice results in fetal death due to impaired liver cell proliferation and increased apoptosis (50). The Wnt/β-catenin signaling pathway is also important for tumor progression because it modulates the differentiation and maintenance of stem cells (2, 21, 63).
Cancer stem cell-like cells (CSCs) display several key characteristics of normal tissue stem cells, such as self-renewal and unlimited proliferative and differentiation capacity. They also possess the intrinsic ability to reproduce all aspects of the parent tumor after metastasis (2). Thus, the “hierarchical model of cancer” considers CSCs to be a tumor's “seed elements,” which are responsible for cancer initiation, reinitiation after metastasis, and local recurrence after therapy (28, 46). Histopathological study of chronic liver disease and experiments in mice support the existence of CSCs in HCC (33, 46). Recent studies indicate that the induction of “stemness” in normal tissues or abnormal self-renewal, differentiation, and proliferation of stem/progenitor cells might be key elements in the generation of CSCs (26, 33).
Multiple transcription factors and signaling pathways, including Wnt/β-catenin, Notch, and Hedgehog, may be involved in the induction and maintenance of “stemness” in fully differentiated cells (50, 60). The induction of pluripotency/multipotency in adult somatic cells can be carried out by the simultaneous expression of several transcription factors (a set of Oct4, Sox2, Klf4, and c-Myc or Oct4, Sox2, Lin28, and Nanog) in recipient adult cells using various delivery vehicles, such as viruses, transposons, and bacterial plasmids (4, 16, 39). Successful generation of induced pluripotent stem cells (iPSCs) has been accomplished by manipulating culture conditions and with a limited number of pluripotency factors (17). The most widely accepted evidence for pluripotency/multipotency is the formation of spheroids and teratomas, unlimited self-renewal of the reprogrammed cells, and differentiation into downstream cell lineages.
Because HCV replicates in fetal hepatocytes and the presence of CSCs has been demonstrated in HCC (2, 20, 33), we investigated whether HCV could induce cancer stem cell-like properties in liver-derived cells. Our study shows that continued HCV expression is linked to the overexpression of a putative stem cell marker, doublecortin and CaM kinase-like-1 (DCAMKL-1), that also affects the abundance of viral RNA and polymerase. Using multiple approaches, including the examination of liver tissues from chronic HCV patients, we further demonstrate that HCV-expressing cells are predisposed to the acquisition of characteristics of CSCs and hepatic progenitor cells.
MATERIALS AND METHODS
Antibodies.
The antibodies used in this study were purchased as indicated below and used in accordance with the manufacturer's suggestions: anti-DCAMKL-1, -CD133, -SOX2, -LGR5, -CK19, -human serum albumin, -α-fetoprotein, and -HCV NS5B polymerase were purchased from Abcam; anti-α-tubulin, -Lin28B, -c-Src, and -actin were from Cell Signaling; anti-c-Myc and anti-Oct3/4 were from Santa Cruz Biotechnologies; and anti-Lin28 was from Protein Tech Group.
Cell culture and transfection assay.
FCA4 cells are derived from the Huh7 hepatoma cell line, and the sustained replication of a subgenomic HCV replicon in this cell line has been characterized previously (1, 14). The GS5 cells are derived from the Huh7.5 cell line (interferon-cured Huh7 cells negative for the HCV replicon [7]) and express an HCV subgenomic replicon that encodes NS5A-green fluorescent protein (GFP) (37). These cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1× penicillin-streptomycin, 1× nonessential amino acids, and 10% fetal bovine serum (all from Invitrogen) and maintained at 37°C and 5% CO2. To cure GS5 cells of the replicon, the cells were cultured and passaged routinely for 45 days in the absence of G418, followed by treatment with 2α interferon (IFN-2α) (100 U/ml; Sigma) plus cyclosporine (3 μg/ml) for 2 weeks. These cells (termed GS5-C) became sensitive to the drug G418 and were found to be negative for GFP, NS5B, and the HCV RNA during subsequent passages. Total lysates of culture cells were prepared according to the protocol described for Pierce M-PER cell lysis buffer (Thermo Fisher Scientific) for Western blotting.
The small interfering RNA (siRNA) that targets nt 308 to 327 (5′GGGAGTGAGAACAATCTAC3′) of both the AS and AL isoforms (short- and long-form A, respectively) of DCAMKL-1 mRNAs (siDCAMKL-1) and scrambled siRNA (SCR) were purchased from Ambion. Incremental amounts of the siDCAMKL-1 or SCR RNAs (0, 20, 50, and 100 nM) were transfected into GS5 cells using the siPORT NeoFX transfection agent (Applied Biosystems) according to the manufacturer's protocol. Similar amounts of the transfection reagent alone were used in control transfections. The cells were harvested 48 h posttransfection to analyze RNA and protein levels.
Plasmids pJFH1 and pJFH1/GND (52) were linearized with XbaI and transcribed with T7 RNA polymerase by using a RiboMax transcription kit (Promega). The RNAs were purified, checked for integrity, and transfected into Huh7 cells using Lipofectamine 2000 (Invitrogen). The pluripotency expression cassette containing plasmid P2PhiC31-LGNSO was used to generate minicircle supercoiled DNA molecules as described by Jia et al. (15). The minicircle DNAs were transfected into Huh7 cells as described above.
Fluorescence-activated cell sorting (FACS).
The DCAMKL-1 antibodies or rabbit IgG (control) was conjugated with DyLight 549 (Thermo Fisher) using the Pierce kit (product no. 53035). The conjugated DCAMKL-1 antibodies or rabbit IgG control was purified and allowed to bind cell surface DCAMKL-1 of Huh7.5 or GS5 cells for 30 min at room temperature as described previously (29, 31). The cells were washed twice with autoMACS rinsing buffer (Miltenyi Biotec), resuspended in the same buffer, and sorted using an Influx-V cell sorter (Cytopeia, Seattle, WA). The instrument was calibrated with unstained cells. The DCAMKL-1-positive and -negative cells were collected separately in DMEM and grown as described above. For the spheroid assay, 100 DCAMKL-positive (DCAMKL+) or DCAMKL-negative (DCAMKL−) cells in 2× DMEM (50 μl) were mixed with Matrigel (50 μl) on an ice bath and plated in nonadherent/ultralow-attachment 96-well plates (BD Biosciences, CA). Fifty microliters of medium was added on top of the gel 2 h later, and the cells were grown as described above. Two weeks later, 100 μl DMEM was added to each well. The spheroids were evaluated and photographed under an inverted light microscope after 4 to 5 weeks.
Real-time reverse transcription-PCR analyses.
