Abstract
The hemagglutinin-neuraminidase (HN) protein of human parainfluenza viruses (hPIVs) both binds (H) and cleaves (N) oligosaccharides that contain N-acetylneuraminic acid (Neu5Ac). H is thought to correspond to receptor binding and N to receptor-destroying activity. At present, N′s role in infection remains unclear: does it destroy only receptors, or are there other targets? We previously demonstrated that hPIV1 and 3 HNs bind to oligosaccharides containing the motif Neu5Acα2-3Galβ1-4GlcNAc (M. Amonsen, D. F. Smith, R. D. Cummings, and G. M. Air, J. Virol. 81:8341–8345, 2007). In the present study, we tested the binding specificity of hPIV2 on the Consortium for Functional Glycomics' glycan array and found that hPIV2 binds to oligosaccharides containing the same motif. We determined the specificities of N on red blood cells, soluble small-molecule and glycoprotein substrates, and the glycan array and compared them to the specificities of H. hPIV2 and -3, but not hPIV1, cleaved their ligands on red blood cells. hPIV1, -2, and -3 cleaved their NeuAcα2-3 ligands on the glycan array; hPIV2 and -3 also cleaved NeuAcα2-6 ligands bound by influenza A virus. While all three HNs exhibited similar affinities for all cleavable soluble substrates, their activities were 5- to 10-fold higher on small molecules than on glycoproteins. In addition, some soluble glycoproteins were not cleaved, despite containing oligosaccharides that were cleaved on the glycan array. We conclude that the susceptibility of an oligosaccharide substrate to N increases when the substrate is fixed to a surface. These findings suggest that HN may undergo a conformational change that activates N upon receptor binding at a cell surface.
INTRODUCTION
The human parainfluenza viruses (hPIVs) are members of the family Paramyxoviridae. They possess two envelope glycoproteins, the hemagglutinin-neuraminidase (HN) and the fusion protein (F). HN binds (H) to N-acetylneuraminic acid (Neu5Ac, sialic acid) on glycoproteins and glycolipids on the host cell and assists viral entry by activating F (10, 26, 30). HN also removes (N) sialic acid from glycans at low pH and low chloride concentrations (24), possibly intracellularly (24), and is presumed to destroy receptors. N is required for paramyxoviruses to form plaques in cell culture (12, 35), but the reason for this requirement is not established.
Previous research suggests several possibilities for the biological role of N. N may facilitate virion release by eliminating receptors that otherwise cause progeny to remain attached to the host cell. hPIV3 N can remove receptors from cells; UV-inactivated virus has been shown to remove measurable sialic acid from infected CV-1 cells (25). Receptor removal from cells affects virion release. The mutant C-28 strain of hPIV3, which has half the neuraminidase activity of wild-type virus, produced HN and nucleoprotein (NP) at the same rate as wild-type virus, as measured by gel densitometry of radiolabeled protein, but produced fewer PFU per unit time (18). The neuraminidase-dead C28a hPIV3 virus showed an even more marked delay in release (12). These findings suggest that infection and protein production are successful but that release of virus from the cells is delayed in the absence of functional N. N has also been shown to affect the type of infectious course that hPIV will take. Treatment of uninfected cells with small amounts of bacterial or hPIV3 neuraminidase removed sufficient receptor to prevent fusion with persistently infected, HN-expressing cells, and treatment of uninfected cells with larger amounts of neuraminidase prevented infection of the cells entirely. N pushed the virus' life course toward persistent rather than lytic infection by prohibiting viral spread via cell-cell fusion (25). Taken together, these findings suggest that N is capable of removing the same receptors that hPIV uses for initial infection of cells and for cell-cell fusion and that removal of these receptors facilitates viral release. However, if N removes receptors too early in the viral life cycle, no infection or limited infection is the result.
N may also regulate overall HN function. Several studies suggest that HN must be properly glycosylated, but not sialylated, for receptor binding to occur. The low-N revertant L1 of a temperature-sensitive Newcastle disease virus (NDV) mutant incorporates sialic acid into itself, as shown by a periodiate-tritiated borohydride treatment that specifically labels sialic acid. The revertant demonstrated an attachment constant that was half of that of the wild type, but neuraminidase treatment restored the wild-type attachment kinetics (34). Similarly, a neuraminidase-null NDV mutant HN protein expressed on the surface of BHK-21 cells incorporated sialic acid into its glycans and was unable to bind red blood cells (RBCs); binding was restored by treatment with exogenous neuraminidase but not by removal of the glycosylation sites from HN (19). The data suggest that HN must be glycosylated but not sialylated for proper folding, binding site accessibility, and/or tetramer assembly; the purpose of the N activity of HN may be to control this function-critical glycosylation.
N could also modulate the relationship between receptor binding and fusion activation. Cells expressing wild-type hPIV3 HN and F fused with red blood cells at pH 8, but most red blood cells were released at pH 5, where N is active. Cells expressing the C28a N-null HN did not show this pH-dependent change in fusion in a reporter-based cell-cell fusion assay (29). In a human airway epithelial cell model, two mutant hPIV3 viruses with higher-than-wild-type fusogenicity or hemagglutinin (HA) avidity, but without a corresponding increase in N, produced lower viral titers than the wild type. Treatment with exogenous neuraminidase partially restored the titers (27). These studies suggest that N may act to balance receptor binding and fusion triggering by limiting the number of receptors that are available for binding on the cell surface and/or by inducing a change in HN that limits fusion triggering.
Structural studies of HN, interestingly, have not clarified the function of N. The crystal structure of NDV HN in complex with a high concentration of the thiosialoside inhibitor Neu5Ac-2-S-α(2,6)Gal1OMe showed the sialic acid moiety of the inhibitor bound at both the neuraminidase active site (site I) and the dimer interface (site II) (44). In contrast, the structures of hPIV5 (43) and hPIV3 (20) showed sialic acid bound only at the neuraminidase active site. However, sialic acid was successfully computationally modeled into the dimer interface of the H552Q mutant of hPIV3 HN (whose binding is resistant to the N inhibitor zanamivir) (28), suggesting that there may be a second site in this mutant HN. In the structures of NDV at pH 6 or higher and in the structure of hPIV3, the monomers in an HN dimer are rotated such that the site I binding pockets face 90° away from each other (9, 20, 32, 44). A pH 4.5 structure of NDV HN, in contrast, has the two binding sites in a 2-fold symmetric arrangement with their active sites pointed 180° away from each other (9); the dimer is evidently capable of assembling in at least two different ways. Stabilization of the 90°-opposed sites in recombinant NDV HN by introduction of engineered disulfide bonds enhanced fusion activity while decreasing N activity, suggesting that fusion triggering does not require a conformational change in HN (23) but that N activity may. In hPIV3, a conformational change in HN could link the receptor-binding and N functions of site I with the fusion-triggering and, potentially, secondary receptor-binding functions of site II. The existing data leave several questions unanswered. What is the relationship between binding specificity and neuraminidase specificity? In what contexts do binding and neuraminidase activity occur?