Total RNAs were isolated from culture cells or tumor xenografts by using an RNeasy isolation kit (Qiagen). The RNA samples were subjected to reverse transcription with Superscript II and random hexanucleotide primers (Invitrogen). In the subsequent step, the cDNAs were used as templates to perform real-time PCR by the SYBR chemistry method (SYBR green I; Molecular Probes). The target (HCV and DCAMKL-1) and control (actin) RNAs were amplified using JumpStart Taq DNA polymerase (Sigma) and the following primers: actin, 5′GGTGATCCACATCTGCTGGAA-3′ (forward) and 5′ATCATTGCTCCTCCTCAGGG3′ (reverse); DCAMKL-1, 5′AGTCTTCCGATTCCGAGTTGAG3′ (forward) and 5′CAGCAACCAGGAATGTATTGGA3′ (reverse); and HCV, 5′CGGGAGAGCCATAGTGG3′ (forward) and 5′AGTACCACAAGGCCTTTCG3′ (reverse).
The crossing threshold values assessed by the real-time PCR were evaluated for the transcripts and normalized to the results for β-actin mRNA. The mRNA levels were expressed as the fold change relative to the level in the control with the plus-or-minus standard error of the mean. In a similar transfection study, an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay (Promega) for cell viability and proliferation was carried out.
Tumor xenografts in a mouse model.
Athymic nude BALB/c mice were purchased from Jackson Laboratory and housed in pathogen-free conditions. The animals were treated in accordance with the guidelines of the American Association for Accreditation of Laboratory Animal Care and the Policy on Humane Care and Use of Laboratory Animals. The studies reported here were preapproved and supervised by the Institutional Animal Care and Use Committee (IACUC). Huh7.5 or GS5 cells (5 × 106) were washed with phosphate-buffered saline (PBS) three times, resuspended in the same buffer, and injected subcutaneously into the dorsal flanks of 4- to 6-week-old mice (3 mice for each cell line). Tumors were measured with calipers, and the volumes were calculated using the formula 0.5 × (length × width2). The tumors were resected 4 to 6 weeks after cell transplantation, and portions of the tumors were preserved in 5% formalin for immunohistochemistry, stored at −80°C in RNase inhibitors for real-time PCR, or directly frozen for Western blot analyses. The control animals were treated similarly but injected with the same volume of the buffer.
Immunohistochemistry.
Heat-induced epitope retrieval was performed on 4-μm formalin-fixed paraffin-embedded sections using a pressurized Decloaking Chamber (Biocare Medical) in citrate buffer (pH 6.0) at 99°C for 18 min. The slides were sequentially treated with 3% hydrogen peroxide, normal serum, and bovine serum albumin (BSA) at room temperature for 20 min. After incubation with primary antibodies and washing with PBS, the slides were incubated using the peroxidase-conjugated EnVision+ polymer detection kit (Dako). Slides were developed with diaminobenzidine (Sigma).
Immunofluorescence and confocal microscopy.
Cells grown on glass coverslips (VWR) were rinsed briefly in PBS, fixed in 4% paraformaldehyde in PBS, pH 7.4, for 20 min at room temperature, washed twice with ice-cold PBS, and permeabilized in ice-cold acetone. Cells were incubated with blocking buffer (10% serum, 0.01% Triton X-100 in PBS, pH 7.4) for 1 h, washed with PBS, and treated with primary antibodies in PBS–0.01% Triton X-100 (PBS-T) containing 1% BSA for 1 to 2 h at room temperature or overnight at 4°C. After a thorough washing with PBS-T, the coverslips were incubated in appropriate Alexa Fluor-conjugated secondary antibodies. The nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.1 to 1 μg/ml PBS). The coverslips were mounted on microscope slides in ProLong gold antifade reagent (Invitrogen) for detection of immunofluorescence using a Nikon 80i or Leica TCS NT (for confocal microscopy).
Human liver tissues and microarrays.
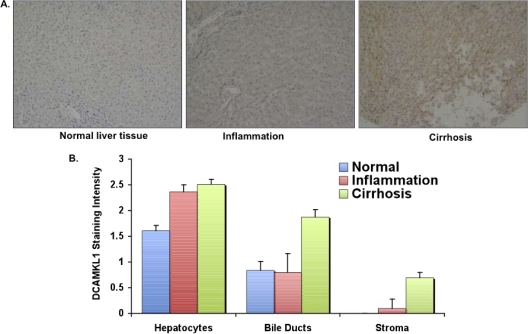
The liver tissue arrays were purchased from US Biomax (Rockville, MD). The array slide contains 30 normal and cancer-adjacent normal liver tissues, 10 cases of inflammation with hepatitis B virus infection (HBV), 9 samples of cirrhosis with HBV, and 31 samples of cirrhosis without HBV infection. The arrays were subjected to immunohistochemical staining as described above using anti-DCAMKL-1 antibodies (ab31704). The intensities of DCAMKL-1 were evaluated and plotted on an arbitrary scale of 0 to 3.
The liver biopsy specimens of the HCV RNA-positive patients with various stages of liver diseases were stained similarly for CK19 and DCAMKL-1. Patient 1 (Pt1) had macronodular cirrhosis, whereas patients 2, 3, 4, and 5 had minimal inflammation and mild fibrosis with chronic hepatitis C. Patient 5 was on dialysis due to renal failure. None of the patients had undergone antiviral therapy prior to liver biopsy. Pt1 was a nonresponder for standard antiviral therapy after the first biopsy (sample 1). The second biopsy specimen (sample 2) from this patient was taken 2 years later.
RESULTS
Hepatoma cells harboring the HCV replicon express a multitude of stem cell-related proteins.
We examined the impact of HCV replicon expression on the status of reprogramming- and hepatic progenitor cell-related proteins using GS5 and FCA4 cell lines (7, 14, 37). The GS5 cells are derived from the Huh7.5 hepatoma cell line that permits efficient replication of HCV. The cells harbor a geneticin (G418)-selectable subgenomic HCV-1b replicon that encodes nonstructural proteins NS3 through NS5B and a functionally active NS5A-green fluorescent protein (GFP) chimera (34, 37). The intensity of the GFP in GS5 cells indicates the replication efficiency of the replicon RNA (37). The cells were cultured in the absence of G418, followed by treatment with IFN-α and cyclosporine. The resulting cells (GS5-C) were considered cured of the replicon because of the absence of HCV RNA, NS5B, and GFP, as well as sensitivity toward G418 during subsequent passages. The FCA4 cells are derived from the parental Huh7 hepatoma cell line and express a subgenomic replicon similar to that in GS5 cells but without GFP. The GS5 and FCA4 cell lines are considered to partially mimic chronic HCV infection in tissue culture conditions because of sustained expression of the replicon over 6 months.
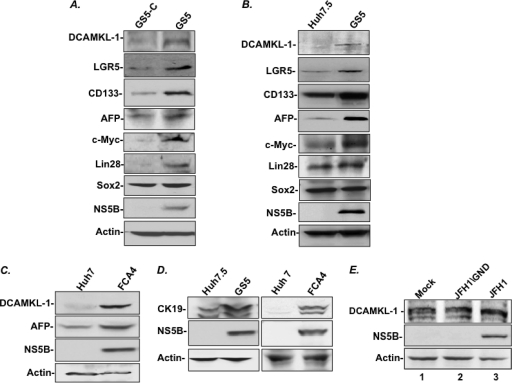
We carried out Western blot analysis of the replicon-expressing cell lines to determine the expression levels of a number of stem cell/pluripotency markers and compared their levels with those in the respective control cells that lack the replicon. As shown in Fig. 1A, the levels of putative stem cell markers DCAMKL-1, LGR5, and CD133 were considerably higher in GS5 cells as than in the cured GS5-C cells. The hepatoblast marker, α-fetoprotein (AFP), and pluripotency-related factors Lin28 and c-Myc but not Sox2 were similarly higher in GS5 cells than in the control GS5-C lysates. In the second set of Western blots, we compared the levels of these factors in GS5 cells with their levels in the parent Huh7.5 cell line that has not been exposed to the HCV replicon. Similar to the above-described observations, the GS5 cells exhibited enhanced expression of DCAMKL-1, Lgr5, CD133, c-Myc, and AFP compared to their expression in Huh7.5 cells (Fig. 1B). The presence of these factors in NS5A-GFP-positive cells was also confirmed by indirect immunofluorescence microscopy (data not shown). Additional evidence for the enhanced DCAMKL-1 and AFP expression in response to the HCV replicon was obtained by Western blot analysis of FCA4 cell lysates (Fig. 1C).
Fig. 1.
Protein expression profiles of putative stem cell, hepatoblast, and hepatic progenitor cell markers in the HCV subgenomic replicon-expressing cells (GS5 and FCA4) compared to those of the corresponding controls (GS5-C, Huh7.5, and Huh7) that lack the HCV replicon. (A to D) A 20-μg amount of total lysates was used in each case for Western blotting. (E) Western blot for determination of the DCAMKL-1 level in the Huh7.5 cells transfected with RNAs (5 μg) representing the wild-type JFH1 infectious clone (lane 3) or the mutant version of UFH1 in which RNA polymerase is inactivated (lane 2). Lane 1, mock transfection without RNA. A 40-μg amount of total lysates was used in each case, and an extensive SDS-PAGE analysis was carried out to resolve closely migrating DCAMKL-1 species.
It has been demonstrated that immature hepatic progenitor cells, such as oval cells and cholangiocytes, in the liver express cytokeratin 19 (CK19) (4, 5). The protein is also considered a marker for aggressive HCC subtypes. Furthermore, the overexpression of CK19 in HCC correlates with the lower rates of patient survival and higher rates of tumor recurrence (40, 41). These observations led us to investigate CK19 levels in the cultured cells. The Western blot of total lysates prepared from both GS5 (Fig. 1D, left) and FCA4 (Fig. 1D, right) cells revealed considerably higher levels of CK19 expression than in their corresponding control cell lysates (Huh7.5 and Huh7, respectively). Collectively, these data revealed higher expression of a host of CSCs and hepatic progenitor cell-related proteins in response to the persistent expression of the HCV replicon. Conversely, curing of the replicon from GS5 cells resulted in diminished expression of these factors (Fig. 1A).
Next, we transfected Huh7.5 cells with full-length RNAs representing wild-type JFH1 (HCV-2b genotype), which has been shown to produce infectious particles, or its replication-deficient version (JFH1/GND). The presence of NS5B in wild-type JFH1 (Fig. 1E, lane 3) and the absence of this protein in the JFH1/GND mutant (lane 2) 72 h posttransfection suggest complete expression of the JFH1 RNA. Since DCAMKL-1 is a phosphorylated protein and is expressed as multiple isoforms, we carried out extensive electrophoresis to resolve closely migrating species. Only a minor change (ratio DCAMKL-1 to actin) and/or a shift of a lower band of DCAMKL-1 was observed in the wild-type cells (lane 3) in comparison to the mutant (lane 2) or mock-transfected (lane 1) cells. Because HCC development is not observed in acute HCV infection, a dramatic increase in the DCAMKL-l level under these experimental conditions is an unlikely scenario (also see Fig. 8 and 9).
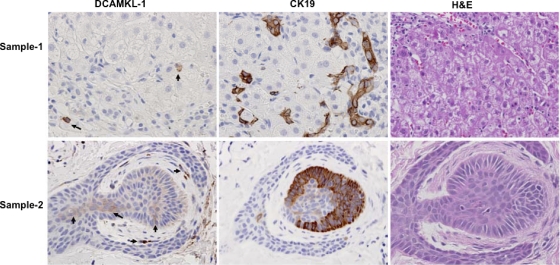
Fig. 8.
Immunohistochemistry for detection of DCAMKL-1 (left) and CK19 (middle) in liver biopsy specimens from a chronic HCV-positive patient (Pt1). Sample 2 (bottom) represents liver tissue of the same patient (Pt1) taken 2 years after the first biopsy (sample 1). Right, hematoxylin and eosin (H&E) staining of similar sections. Arrows show intense staining for DCAMKL-1.
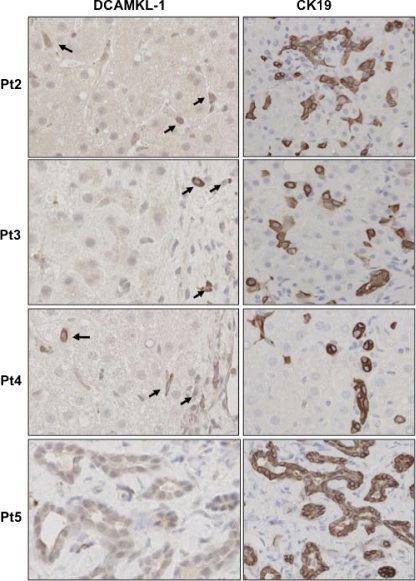
Fig. 9.
Immunohistochemistry for expression of DCAMKL-1 (left) and CK19 (right) in liver biopsy specimens of chronic HCV-positive patients at various stages of liver diseases (see Materials and Methods).
Enhanced expression of DCAMKL-1 and CK19 in Huh7 cells transfected with reprogramming factors.