To begin to answer these questions, we determined the specificities of both H and N. We previously reported that hPIV1 (C35) bound to structures containing the minimal motif Neu5Acα2-3Galβ1-4GlcNAc, including when the motif is sulfated or with GalNAc on Gal, fucosylated on GlcNAc (sialyl-Lewisx), or attached to an extended polylactosamine or mannose chain. At pH 5, it also binds to some Neu5Acα2-8 and Neu5Acα2-3Galβ1-3 glycans. hPIV3 binds a restricted set of structures containing the same minimal motif with or without fucosylation and requires the addition of at least one additional galactose to the chain for binding. We have now studied the binding of these together with hPIV2 and recent clinical isolates on newer, expanded glycan arrays. We also examined the susceptibilities of substrates to N on the glycan array, red blood cells, and soluble small molecules and glycoproteins. We propose that N activity is facilitated by the fixation of substrates to a surface, perhaps mediated by a conformational change in HN.
MATERIALS AND METHODS
Virus culture and purification.
hPIV1 strain C35 and hPIV3 strain C243 were from ATCC. Clinical isolates Oklahoma/4409/2010 (hPIV1), Oklahoma/3955/2005 (hPIV2), Oklahoma/94/2009 (hPIV2), Oklahoma/283/2009 (hPIV2), Oklahoma/410/2009 (hPIV3), and Oklahoma/754/2009 (hPIV3) were obtained from the virology laboratory at the University of Oklahoma Children's Hospital. LLC-MK2 (MK2) cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco) with 10% supplemented calf serum (HyClone). MDCK cells were maintained in DMEM supplemented with 1% glutamine (200 mM; Gibco), 2% amino acids (50×; Gibco), 1% vitamins (MEM, 100×; Gibco), and 3% sodium bicarbonate (7.5%; Gibco). All hPIV strains were propagated in MK2 cells in DMEM/F12 infection medium with 1% ITS+ as previously described (4). Trypsin (tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK] treated; Worthington) was added at 1 μg/ml to the infection medium for hPIV2 as for hPIV1. Influenza A virus strains A/Oklahoma/447/2008 (seasonal H1N1), A/Oklahoma/2952/2009 (2009 pandemic H1N1), and A/Oklahoma/483/2008 (H3N2) were propagated in MDCK cells in the same infection medium but with 0.5 μg/ml trypsin. Viruses were purified as previously described (4).
Determination of ratios of viral proteins using amino acid analysis.
Three samples of purified virus were subjected to 9% sodium dodecyl sulfate-polyacryl-amide gel electrophoresis (SDS-PAGE) and then blotted onto a polyvinylidene diflu-oride (PVDF) membrane. Blots were stained with Coomassie blue (from concentrate; Sigma). All visible bands were excised and quantified by amino acid analysis at the University of Oklahoma Molecular Biology Proteomics Facility, which also confirmed their identity. For hPIV2, the identities of bands were determined by mass spectrometry prior to amino acid analysis. The amount of each protein in a sample, and therefore the ratios of proteins to each other, was determined using the following formula: mol protein = mol amino acid in sample/(mol amino acid/mol protein). The result was averaged for amino acids whose prevalence in the sample was close to the expected value; the set of amino acids chosen was different for each sample, but Ala, Met, Gly, Ser, and Thr were avoided. The percentage of total viral protein contributed by each component was calculated as the amount of each protein divided by the sum of the five gel bands.
The P and F bands of hPIV2 could not be separated by gel electrophoresis. Therefore, these two bands were calculated together, and the proportions of P and F were derived from the average P/F ratio for hPIV1 and -3.
Determination of total viral protein by gel electrophoresis.
Virus was subjected to 9% SDS-PAGE followed by Coomassie blue staining and visual comparison of the NP band intensity to known amounts of bovine serum albumin (BSA) (Pierce). The amount of NP in each sample was multiplied by the percentage of total viral protein that is NP to yield the total viral protein. The amount of HN in a sample was calculated by multiplying the total viral protein by the percentage that is HN.
Sequencing of HN.
RNA was extracted from purified virus using the QIAamp viral RNA minikit (Qiagen). Reverse transcriptase PCR was performed using the OmniScript RT kit (Qiagen) and the HN forward primers for each virus as described below. PCR was performed using the Platinum Taq DNA polymerase kit (Invitrogen). PCR of the HN gene used the following primers: hPIV1 forward, 5′-AACGATGGCTGAAAAAGGG-3′; hPIV1 reverse, 5′-CTATTTGTCATATAAATGTCTATTC-3′; hPIV2 forward, 5′-CAGCTTAATCCACTCAACATATAA-3′; hPIV2 reverse, 5′-AGCCAAAACGTATAACTATTGCAT-3′; hPIV3 forward, 5′-ATGGAATACTGGAAGCACAC-3′; and hPIV3 reverse, 5′-TTAACTGCAGCTTTTTGGAA-3′. PCR was performed in an MJ Research programmable thermal controller using the following program: 94°C for 5 min; 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; and 72°C for 5 min. Sequencing was performed at the Oklahoma Medical Research Foundation Sequencing Core Facility. Additional primers used for sequencing were as follows: hPIV1 Fwd 451, 5′-AGGAATTGGCTCAGATATGC-3′; hPIV1 Fwd 1024, 5′-AGTGTAGGAAGTGGGATAAAG-3′; hPIV1 Fwd 1463, 5′-TCCGAGAGAATGCATATCAG-3′; hPIV1 Rev 1527, 5′-TACAGTGTGGTTGTAGCAAC-3′; hPIV1 Rev 1081, 5′-CCTTGGAGCGGAGTTGTTA-3′; hPIV1 Rev 608, 5′-GTGGTTGATCCAGAAAGTAGA-3′; hPIV2 Fwd 496, 5′-CATACAATGGGACGCCTAAATATG-3′; hPIV2 Fwd 1045, 5′-AGCAGTCCTCACGCTATTTTAT-3′; hPIV2 Fwd 1452, 5′-TGCCCTGCTAATTGCATCA-3′; hPIV2 Rev 545, 5′-GAGATGTTGCTGAGGGGATA-3′; hPIV2 Rev 968, 5′-CCTAGATGATAGATCCCGCTT-3′; hPIV2 Rev 1382, 5′-GGTACCCATTGAGCCTCA-3′; hPIV3 Fwd 488, 5′-CCACAAAGAATAACACATGATG-3′; hPIV3 Fwd 959, 5′-GATGGTTCAATCTCAACAACA-3′; hPIV3 Fwd 1457, 5′-GAATGTCCATGGGGACATTC-3′; hPIV3 Rev 482, 5′-TGTTATTCTTTGTGGCAGCAC-3′; hPIV3 Rev 843, 5′-TGTATTTAGGAGTGCTAGAG-3′; and hPIV3 Rev 1353, 5′-ATTGTAACTTGCTATGCCAAC-3′. Completed sequences were analyzed for their phylogenetic relationships to each other and to GenBank sequences for hPIV1, hPIV2, and hPIV3 using MultAlin (8) and CLUSTALW with PHYLIP (39) using default settings.
Neuraminidase assay and determination of neuraminidase activity on soluble substrates.