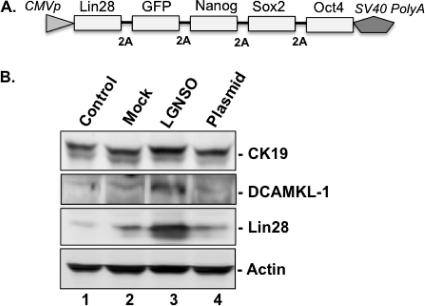
The P2PhiC31-LGNSO vector has been used by Jia et al. (15) to generate minicircle supercoiled DNA molecules that encode a set of four reprogramming/pluripotency factors (Lin28, Nanog, Sox2, and Oct4) and a GFP reporter under the control of the cytomegalovirus (CMV) promoter (Fig. 2A). The presence of the “self-cleavage” 2A sequence motif between the cistrons facilitates the translation of each of the reprogramming proteins, as well as GFP, in equal amounts from a single mRNA (Fig. 2A). Using the LGNSO system, these investigators (15) successfully induced pluripotency in human adipose cells following transfection of the minicircle DNA.
Fig. 2.
(A) Genetic organization of minicircle DNA cassette expressing four pluripotency factors, Lin28, Nanog, Sox2, and Oct4, and a reporter, GFP. CMVp, CMV promoter/enhancer; SV40 PolyA, simian virus 40 polyadenylation signal sequence. This expression cassette is produced as a closed minicircle and transcribed into a single mRNA in the transfected cells. The open reading frames are spaced with the 2A peptide sequence for producing multiple individual proteins from a single mRNA. (B) Enhanced expression of CK19 and DCAMKL-1 in Huh7 cells that were transiently transfected with minicircle DNA (lane 3, indicated as LGNSO) or an empty vector (lane 4, plasmid). Mock, cells transfected with Lipofectamine alone (lane 2) or not transfected (lane 1, Control). The protein bands detected by Western blotting are indicated.
We carried out Western blot analysis using lysates of the Huh7 cells transfected with the purified minicircle DNA for 72 h. A considerable increase in the expression of Lin28 was observed only in the minicircle-transfected cells (Fig. 2B, lane 3) and not in the empty vector or mock-transfected controls (lanes 1, 2, and 4). In addition, GFP expression was observed in ∼10 to 12% of minicircle-transfected cells by live-cell microscopy (not shown). These observations confirmed the successful expression of minicircle DNA in Huh7 cells. Interestingly, a concomitant increase in the DCAMKL-1 and CK19 levels was observed due to the overexpression of pluripotency factors (Fig. 2B, lane 3) compared to their expression in the controls (lanes 1, 2, and 4).
Localization of DCAMKL-1 in HCV replicon-expressing cells.
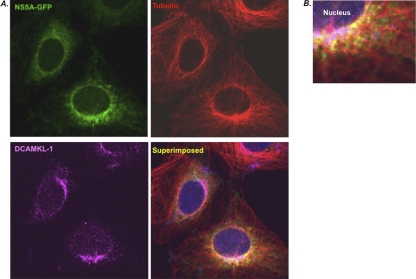
Previous studies have shown that HCV replication complexes associate with microtubules through NS5A and NS3 proteins and that their saltatory movements are fully dependent on the intact microtubule filaments (19, 54). Since DCAMKL-1 catalyzes polymerization of tubulins into microtubules (18, 24), we carried out confocal microscopy of GS5 cells that were stained for DCAMKL-1 and α-tubulin. As expected, considerable amounts of NS5A-GFP were colocalized with the microtubule filaments (MTFs) (Fig. 3A and B, yellow). Interestingly, DCAMKL-1 staining was more pronounced in the perinuclear regions where NS5A-MTF showed higher intensities. The occasional white spots in this location (Fig. 3B) appear to be due to colocalization of DCAMKL-1 with MTFs, as expected. However, it is difficult to ascertain whether a fraction of NS5A makes direct contact with DCAMKL-1 in this region.
Fig. 3.
Confocal microscopy for visualization of NS5A-GFP (green) and DCAMKL-1 (magenta) in the GS5 cells. The microtubule filaments (red) were stained using monoclonal anti-α-tubulin antibody (A). The relative localization and colocalization of these proteins are shown in the superimposed (A) and in the magnified figure (B). Blue, nuclear stain.
GS5 and Huh7.5 cells expressing DCAMKL-1 on the membrane surface exhibit cancer stem cell-like properties.
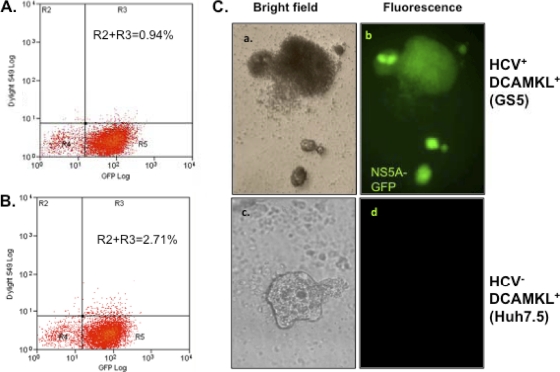
We have previously demonstrated that tumor cells expressing DCAMKL-1 on their membrane surface can be isolated by using FACS (29–31). The isolated DCAMKL-1+ cells form spheroids in Matrigel (BD Biosciences) and display CSC properties. We used this established method to isolate DCAMKL-1 and HCV NS5A-GFP double-positive cells (HCV+ DCAMKL-1+) from GS5 cells using specific anti-DCAMKL-1 antibodies (Fig. 4B) and nonspecific IgG (Fig. 4A, control) conjugates. The FACS data suggest that nearly 1.5 to 1.9% (calculated values were obtained after subtraction from the control results presented in Fig. 4A) of cells were positive for surface DCAMKL-1 expression. These cells formed spheroids of various shapes and sizes (Fig. 4C, top). Using a similar strategy, we found that 0.5 to 0.9% of cells in the Huh7.5 population also expressed DCAMKL-1 on the surface (data not shown), and these cells were capable of forming spheroids even though they lack HCV (Fig. 4C, bottom). The intensity of NS5A-GFP in GS5 spheroids was higher in certain areas (e.g., budding areas) in some cases. The DCAMKL-1− pools of these cell lines did not form spheroids under similar conditions (data not shown). These observations reinforce our previous findings on colon and pancreatic cancers that surface expression of DCAMKL-1 is related to CSC properties irrespective of the origin of malignancies (31, 48).
Fig. 4.
DCAMKL-1-positive cells show CSC traits. (A, B) The GS5 cells were treated with DyLight 549-normal IgG (A, control) or Dylight 549–anti-DCAMKL-1 (B) conjugate and subjected to FACS analysis using lasers that excite both GFP and DyLight 549. (C) Spheroid assay using isolated HCV− DCAMKL-1+ cells from the Huh7.5 (bottom) or HCV+ DCAMKL-1+ cells from the GS5 (top) cell line. The cells (∼100 from each sample) were plated with culture medium containing 50% Matrigel in a 96-well plate. The cells were grown in standard culture conditions, and spheroids were photographed after 4 weeks (a to d).
Distinct tumor phenotypes are initiated by GS5 and Huh7.5 cells in athymic nude mice.
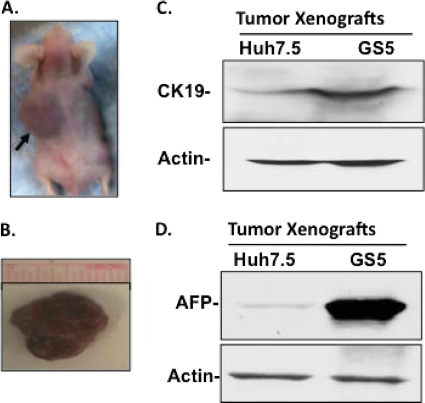
We isolated the top 26% of cells with high GFP expression from the GS5 population by using FACS. These cells were then expanded in tissue culture and injected into the dorsal flanks of three nude mice. In parallel, similar numbers of Huh7.5 cells were injected into three control mice, whereas three additional mice (control) were injected with buffer alone. Since both cell lines contain DCAMKL-1+ cells to various extents, the transplanted cells formed tumors (3 of 3 for GS5 cells and 2 of 3 for Huh7.5 cells) over a period of 3 to 4 weeks. The control mice did not develop tumors at all. An example of a GS5 tumor in one of the mice is shown in Fig. 5A (indicated by an arrow). The resected tumors (Fig. 5B) were analyzed for the expression of DCAMKL-1 and hepatic markers. Both the GS5 and Huh7.5 tumors expressed human albumin, suggesting that these tumors were originated from the transplanted cells. The expression levels of AFP and CK19 were found to be significantly higher in GS5 tumors than in Huh7.5 tumors, as shown by Western blotting (Fig. 5C and D) and immunohistochemistry (Fig. 6). The band intensity of AFP in the GS5 tumor lane (Fig. 5D) appears to be extremely high because the lysates prepared from the solid mass of the tumor contain this protein from both intracellular and secreted (in the intercellular spaces) regions. However, the cell culture lysates contain only intracellular AFP (Fig. 1). Interestingly, hepatic progenitor-like cells as marked by the coexpression of AFP with CK19 were observed only in GS5 tumors (Fig. 6D, indicated with arrows).
Fig. 5.
Tumor xenograft of GS5 cells. The HCV subgenomic replicon-expressing cells maintain their distinct phenotypes (enhanced CK19 and AFP expression) in tumor xenografts. (A, B) An example of a GS5 tumor within the animal (A, arrow) and the resected tumor (B) from the animal. The tumors developed by Huh7.5 (not shown) had various sizes but were usually smaller than the GS5 tumors. (C, D) Western blot analysis was performed for the expression of cytokeratin 19 (C, CK19) and α-fetoprotein (D, AFP) in the tumors generated by Huh7.5 and GS5 cells.
Fig. 6.
Coexpression of CK19 and AFP in GS5-derived (B) or Huh7.5-derived (A) tumor xenografts. (A, B) Images show staining for CK19 (brown). (C, D) Similar sections were costained for both CK19 (purple) and AFP (brown). The AFP+ CK19+ double-positive cells represent hepatic progenitor-like cells and are indicated with arrows in panel D. These cells are absent in panel C.
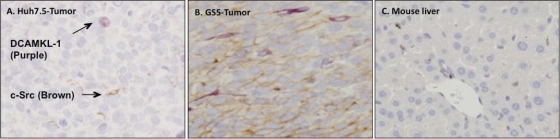
A significant number of DCAMKL-1-positive cells were localized in certain areas of GS5 tumors in which most cells exhibited a high level of activated c-Src expression, indicating that these tumors possess metastatic potential (Fig. 7B). Only a few c-Src- and DCAMKL-1-expressing cells were observed in the Huh7.5 tumors (Fig. 7A), whereas these proteins were not detected in the liver of the same mouse (Fig. 7C). Collectively, these data indicate that both cell types contain phenotypically distinct CSC populations and that their characteristics are most probably preserved in the respective tumors due to self-renewal.
Fig. 7.
(A, B) Costaining by immunohistochemical methods of DCAMKL-1 and active c-Src in the tumor tissues initiated by Huh7.5 (A) and GS5 (B) cells. Purple, DCAMKL-1; brown, active c-Src (pTyr418 c-Src). (C) Control, stained liver section of a mouse with a GS5 tumor as shown in Fig. 5A, showing that both antibodies were not reactive to the animal liver tissues.
Overexpression of DCAMKL-1 and CK19 in the livers of HCV-positive patients.
Liver biopsy samples from five HCV RNA-positive patients were examined for the expression of DCAMKL-1 and CK-19. The liver tissues from patient 1 (Pt1), without cirrhosis (Fig. 8, sample 1), showed sporadic DCAMKL-1+ cells mostly in the area of inflammation, whereas CK19 staining was observed both in sporadic cells and in the disorganized group of cells. Sample 2 (Fig. 8, bottom) represents a liver biopsy specimen from the same patient (Pt1) taken 2 years after the first biopsy. At this stage, the patient was nonresponsive to the standard antiviral treatment and had persistent HCV infection with extensive cirrhosis (stage 4). The dysplastic regenerative nodules in the specimen showed intense staining for DCAMKL-1 and large numbers of DCAMKL-1+ cells in both apical and stem regions. Interestingly, the nodules also showed massive expression of CK19. Other patients, Pt2 through Pt5 (Fig. 9), were also positive for HCV infection and had mild to moderate inflammation and/or cirrhosis (early stage 1 to 2). These patients showed sporadic DCAMKL-1+ cells and intense staining for CK19 in the cells that are not part of the biliary tract. Primary human hepatocytes purchased from BD Bioscience and maintained in Hepato-STIM medium (BD Bioscience) on collagen I-coated coverslips did not exhibit staining for DCAMKL-1 (not shown).
To further investigate the status of DCAMKL-1 in various liver diseases, we used liver tissue arrays that represented 80 human liver specimens representing various liver diseases (inflammation, HBV infection, and cirrhosis) or normal liver tissues (details in Materials and Methods). The analysis of these specimens suggested that as the liver progresses on a path from normal (Fig. 10A, left) to the presence of inflammation and further advances to cirrhosis, DCAMKL-1 expression tends to increase in the hepatocytes (Fig. 10B). Its expression also increases in bile ducts and the stroma during cirrhosis.