2′-(4-Methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUN), 3′-N-acetylneuraminyllactose (3′NANL), human α1-acid glycoprotein (hAGP), human transferrin, bovine submaxillary mucin, hen ovomucoid, and human glycophorin A were from Sigma. Gangliosides GM1, GM2, and GM3 were from Calbiochem. Bovine fetuin (bFetuin) was purified from fetal calf serum (Sigma) by the method of Spiro (36). The sialic acid concentration in the substrates was measured as amount released after overnight incubation with excess hPIV1 and -2 virus mixture (cleavable sialic acid) or Arthrobacter ureafaciens (Sigma) or Micromonospora viridifaciens neuraminidase (total sialic acid). A proteinase K digest of bFetuin (fetuin peptides) was prepared by incubating a solution of 40 mg/ml bFetuin and 3 to 4 mg/ml proteinase K (Invitrogen) in neuraminidase buffer (50 mM Na acetate, 4 mM CaCl2, pH 5.5) at 50°C for 2 h.
Neuraminidase activity on MUN was assayed by the fluorescence method of Potier et al. (31) adapted for 96-well plates. Activity on all other substrates was assayed by the thiobarbituric acid assay of Warren (41) with the following modifications for a 96 well plate: Na2SO4 and H2SO4 were omitted from the arsenite reagent, H2SO4 was omitted from the thiobarb reagent, the plate was heated on a dry heating block at 85°C, and dimethyl sulfoxide (DMSO) was used to stabilize the color in place of the butanol extraction. Virus, buffer, and substrate were mixed to a starting volume of 30 μl in a 96-well flat-bottom enzyme-linked immunosorbent assay (ELISA) plate (Microlon; Greiner Bio-One). The reaction mixture was incubated at 37°C for 10 min (for enzyme activity analysis) or 60 min (for determination of substrate susceptibility). Development reagents were added as in Warren's assay, using 5 μl periodate, 50 μl arsenite, and 100 μl thiobarb. After heating (85°C for 20 min), 50 μl DMSO was added. If necessary, protein precipitates were spun down for 5 min at 2,000 rpm in a GS-6R (Beckman) tabletop centrifuge, and then 150 μl supernatant was transferred to a new plate. The plate was read at 550 nm in a SpectraMax M2 microplate reader using SoftMax Pro software. For glycoprotein substrates, the initial 37°C incubation was followed by deactivation and denaturation of the neuraminidase at 80°C for 5 min and then addition of 10 μl of 3 to 4 mg/ml proteinase K and incubation at 50°C for 20 min to avoid a large protein precipitate. Periodate was then added and the procedure continued as described above.
Enzyme activity assays were set up such that all wells contained the same amount of viral protein and a variable amount of the substrate to be tested. A substrate-only blank was included for each substrate concentration. A 4-methylumbelliferone (Sigma) or N-acetylneuraminic acid (Sigma) standard curve was included as appropriate for each assay.
The enzyme activity parameters Km (Michaelis-Menten constant) and Vmax (maximum initial velocity, used to derive the specific activity [in μmol sialic acid released per min per mg total viral protein]) were calculated using Prism 4 software with the nonlinear regression equation “Michaelis-Menten.” kcat was calculated as follows: kcat (s−1) = Vmax (μmol/s)/HN in sample (μmol).
Hemagglutination assays and determination of neuraminidase activity on red blood cells.
Human blood testing samples were obtained from the Oklahoma Blood Institute. Whole chicken and turkey blood were from Lampire (Pipersville, PA). Red blood cells (RBCs) were washed four times with cold phosphate-buffered saline (PBS) to remove serum and then suspended to ∼50% in Alsever's solution (Sigma). Human red blood cells were made to a final concentration of 0.8% and avian cells to a concentration of 0.5% in PBS or acetate buffer (100 mM NaCH3COOH, 2 mM CaCl2, 2 mM MgCl2, pH 5.5) as needed. Viruses were serially diluted in 50 μl PBS or acetate buffer in a round-bottom microtiter plate, and then 50 μl red blood cells was added. The assay mixture was incubated at 4°C for 30 to 60 min and the titer read. To test for neuraminidase action on red blood cells, the initial binding was performed in acetate buffer. The red blood cells were then allowed to elute at room temperature. Cells were gently resuspended by pipetting and the plate reincubated at 4°C for 30 to 60 min. In the event that no rebinding was observed, an amount of purified virus sufficient for agglutination was added to each well of the assay plate, the cells resuspended by pipetting, and the plate incubated at 4°C for 30 to 60 min. Hemagglutination inhibition assays were performed by incubating 4 hemagglutinating units (HAU) of virus with 2-fold dilutions of glycoprotein (see materials listed in “Neuraminidase assay and determination of neuraminidase activity on soluble substrates” above) for 30 min at 4°C and then adding red blood cells and incubating as described above.
Fluorescent labeling of viruses and glycan array analysis.
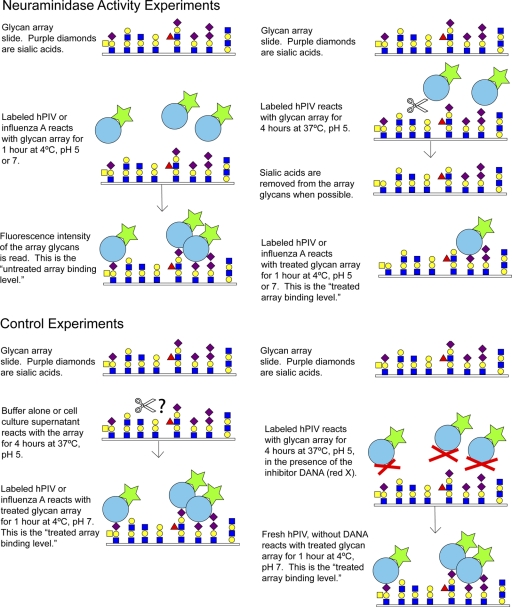
Purified virus was labeled with Alexa Fluor 488 (Molecular Probes) and bound to the glycan array as previously described (4). Viruses sent to the array had hemagglutinin titers on the order of 104 HAU/ml and total protein of 0.5 to 1 mg/ml and were diluted 5- to 200-fold depending on labeling efficiency. Three concentrations in this dilution range were tested for each virus. High-affinity ligands were defined as those glycans bound at >10% of the level for the highest-binding species at all concentrations tested. For neuraminidase specificity studies, Alexa 488-labeled virus was diluted in acetate buffer with 1% BSA and 0.05% Tween 20 at pH 5.5 and incubated with the glycan array for 1 to 4 h at 37°C to allow neuraminidase to cleave structures on the array. The temperature was then lowered to 4°C and labeled virus was allowed to bind to the array. The slide was dried and read. In some experiments, the original virus was washed off and fresh virus added at pH 7 and 4°C and left for 1 h. A structure was considered to have been cleaved if its binding after N treatment was less than 20% of its original binding level or to have been partially cleaved if its binding after N activity was between 20% and 60% of its original binding level. For inhibition experiments, 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (DANA) (Sigma) was used to block neuraminidase activity. For some samples, Alexa-labeled influenza A virus was subsequently bound to the same array.
Use of photographic processing software in construction of figures.
Adobe Photoshop CS3 was used to annotate the sequence alignment in Fig. 1, to annotate the subgroup labels in Fig. 2, to resize and move text labels for improved legibility in Fig. 2, and to crop three wells for presentation from each 8- or 12-well hemagglutination assay in Fig. 4. In all other figures, processing software was used only for assembly of data into a single image file.
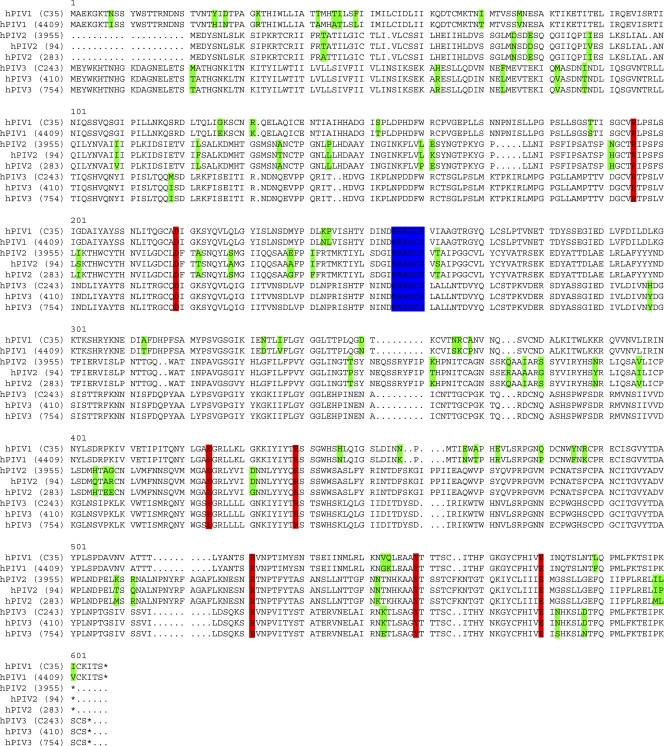
Fig. 1.
Protein sequence alignment of the hPIV1, -2, and -3 strains used in this study and described in Materials and Methods. Residues highlighted in red make contacts with sialic acid in the active site (7). Residues highlighted in blue are residues that have demonstrated structural importance; these positions did not change in a panel of escape mutants, and intentional mutation of these residues in a recombinant HN eliminated both H and N activity (18, 20). Residues in green are differences between strains of a single hPIV. Note that no changes occur in active-site residues or in structural components from strain to strain.
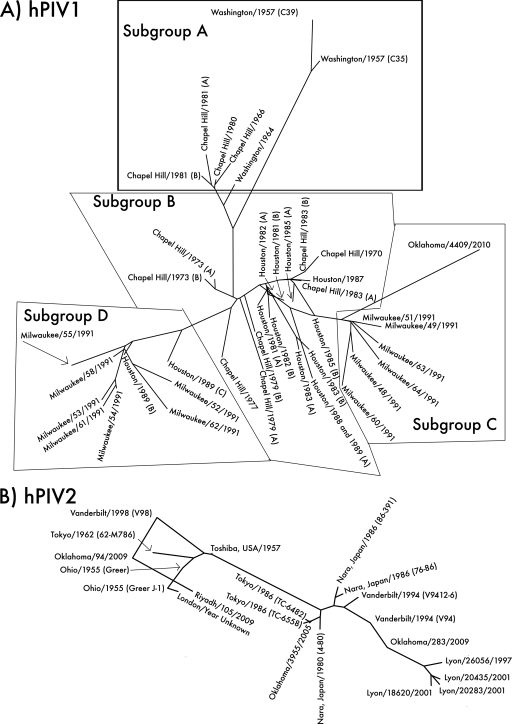
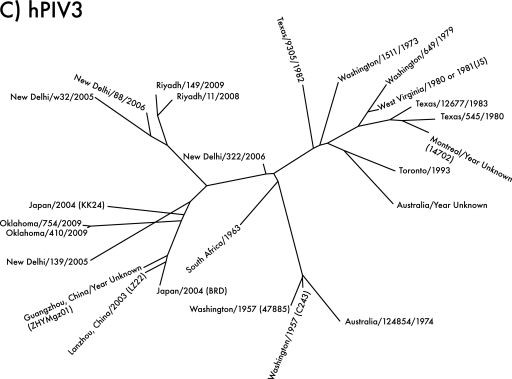
Fig. 2.
Unrooted phylogenetic trees of hPIV1, -2, and -3, based on sequences of type strains and local clinical isolates and clinical isolate sequences from GenBank. (A) Phylogeny of hPIV1. Local isolate Oklahoma/4409/2010 (far right) shares 93.7% amino acid sequence identity with the type strain Washington/1957 (C35) (top) and groups with the Milwaukee/1991 isolates previously identified as belonging to antigenic subgroup C (15). Assignment of sequences to subgroups is based on the Chapel Hill and Milwaukee isolates (14, 15). (B) Phylogeny of hPIV2. Local isolates Oklahoma/3955/2005, Oklahoma/94/2009, and Oklahoma/283/2009 are 94 to 96% identical in amino acid sequence to the type strain Ohio/1955 (Greer). Each Oklahoma isolate represents a different lineage, despite having originated in the same hospital over the course of only 4 years. hPIV2 strains tend to group geographically; note that the four Lyon isolates (lower right) group together, as do most of the isolates from Japan (center). However, closely related strains can also be isolated in different places at different times; for example, Ohio/1955, Tokyo/1962, Vanderbilt/1998, and Oklahoma/94/2009 share a branch of the tree. (C) Phylogeny of hPIV3. Local isolates Oklahoma/410/2009 and Oklahoma/754/2009 are 97 to 98% identical in amino acid sequence to the type strain Washington/1957 (C243). hPIV3 strains appear to group both temporally and geographically; strains from the New World and Australia before 1995 form one major branch of the tree (right), while strains from Asia after 1995 form the other (left). The Oklahoma/2009 isolates may be Asian in origin.
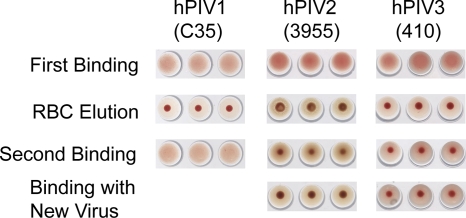
Fig. 4.
N action on chicken (hPIV1 and -2) or turkey (hPIV3) red blood cells. All assays were conducted in pH 5 acetate buffer. hPIV1 elutes from RBCs but is able to rebind them, indicating that elution is due to Brownian motion at higher temperature. hPIV2 and hPIV3 do not rebind RBCs after elution, and addition of new virus does not restore binding, indicating that receptors have been removed by N activity. The three wells shown represent three 2-fold dilutions of virus.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new HN sequences are as follows: Oklahoma/4409/2010 (hPIV1), JN089925; Oklahoma/3955/2005 (hPIV2), JF912196; Oklahoma/94/2009 (hPIV2), JF912194; Oklahoma/283/2009 (hPIV2), JF912195; Oklahoma/410/2009 (hPIV3), JF912197; Oklahoma/754/2009 (hPIV3), JF912198; and Washington/1957 C243 (hPIV3), JN089924.
RESULTS
Sequencing and phylogeny of hPIV clinical isolates.