Fig. 10.
DCAMKL-1 staining results of the human liver tissue array. The array was processed for immunohistochemical staining. (A) Examples of DCAMKL-1 intensities in different liver diseases. (B) Evaluation of DCAMKL-1 intensities in different cell populations on the array on an arbitrary scale of 0 to 3. Error bars represent standard errors of the means.
DCAMKL-1 affects the abundance of HCV RNA and NS5B polymerase.
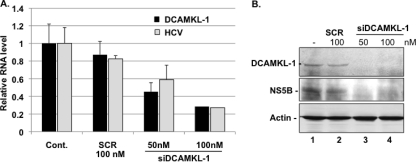
HCV replication complexes associate with microtubule MTFs and use the MTF tracks for their movement and transport (19, 44, 54). Since DCAMKL-1 protein is known to bind, stabilize, and polymerize tubulins into MTFs (24), we examined the effect of DCAMKL-1 on the HCV replicon expression. GS5 cells were transfected with scrambled siRNA (SCR; control) or siRNA against DCAMKL-1 (siDCAMKL-1) (48). This siRNA targets nt 308 to 327 (the predicted site) in the DCAMKL-1 mRNAs encoding both the AL (long form) and AS (short form) isoforms (9). As shown in Fig. 11A, siDCAMKL-1 transfection in GS5 cells resulted in the inhibition (75% on average at 100 nM) of DCAMKL-1 mRNA, whereas the same concentration of the SCR had minimal effect. This decrease in the mRNA was accompanied by the disappearance of the DCAMKL-1 protein in total lysates, suggesting that the siRNA had effectively inhibited DCAMKL-1 expression (Fig. 11B, top, lanes 3 and 4). The DCAMKL-1 depletion in these cells was also accompanied by a significant decrease (70 to 80%) of both the HCV RNA (Fig. 11A) and NS5B polymerase levels (11B, middle, lanes 3 and 4). The MTT assay results suggested that siRNA transfection did not affect cell proliferation under these experimental conditions (not shown).
Fig. 11.
siRNA against DCAMKL-1 reduces the expression of the HCV replicon in GS5 cells. (A) Inhibition of the HCV RNA abundance by siRNA against DCAMKL-1 (siDCAMKL-1). siDCAMKL-1 (50 nM and 100 nM) or scrambled siRNA (SCR, 100 nM) were transfected into GS5 cells in triplicates. Total RNAs were extracted and subjected to real-time reverse transcription-PCR. The levels of HCV and DCAMKL-1 RNAs in control untransfected GS5 cells (Cont.) were set as 1 (or 100%). Actin mRNA in each sample was used as internal control for the PCR. The transfections were repeated twice to confirm the results. Error bars represent standard errors of the means. (B) Western blot analysis of cell lysates from siDCAMKL-1-transfected (lanes 3, 4) or untransfected (lane 1) cells as indicated. Lane 2, sample was transfected with scrambled siRNA (SCR).
DISCUSSION
We have used subgenomic-replicon-expressing cell lines, liver biopsy specimens from patients with chronic HCV infection, liver tissue microarrays, siRNAs, an iPSC expression cassette, and a mouse tumor model to investigate HCV-induced pathological changes in liver cells. Several lines of evidence presented here implicate HCV in the enhanced expression of proteins involved in cellular reprogramming and tumorigenesis. Among the putative stem cell-related proteins, DCAMKL-1 appears to play an important role during HCV-induced liver carcinogenesis. We further demonstrated that curing of the HCV replicon leads to marked reductions in the levels of DCAMKL-1 and reprogramming factors. Similarly, knockdown of DCAMKL-1 results in a considerable decrease in the abundance of HCV RNA. The expression of hepatic progenitor markers in cells supporting HCV replication and DCAMKL-1 overexpression in regenerative cirrhotic nodules of the HCV-positive patients reiterated the association of HCV with DCAMKL-1 expression in chronic infection. Together, the studies presented here advocate in favor of HCV-induced acquisition of CSC traits in liver cells during chronic infection.
Our studies suggest a novel role of DCAMKL-1 in maintaining HCV RNA abundance. The basal expression level of DCAMKL-1 is found to be extremely low or absent in normal hepatocytes. However, active HCV replication, the expression of pluripotency factors, and inflammation and cirrhosis of the liver contribute to its elevated expression. Visualization of DCAMKL-1 by confocal microscopy suggests that this protein is highly enriched in the perinuclear region where colocalization of NS5A-GFP with MTFs is prevalent. While DCAMKL-1 may or may not be directly in contact with replication complexes, our siRNA-based experiments showed that it exerts profound influence on the abundance of both viral RNA and NS5B polymerase. MTFs are polarized polymers of α- and β-tubulin heterodimers that undergo phasic polymerization and depolymerization, and this process is required for cellular transport and cell division (44). It is conceivable that DCAMKL-1, by virtue of its MTF polymerizing and stabilizing activity, may provide microtubule (MT)-dependent transport and fast saltatory movements of replication complexes (RCs) over long distances (14, 15). By promoting RC movement, DCAMKL-1 is likely to increase HCV's replication efficiency. Such a pleiotropic effect may also compensate for architectural distortion created by HCV-induced membranous weblike structures in the cells. Roohvand et al. (44) demonstrated that dynamic polymerization/depolymerization of MTFs also affects postfusion steps of the HCV life cycle. It remains to be investigated whether DCAMKL-1 affects these steps.
Similar to many solid tumor cell lines, the hepatoma cells used in these studies are expected to contain a subpopulation of CSCs (36). We showed that both GS5 and Huh7.5 cells were able to form solid tumors in athymic nude mice. According to the hierarchical model of cancer, the original phenotypes of CSCs represented in the culture pool must also be maintained in their respective tumors due to their self-renewal. The Western blot and immunohistochemical analyses of tumor xenografts validated this assumption. Each tumor type reflected the original phenotype of the parent culture cells. For example, GS5 cells and their tumor xenografts exhibited higher expression of DCAMKL-1, CK19, and AFP than the control cells or tumors (Fig. 5, 6, and 7). The GS5 tumors also showed very high populations of CK19+ AFP− cells (hepatic stem cell-like cells) and CK19+ AFP+ cells (hepatoblast and transit-amplifying cells [61]). These HCV-induced stem cell-like features were clearly absent in Huh7.5 culture cells and their tumor xenografts. Higher expression of these proteins was also detected in the HCV replicon-expressing FCA4 cell line and the liver tissues obtained from HCV-infected patients. Thus, the retrodifferentiation or acquisition of CSC properties is most likely attributed to HCV rather than mere clonal features of the GS5 and FCA4 cell lines. In fact, overexpression of the reprogramming cassette in the heterogeneous population of Huh7 caused modest increases in both DCAMKL-1 and CK19 within 72 h. A considerable reduction (Fig. 1) in the level of these factors in cured GS5 cells (GS5-C) reiterated this observation. Zhao et al. (62) also observed CK19+ oval cells in HCV-positive liver cirrhosis (47) and coexpression of CK19 and AFP in hepatic progenitor cells.