The HN protein of human parainfluenza viruses is the primary antigenic target for both humoral and T-cell responses (7, 38). HN can therefore be expected to change over time as hPIV responds to selective pressure. The prototype strains of hPIV were isolated in the 1950s, and due to their age they may not accurately represent the properties of current hPIVs. In addition to the ATCC type strains hPIV1 C35 and hPIV3 C243, we obtained local clinical isolates as examples of currently circulating hPIV strains.
Sequence analysis of local isolates showed that active-site residues and residues thought to be structurally important are conserved (19, 21) (Fig. 1). In accord with this observation, we found that the enzyme activities and red blood cell specificities of hPIV2 and -3 clinical isolates are essentially identical to those of the type strains and that their glycan array binding profiles are a close although not exact match (see below). hPIV1 strain 4409 grew poorly in MK2 cells and therefore was not tested for enzyme activity and glycan array binding. Henceforth, “hPIV2” and “hPIV3” refer to the aggregate behavior of the strains used.
The phylogeny of local clinical isolates compared to sequences in GenBank is shown in Fig. 2.
Activity of neuraminidase on soluble substrates.
The biological function of N presumably requires a precisely calibrated enzyme activity; HN must destroy enough of its target in the available time to facilitate virion release, remove sialic acids from other HN molecules, or modulate binding strength without altering attachment to new cells (18, 19, 25, 27). The activity of N, therefore, is expected to reflect its role in the life cycle of hPIV. In order to determine the efficiency of neuraminidase activity on a variety of structures, we tested the enzyme activities of hPIV1, -2, and -3 on soluble substrates. We tested small molecules, which are expected to be easily cleavable by N without concern for any effects due to substrate size or shape, and glycoproteins, whose complex carbohydrate structures are expected to more closely resemble those of the glycans that hPIV might encounter on host cells.
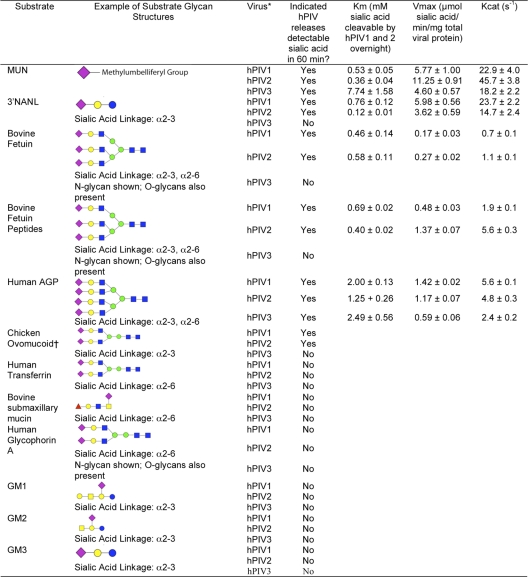
We first identified potential substrates by testing the ability of hPIVs to release sialic acid from a panel of small molecules and commercially available glycoproteins. hPIV1 and -2 released quantifiable amounts of sialic acid from MUN, 3′NANL, bovine fetuin (bFetuin), human AGP (hAGP), and chicken ovomucoid during a 1-hour incubation. hPIV3 released measurable sialic acid from MUN and hAGP (Table 1). None of the viruses tested released detectable sialic acid from 6′NANL, human transferrin, bovine submaxillary mucin, human glycophorin A, or ganglioside GM1, GM2, or GM3.
Table 1.
Enzyme activity parameters of hPIVs on soluble substratesa
Symbols: purple diamonds, N-acetylneuraminic acid; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose; red triangles, fucose; blue circles, glucose; yellow squares, N-acetylgalactosamine. *, measurements were made using strains C35 (hPIV1), Oklahoma/3955/2005 (hPIV2), and C243 (hPIV3). Clinical isolates Oklahoma/283/2009 (hPIV2), Oklahoma/94/2009 (hPIV2), Oklahoma/754/2009 (hPIV3), and Oklahoma/410/2009 (hPIV3) gave results similar to the values for hPIV2 and -3 values reported here. †, chicken ovomucoid was not selected for kinetics analysis due to low solubility and sialic acid content.
Based on these findings, we selected MUN, 3′NANL, bFetuin, and hAGP as substrates and quantified N activity using a Michaelis-Menten assay. MUN and 3′NANL are small molecules; bFetuin and hAGP are 40- to 50-kDa glycoproteins that contain sialylated glycans with up to three (bFetuin) or four (hAGP) branches on each N-linked glycan (11, 16). A proteinase K digest of bovine fetuin (bFetuin peptides) was also included as a test for the effect of protein size or folding on neuraminidase susceptibility. Chicken ovomucoid was cleaved by N, but its low sialic acid content and lower water solubility did not allow us to achieve a sufficient concentration in the assay to measure Km and Vmax.
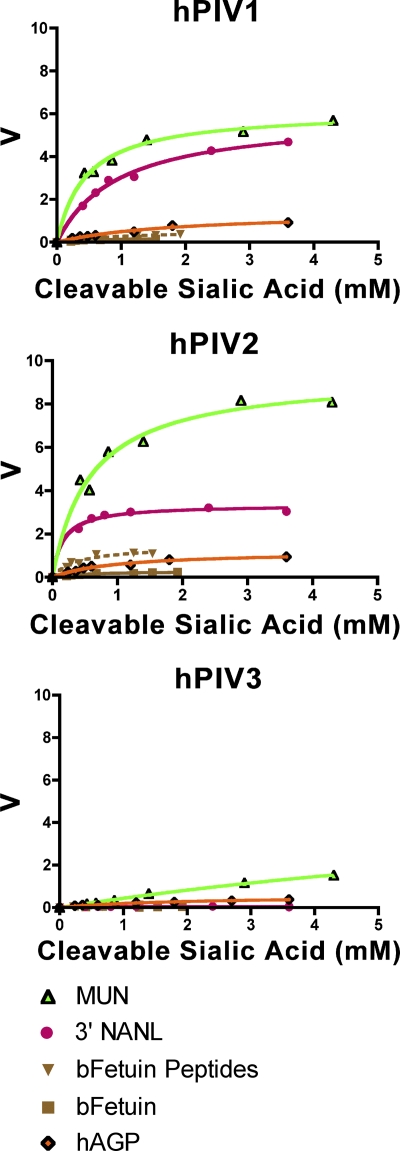
hPIV1 and -2 cleaved all five susceptible substrates with various efficiencies (Fig. 3; Table 1). The Michaelis-Menten constant (Km), often considered a measure of the affinity of the enzyme for substrate, ranged from 0.5 to 2.0 mM for the various substrates. However, the reaction rate per milligram of viral protein (specific activity derived from Vmax) and turnover rate (kcat) were at least 2-fold higher on small-molecule substrates than on glycoproteins or glycopeptides. hPIV3 cleaved only MUN and hAGP to detectable levels. On MUN, hPIV3 had a 10-fold lower affinity and at least a 2-fold lower activity and turnover rate than hPIV1 and -2; on hAGP, its kinetic parameters were similar to those of hPIV1 and -2. The activity of hPIV3 on MUN was 3-fold higher than its activity on AGP. hPIV3 has previously been shown to have lower relative activity than hPIV1 or -2 when allowed to cleave sialyllactose over a period of hours (1). Our results demonstrate that this pattern holds for other substrates and quantify the activities of all three viruses (Table 1).
Fig. 3.
Enzyme activities of hPIV1, -2, and -3 HN (as whole virus) on soluble substrates. Examples of Michaelis-Menten curves for each virus are shown; results of one representative experiment are shown for each substrate. V is measured as μmol sialic acid released/min/mg total viral protein. Note that curves for small molecules (MUN and/or NANL) exhibit higher Vmax than those for glycoproteins or peptides for all three viruses. The maximum sialic acid concentration was limited by substrate solubility for glycoproteins.
To control for the possibility that the low N susceptibility of glycoproteins is the result of the protein itself, we digested bFetuin with proteinase K. Digestion of bFetuin increased the Vmax of hPIV1 and -2 by 3- to 5-fold, but even after digestion the Vmax was only 10 to 30% of that with 3′NANL or 10% of that with MUN; protease digestion did not affect the activity of hPIV3 on bFetuin (Fig. 3; Table 1).
To see if the glycoprotein substrates are less efficiently cleaved as a result of failure of the viruses to bind them, we tested the abilities of hAGP and bFetuin to act as hemagglutination inhibitors. Both proteins were able to inhibit hemagglutination by all three viruses (Table 2), with inhibitory concentrations ranging from 2-fold to 10-fold lower than the Km values.
Table 2.
Hemagglutination inhibition of hPIVs by substrate proteins
| Virus | Substrate | Inhibitory concn (mM sialic acid) |
|---|---|---|
| hPIV1 | AGP | 0.04 |
| Fetuin | 0.09 | |
| hPIV2 | AGP | 0.14 |
| Fetuin | 0.19 | |
| hPIV3 | AGP | 0.16 |
| Fetuin | 0.19 |
To test whether activity increased when glycoproteins were fixed to a surface, we bound hAGP and bFetuin to an ELISA plate. The thiobarbituric acid assay could not detect sialic acid released from the small amount of protein that bound the well, even if glycoproteins were digested overnight with a bacterial neuraminidase.
Composition of hPIV.
To calculate the Vmax and kcat of HN, it is necessary to know the concentration of HN in a given sample. We determined the proportion of viral protein that is HN using amino acid analysis, as described in Materials and Methods. Five proteins (P, HN, NP, F, and M) were detectable by 9% SDS-PAGE with Coomassie blue staining. Amino acid quantification showed HN monomer to comprise 16.2% ± 7.7%, 23.9% ± 0.6, and 23.2% ± 3.6% (wt/wt) of the total protein of hPIV1, -2, and -3, respectively, measured as the sum of the 5 protein gel bands. Bousse and Takimoto estimated from gel densitometry that HN constitutes 19% of hPIV1, 18% of hPIV2, and 28% of hPIV3 (5); while our findings are somewhat higher for hPIV2 and lower for hPIV3, both sets of measurements suggest that any given hPIV is about 20% HN. hPIV1 exhibited an HN/F ratio of 1.7 ± 0.9. hPIV2 had the highest HN/F ratio of the three virus types tested at 5.0 ± 1.1, and hPIV3 had an HN/F ratio of 2.0 ± 0.5. NP makes up 43.3% ± 6.0% of hPIV1, 43.2% ± 6.9% of hPIV2, and 33.5% ± 5.9% of hPIV3 by mass.
Effect of neuraminidase on red blood cells.
Two of the three proposed functions of HN, removal of receptors to facilitate virion release and modulation of binding and fusion, require N to destroy targets on cell membranes. We used RBCs, which have sialylated glycans that are ligands for HN binding, as a model system to determine the susceptibility of cell surface ligands to N. Our solution assays demonstrate that hPIVs prefer small molecules to glycoproteins or glycolipids as N substrates in solution, so we investigated the ability of N to cleave glycoproteins or glycolipids in the context of a cell membrane. We previously reported that hPIV1 rebinds eluted chicken RBCs at both pH 7 and pH 5, while hPIV3 rebinds eluted human or guinea pig RBCs at pH 7 but not at pH 5 (4). We expanded this experiment and included hPIV2.
N is active at pH 5 at temperatures from 25 to 50°C (24), but there is no detectable activity at 4°C (data not shown). Under conditions not conducive to N (pH 5, 4°C), hPIV1 and -2 bound chicken RBCs, and hPIV3 bound turkey RBCs. Raising the temperature of the assay to 25°C caused complete elution of bound RBCs from all three viruses. Elution can be caused by either disruption of weak interactions by increased Brownian motion or receptor destruction by N. RBCs eluted as the result of a temperature effect maintain intact glycans and can be re-bound by virus, but RBCs eluted as the result of neuraminidase activity have their receptors destroyed and cannot be rebound. hPIV1 rebound RBCs upon resuspension of the red cells followed by a second incubation at 4°C, even if the assay mixture was allowed to remain at an N-permissive temperature overnight; RBCs therefore elute from hPIV1 due to temperature effects. hPIV2 and -3 did not rebind RBCs. Addition of fresh virus did not restore binding, indicating that RBC receptors had been destroyed and that the lack of binding was not the result of loss of viral H activity due to assay conditions (Fig. 4).
Binding profiles of HN.
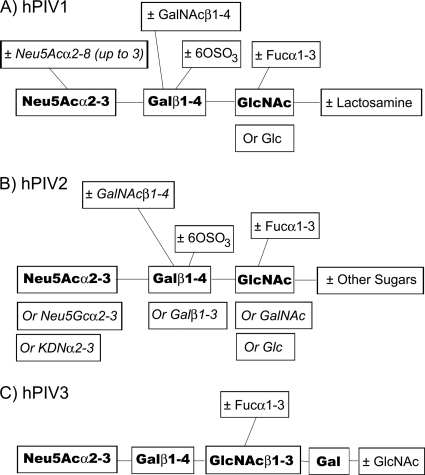
In order to assemble a more complete picture of the potential receptor structures of the hPIVs, we extended binding experiments done with the Consortium for Functional Glycomics' glycan array v. 2.0 (4) to the expanded arrays v. 3.2 (406 glycans), 4.0 (442 glycans), 4.1 (465 glycans), and/or 4.2 (511 glycans) at both pH 5 and pH 7. The binding motifs of hPIV1 and -3 remained as reported earlier (4). On the newer arrays, hPIV1, but not hPIV3, recognized its binding motif in the context of a glycan with two branches; the array did not contain any tri- or tetra-antennary structures. hPIV2 recognized the same minimal binding motif, Neu5Acα2-3Galβ1-4GlcNAc, as hPIV1 (Fig. 5). Like hPIV1, hPIV2 bound fucosylated (sialyl-Lewisx) or sulfated motifs; unlike hPIV1, it did not bind to structures with GalNAc. At pH 5, hPIV2 bound the glycan array with considerably broader specificity; it accepted structures with Galβ1-3 as the second sugar, Neu5Gcα2-3 or KDNα2-3 as the first sugar, and GalNAc as the third sugar. Binding motifs did not differ significantly between high and low virus concentrations. Some structures, such as glycans with Neu5Acα2-8 linkages at pH 7 and glycans with Galβ1-3 linkages, bound to hPIV1 with low affinity (that is, weakly at high concentration but not detectably at low concentration); glycans with Neu5Gcα2-3 or KDNα2-3 sugars were low-affinity targets for hPIV2. hPIV3 bound the same limited glycan set regardless of virus concentration.