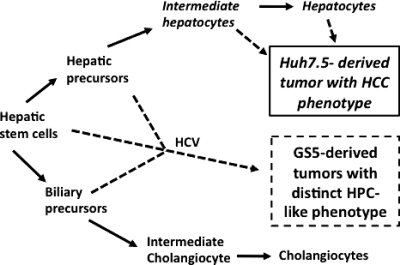
Tsamandas et al. (51) demonstrated that hepatic progenitor cells are frequently present in liver tissues of hepatitis C patients and that their number tends to increase as the disease advances to cirrhosis, a known risk factor for HCC initiation. It was further demonstrated that a large group of patients who do not respond to the standard HCV treatment show higher expression of CK19 and AFP than responders; moreover, these proteins can be used as reliable markers for detecting hepatic progenitor cells in these patients. Using extensive gene expression profiling of HCC tissues, other investigators independently reached the conclusion that HCCs originating from hepatic progenitor cells or hepatic progenitor cell-like cells express high levels of stem cell signatures such as CK19 and AFP (22, 55). Our analyses further provide mechanistic insights that high levels of expression of DCAMKL-1, CK19, and AFP are related to the HCV-induced retrodifferentiation of hepatic cells, leading to the acquisition of a hepatic progenitor cell-like phenotype. We also observed that multiple DCAMKL-1-positive cells were embedded in the areas containing active c-Src in GS5 tumors (Fig. 7). It is known that both the expression and activation of c-Src are enhanced under various stress conditions that favor tumor growth and metastasis (3). In these aspects, the GS5 tumor traits are reminiscent of HCCs with stem cell-like features (8, 22). Thus, it is conceivable that HCV-induced reprogramming of liver cells may contribute to hepatocarcinogenesis (a model is proposed in Fig. 12).
Fig. 12.
Proposed “retrodifferentiation model” for HCV-induced liver carcinogenesis. Solid arrows, pathways of normal hepatic stem cell differentiation; arrows with broken lines, possible sources of Huh7.5- and GS5-derived tumors. The majority of Huh7.5 culture/tumor cells lack or show very low levels of hepatic/biliary precursor markers. Therefore, they represent intermediate or differentiated hepatocytes. On the other hand, GS5 culture/tumor cells contain stem cell/progenitor cell markers and high populations of CK19+ AFP− (hepatic stem cells) and CK19+ AFP+ (hepatoblasts, transit-amplifying cells, or hepatic/biliary precursors) cells. The expression levels of DCAMKL-1, CK19, and AFP in these cells are remarkably higher than those in Huh7.5 cells. These acquired distinctions can probably be attributed to retrodifferentiation of the parent Huh7.5 cells into the GS5 phenotype that was induced by sustained expression of the HCV replicon. HCC, hepatocellular carcinoma; HPC, hepatic progenitor cell.
In conclusion, our data suggest a possible cellular target for the evaluation of small-molecule inhibitors against HCV-induced liver cancer. The analyses of our data revealed an HCV-(DCAMKL-1)-MTF-CSC axis in the hepatoma cell lines. Using both in vitro and in vivo models, our data provide evidence that this collaboration may possibly promote HCV replication and HCV-induced hepatocarcinogenesis during chronic infection.
ACKNOWLEDGMENTS
We thank Stan A. Lightfoot for the evaluation of DCAMKL-1 intensities in liver tissues, Hengli Tang (FSU) for GS5 cells, and the AIM Core (Advanced Immunohistochemistry & Morphology Core Facility at this institution) for immunohistochemistry. We also thank Mark A. Kay and Zhi Ying Chen (Stanford) for the plasmid P2PhiC31-LGNSO.
This work was supported by NIH grants U01DK085508 and R21CA13748, a Veterans Affairs Merit Award, grants AR101-030 (Oklahoma Center for the Advancement of Science and Technology) to C.W.H. and NCI R01CA135559 to S.A., and start-up funds from OUHSC to S.U. H.A. is supported by a student fellowship from the Egyptian Government.
Footnotes
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Ali N., Tardif K. D., Siddiqui A. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001–12007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alison M. R. 2005. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 1:253–260 [DOI] [PubMed] [Google Scholar]
- 3. Allam H., Ali N. 2010. Initiation factor eIF2-independent mode of c-Src mRNA translation occurs via an internal ribosome entry site. J. Biol. Chem. 285:5713–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aoi T., et al. 2008. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321:699–702 [DOI] [PubMed] [Google Scholar]
- 5. Bartenschlager R., Cosset F. L., Lohmann V. 2010. Hepatitis C virus replication cycle. J. Hepatol. 53:583–585 [DOI] [PubMed] [Google Scholar]
- 6. Bartosch B., Thimme R., Blum H. E., Zoulim F. 2009. Hepatitis C virus-induced hepatocarcinogenesis. J. Hepatol. 51:810–820 [DOI] [PubMed] [Google Scholar]
- 7. Blight K. J., McKeating J. A., Rice C. M. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding S. J., et al. 2004. From proteomic analysis to clinical significance: overexpression of cytokeratin 19 correlates with hepatocellular carcinoma metastasis. Mol. Cell. Proteomics 3:73–81 [DOI] [PubMed] [Google Scholar]
- 9. Engels B. M., Schouten T. G., van Dullemen J., Gosens I., Vreugdenhil E. 2004. Functional differences between two DCLK splice variants. Brain Res. Mol. Brain Res. 120:103–114 [DOI] [PubMed] [Google Scholar]
- 10. Farazi P. A., DePinho R. A. 2006. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 6:674–687 [DOI] [PubMed] [Google Scholar]
- 11. Fried M. W., et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 12. Fukasawa M. 2010. Cellular lipid droplets and hepatitis C virus life cycle. Biol. Pharm. Bull. 33:355–359 [DOI] [PubMed] [Google Scholar]
- 13. Gale M., Jr., Foy E. M. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939–945 [DOI] [PubMed] [Google Scholar]
- 14. Guo J. T., Bichko V. V., Seeger C. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia F., et al. 2010. A nonviral minicircle vector for deriving human iPS cells. Nat. Methods 7:197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaji K., et al. 2009. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458:771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J. B., et al. 2009. Direct reprogramming of human neural stem cells by OCT4. Nature 461:649–653 [DOI] [PubMed] [Google Scholar]
- 18. Kim M. H., et al. 2003. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat. Struct. Biol. 10:324–333 [DOI] [PubMed] [Google Scholar]
- 19. Lai C. K., Jeng K. S., Machida K., Lai M. M. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lazaro C. A., et al. 2007. Hepatitis C virus replication in transfected and serum-infected cultured human fetal hepatocytes. Am. J. Pathol. 170:478–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee H. C., Kim M., Wands J. R. 2006. Wnt/Frizzled signaling in hepatocellular carcinoma. Front. Biosci. 11:1901–1915 [DOI] [PubMed] [Google Scholar]