Fig. 5.
Motifs bound by hPIV1, -2, and -3. Bold, minimal binding motif. Italic, substituents and modifications to the motif that bound at pH 5 but not at pH 7.
To test for changes in binding profiles in recent clinical isolates, we tested hPIV3 strain Oklahoma/410/2009 at both pH 5 and 7 and hPIV2 strain Oklahoma/283/2009 at pH 7 on the glycan array. Strain 410 did not bind the fucosylated glycan bound by the type strain C243, but it did share the requirement that the glycan chain be at least four sugars long. Strain 3955 bound biantennary sialylated glycans, but strain 283 did not. However, the two hPIV2 strains and the two hPIV3 strains recognized the same Neu5Acα2-3Galβ1-4GlcNAc minimal binding motif, and both hPIV2 strains recognized fucosylated and sulfated motifs.
Neuraminidase specificity of HN on the glycan array.
The structures bound by hPIV on the glycan array represent terminal oligosaccharide motifs, or more complete N- or O-linked glycan structures, that are potential receptors for the virus. We hypothesized that if the biological function of N involves primarily destruction of receptors, then the same structures that are bound on the glycan array should also be destroyed under N-active conditions.
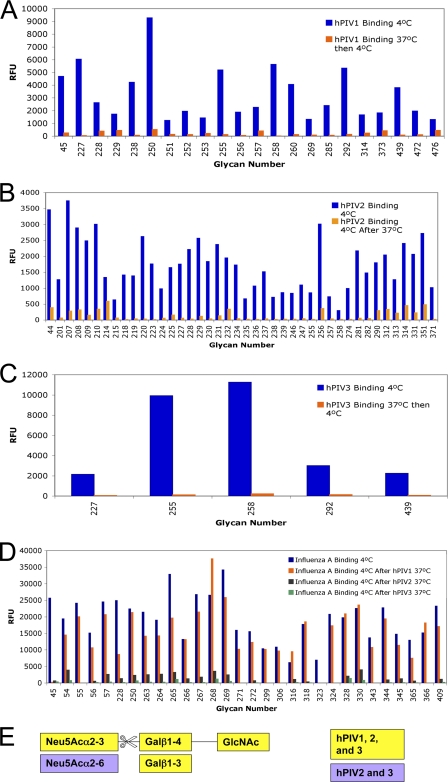
Treatment of the array with hPIV1, -2, or -3 under N-active conditions (4 h at 37°C and pH 5) followed by binding of the same or a new virus to the treated array (1 h at 4°C and pH 5 or 7) revealed that hPIV binds the treated array at a lower level than an untreated array. All three hPIVs reduced their high- and low-affinity ligand glycans by 80% or more compared to the untreated array (Fig. 6A to C; see Table S2 in the supplemental material). None of the three viruses had completely resistant ligands. hPIV1 did have one ligand, Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAcβ-Sp8, that was “partially” cleaved (hPIV1 binding was reduced by 70 to 80%), although other ligands with GalNAc were completely cleaved. We did not find any comparable “partially” cleaved structures for hPIV2 or 3. hPIV1, -2, and -3 cleaved the Neu5Acα2-3Gal bond, regardless of the remainder of the glycan structure.
Fig. 6.
Neuraminidase specificity of HN on the glycan array. In all experiments, a slide incubated with the probe virus at 4°C (untreated array) is compared to a slide incubated with the cleaving virus at 37°C and pH 5 and then the probe virus at 4°C (treated array). (A to C) hPIV1 (A), -2 (B), and -3 (C) bind a treated array at a lower level than an untreated array, indicating destruction of their ligands. Array v. 4.2 was used for panels A and C and v. 3.2 for panel C. (D) Treatment of the array with hPIV1 N does not reduce the binding (i.e., cleave the ligands of) influenza A virus (seasonal H1N1), but treatment with hPIV2 or -3 N does, indicating that hPIV2 and -3 cleave α2-6-linked sialic acids but hPIV1 does not. Array v. 4.1 was used. (E) Motifs found in cleaved structures. For structures corresponding to array glycan numbers, see Table S2 in the supplemental material.
To determine whether hPIV can also cleave glycans that are not ligands, we first treated the array with hPIV under N-active conditions (4 h at 37°C and pH 5) and then washed the slide and bound influenza A virus (1 h at 4°C and pH 7). After treatment with hPIV1, influenza A virus bound the array at levels comparable to those for an untreated array; hPIV1 did not cleave the Neu5Acα2-6Gal bonds recognized by influenza A virus. However, treatment with hPIV2 and -3 N reduced influenza A virus binding to 20% or less of the untreated array level; hPIV2 and -3 have the ability to cleave Neu5Acα2-6 in the context of the array but not 6′NANL (Fig. 6D; see Table S2 in the supplemental material). hPIV2 cleaved glycans containing the motif Neu5Acα2-6Galβ1-4GlcNAc attached to Sp0, and this motif attached to extended polylactosamine or mannose-containing glycans (Fig. 6D, blue and gray bars; see Table S2 in the supplemental material). hPIV3 cleaved all of influenza A virus's ligands, including sulfated and fucosylated Neu5Acα2-6 motifs (Fig. 6D, blue and green bars; see Table S2 in the supplemental material).
We tested the effects of the buffer and cell culture system on the array in order to ensure that these results were due to N activity. We treated the glycan array under the N-permissive conditions (4 h at 37°C and pH 5) without virus. We also treated the array under the same conditions but added an MK2 or MDCK cell culture supernatant. We found that hPIV1 or influenza A virus binding was reduced by an average of ≤10% of the untreated array binding intensity. Only two glycans, Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ-Sp8 and Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ-Sp8, bound hPIV1 or influenza A virus at less than 20% of the untreated array level after treatment. We concluded that the array decays only mildly as the result of pH 5 treatment or contaminants from the cell culture system; we defined “cleaved” as reduced by 80% or more of the untreated array level to ensure that any such decay is not mistakenly identified as N activity.
To further ensure that the difference between treated and untreated array binding was the result of neuraminidase activity, we mixed labeled hPIV1 with 5 mM DANA, a concentration that completely blocks N activity on MUN and also blocks hPIV1 binding to the array, and then incubated this mixture with the array under N-permissive conditions (4 h at 37°C and pH 5). We then washed the array to remove DANA and added fresh hPIV1, without DANA, at 4°C and pH 7. hPIV1 bound the treated array at an average of 37% of the untreated array level, suggesting that the inhibitor blocks neuraminidase activity (although incompletely) in the context of the glycan array. That the presence of an inhibitor makes any difference in apparent cleavage activity suggests that the neuraminidase is indeed responsible for the binding reductions observed. The lower 50% inhibitory concentration (IC50) of DANA against the glycan array than against the small substrates may reflect a higher affinity of HN for glycan attached to the solid array (i.e., a slow off rate). Due to the complexity of these experiments, we have included a schematic of the experimental design (Fig. 7).
Fig. 7.
Schematic of the glycan array neuraminidase specificity experiments.
All glycan array experiments were performed using labeled virus. The presence of the label does not affect the specificity of HN; glycan arrays of hPIV1 performed using an antibody detection system yield binding specificities that do not significantly differ from ours (3).