- 22. Lee J. S., et al. 2006. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 12:410–416 [DOI] [PubMed] [Google Scholar]
- 23. Levrero M. 2006. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 25:3834–3847 [DOI] [PubMed] [Google Scholar]
- 24. Lin P. T., Gleeson J. G., Corbo J. C., Flanagan L., Walsh C. A. 2000. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J. Neurosci. 20:9152–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindenbach B. D., Rice C. M. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938 [DOI] [PubMed] [Google Scholar]
- 26. Lowry W. E., et al. 2008. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 105:2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manns M. P., et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 28. Marquardt J. U., Factor V. M., Thorgeirsson S. S. 2010. Epigenetic regulation of cancer stem cells in liver cancer: current concepts and clinical implications. J. Hepatol. 53:568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. May R., et al. 2008. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26:630–637 [DOI] [PubMed] [Google Scholar]
- 30. May R., et al. 2009. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells 27:2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. May R., et al. 2010. Identification of a novel putative pancreatic stem/progenitor cell marker DCAMKL-1 in normal mouse pancreas. Am. J. Physiol. Gastrointest. Liver Physiol. 299:G303–G310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mellor J., Holmes E. C., Jarvis L. M., Yap P. L., Simmonds P. 1995. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. The International HCV Collaborative Study Group. J. Gen. Virol. 76(Pt. 10):2493–2507 [DOI] [PubMed] [Google Scholar]
- 33. Mishra L., et al. 2009. Liver stem cells and hepatocellular carcinoma. Hepatology 49:318–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moradpour D., et al. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 36. Nakanishi T., et al. 2010. Side-population cells in luminal-type breast cancer have tumour-initiating cell properties, and are regulated by HER2 expression and signalling. Br. J. Cancer 102:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson H. B., Tang H. 2006. Effect of cell growth on hepatitis C virus (HCV) replication and a mechanism of cell confluence-based inhibition of HCV RNA and protein expression. J. Virol. 80:1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park C. Y., et al. 2009. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J. Hepatol. 51:853–864 [DOI] [PubMed] [Google Scholar]
- 39. Park I. H., et al. 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451:141–146 [DOI] [PubMed] [Google Scholar]
- 40. Parkin D. M. 2001. Global cancer statistics in the year 2000. Lancet Oncol. 2:533–543 [DOI] [PubMed] [Google Scholar]
- 41. Poenisch M., Bartenschlager R. 2010. New insights into structure and replication of the hepatitis C virus and clinical implications. Semin. Liver Dis. 30:333–347 [DOI] [PubMed] [Google Scholar]
- 42. Qu L., Lemon S. M. 2010. Hepatitis A and hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses. Semin. Liver Dis. 30:319–332 [DOI] [PubMed] [Google Scholar]
- 43. Raychoudhuri A., et al. 2010. Hepatitis C virus infection impairs IRF-7 translocation and alpha interferon synthesis in immortalized human hepatocytes. J. Virol. 84:10991–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roohvand F., et al. 2009. Initiation of hepatitis C virus infection requires the dynamic microtubule network: role of the viral nucleocapsid protein. J. Biol. Chem. 284:13778–13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosen H. R. 2011. Clinical practice. Chronic hepatitis C infection. N. Engl. J. Med. 364:2429–2438 [DOI] [PubMed] [Google Scholar]
- 46. Shi G. M., et al. 2008. Identification of side population cells in human hepatocellular carcinoma cell lines with stepwise metastatic potentials. J. Cancer Res. Clin. Oncol. 134:1155–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun C., Jin X. L., Xiao J. C. 2006. Oval cells in hepatitis B virus-positive and hepatitis C virus-positive liver cirrhosis: histological and ultrastructural study. Histopathology 48:546–555 [DOI] [PubMed] [Google Scholar]
- 48. Sureban S. M., et al. 2009. Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology 137:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tardif K. D., Waris G., Siddiqui A. 2005. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 13:159–163 [DOI] [PubMed] [Google Scholar]
- 50. Thompson M. D., Monga S. P. 2007. WNT/beta-catenin signaling in liver health and disease. Hepatology 45:1298–1305 [DOI] [PubMed] [Google Scholar]
- 51. Tsamandas A. C., et al. 2006. Potential role of hepatic progenitor cells expression in cases of chronic hepatitis C and their relation to response to therapy: a clinicopathologic study. Liver Int. 26:817–826 [DOI] [PubMed] [Google Scholar]
- 52. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waris G., Tardif K. D., Siddiqui A. 2002. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem. Pharmacol. 64:1425–1430 [DOI] [PubMed] [Google Scholar]
- 54. Wolk B., Buchele B., Moradpour D., Rice C. M. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Woo H. G., et al. 2010. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 70:3034–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu P. C., et al. 1996. Classification of hepatocellular carcinoma according to hepatocellular and biliary differentiation markers. Clinical and biological implications. Am. J. Pathol. 149:1167–1175 [PMC free article] [PubMed] [Google Scholar]
- 57. Wurmbach E., et al. 2007. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45:938–947 [DOI] [PubMed] [Google Scholar]
- 58. Yang J. D., Roberts L. R. 2010. Epidemiology and management of hepatocellular carcinoma. Infect. Dis. Clin. North Am. 24:899–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang J. D., Roberts L. R. 2010. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol. 7:448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yin L., Velazquez O. C., Liu Z. J. 2010. Notch signaling: emerging molecular targets for cancer therapy. Biochem. Pharmacol. 80:690–701 [DOI] [PubMed] [Google Scholar]
- 61. Zhang L., Theise N., Chua M., Reid L. M. 2008. The stem cell niche of human livers: symmetry between development and regeneration. Hepatology 48:1598–1607 [DOI] [PubMed] [Google Scholar]
- 62. Zhao D., et al. 2009. Derivation and characterization of hepatic progenitor cells from human embryonic stem cells. PLoS One 4:e6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zulehner G., et al. 2010. Nuclear beta-catenin induces an early liver progenitor phenotype in hepatocellular carcinoma and promotes tumor recurrence. Am. J. Pathol. 176:472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]