DISCUSSION
hPIV2 and -3 distribute geographically and temporally, with cocirculation of strains as seen in hPIV1.
Both the HN protein of the human parainfluenza viruses and the HA protein of influenza virus change in response to their environment. HA demonstrates antigenic drift over time in a remarkably linear fashion: it accumulates amino acid changes that persist from season to season. While HN is not known to undergo progressive antigenic drift, it has been shown to accumulate changes in mapped antigenic sites (13). hPIV1 has been previously classified into four subgroups (A to D) (14, 15); subgroup A has the fewest changes from the type strain (Fig. 2A). The number of changes does not increase with time, suggesting that strains persist over multiple seasons (14). Instead, strains appear to be geographically restricted, so different strains are found in different places in a given season (17). hPIV2 and -3 also exhibit cocirculation of strains, with distantly related isolates appearing in a single year in the same place (Fig. 2B and C). While hPIV3 appears to group temporally and geographically and hPIV2 appears to show some geographic distribution, these patterns could simply be artifacts of the limited GenBank sequence set. Overall, the evidence favors divergence into multiple strains that remain stable over time, rather than progressively evolving lineages as seen with influenza virus.
The glycan array can be a useful tool for understanding protein functions.
Although the glycan array does not match the glycome of any particular cell or tissue type or resemble a cell surface, the array does contain complete N-linked glycans, complete O-linked glycans, complete carbohydrate components of glycolipids, and shorter oligosaccharides that can be found on multiple types of glycosylated biomolecules. Therefore, glycans on the array that are ligands for HN are expected to correspond to oligosaccharides that the physiological receptors or targets for N contain, and, since there are spacer molecules between the glycan and the array matrix, physical interactions of HN with glycan are likely to be the same in vivo and in vitro. Accordingly, any glycan that is destroyed by N on the array is susceptible to N, assuming that there are no contributions from the cell membrane itself that the glycan array cannot replicate. The fact that N-susceptible substrates can gain resistance in certain contexts highlights the presentation dependence of N activity.
The susceptibility of substrates to neuraminidase activity is regulated by glycan presentation.
Our findings demonstrate that while all three hPIVs cleave the Neu5Acα2-3Gal and/or Neu5Acα2-6 bond, glycans that are small molecules or are fixed to a surface are more susceptible to N than glycans on glycoproteins.
One possible hypothesis is that N-linked glycans with three and four branches are less accessible to HN than those with one or two branches. The observation that bFetuin, which has most of its Neu5Ac on three-branched glycans (11, 37), and hAGP, which has most of its Neu5Ac on two- to four-branched glycans (16), are less susceptible to N than MUN or sialyllactose supports this hypothesis. However, glycan complexity alone does not account for the observation that some one- or two-branched glycans are susceptible to N on the array but not on a glycoprotein or glycolipid in solution.
A second possibility is that N requires substrates to be spaced such that multiple active sites on a tetramer can be engaged, as has been deduced for some strains of NDV from the cooperativity of N cleavage (22). The observation that molecules that are small compared to the HN tetramer, or that are fixed to the glycan array at a spacing of approximately 0.5 nm (calculated from reference 2), are more susceptible to N than glycoproteins of a size comparable to HN whose glycans may not be anchored to the protein at distances that allow engagement of two or more active sites (calculated from Protein Data Bank [PDB] 3BX6 [33]) supports this idea. However, the observation that digestion of bFetuin with proteinase K does not restore small-molecule-like susceptibility suggests that other factors are also in effect.
A third possibility is that interaction of HN with surface-fixed substrates induces a conformational change that activates N. Previous studies suggest that conformational changes in HN are possible and can affect the neuraminidase activity. Crennell et al. noted that the pH 4.6 structure of NDV has a minimal dimer interface and an active-site structure conducive to N activity, while the pH 6.5 structure has a more extensive dimer interface and active-site residues rotated away from their catalytic positions (9). Ryan et al. subsequently proposed, based on structures of NDV with inhibitors, that the active site can rearrange in response to ligand binding (32). Mahon et al. found that while a complete rearrangement of the interface is not necessary for HN function, preventing conformational changes by introducing disulfide bonds can affect both N and fusion activity (23). Finally, Yuan et al. observed that interactions between HN monomers in the tetrameric structure are weak, and therefore the tetrameric structure itself may be subject to rearrangements (43). They suggested that these rearrangements may involve the stalk region of the protein as well, as mutations in the stalk of NDV HN had previously been shown to affect N activity (40).
We propose that the monomers of an HN dimer can potentially form any of several different types of interactions, including changes to the dimer interface and rearrangement of the tetrameric structure, and that conversion between conformations upon receptor engagement increases the neuraminidase activity. For example, if substrates are attached to a matrix and are therefore in the same plane, an HN tetramer might engage the substrate using two of its four active sites simultaneously (the two that face 180° away from the virion surface in the pH 6.5 structure described by Crennell et al. [9]). Receptor binding could trigger slight changes in the active site of the molecule under the proper conditions; such changes could be transmitted to the dimer or tetramer interface, leading to rearrangement of the HN molecule that triggers F and increases N.
hPIV has broader N specificity than H specificity if the substrate is fixed to a surface.
The N active sites of hPIV2 and -3 are apparently flexible enough to accommodate Neu5Acα2-6 linkages, which are cleaved from the glycan array although they are not held with sufficient affinity for binding to be measurable. In contrast, all three hPIVs have been previously reported to have at least 8-fold-higher activity, and as high as 1,000-fold-higher activity, on soluble α2-3 sialyllactose than on α2-6 sialyllactose (1), and if hPIV1 and -2 are allowed to cleave bFetuin or hAGP overnight, they release only half the amount of sialic acid released by a nonspecific bacterial neuraminidase, suggesting that only one of the two available linkages is cleaved. They release 100% of the sialic acid from 3′NANL under the same conditions. These data are consistent with a surface-induced conformational change in HN; changes in the active site upon binding to a fixed receptor enable HN to accommodate Neu5Acα2-6. While the flexibility of the active site is likely important for overall regulation of N, the ability to cleave Neu5Acα2-6 does not have an obvious function. hPIV1 does not cleave Neu5Acα2-6, yet it is able to replicate just as effectively as hPIV2 and -3. This is the first report of extensive cleavage of α2-6-linked glycans by hPIV2 and -3.
The specificity of N is broader on surface-fixed substrates than on soluble substrates.
The specificity of hPIV on the array suggests that hPIV1 should cleave GM3 and hPIV2 should cleave transferrin (42) (structures are shown in Table 1), and hPIV3's ability to cleave human RBCs (4) suggests that it should cleave human glycophorin A, a major source of glycans on RBC membranes (6), yet none of these were susceptible in solution. 3′NANL, bFetuin, and hAGP all contain motifs that are cleaved on the glycan array (11, 16, 37), yet 3′NANL is more easily cleaved by hPIV1 and -2 than either glycoprotein. Neither substrate size nor the ability of HN to bind substrate was responsible for the specificity mismatch, leaving only the presentation of glycans as a possible explanation. The resistant RBC ligands of hPIV1 are the only instance we found of surface-fixed substrates that are not N susceptible. These ligands may be structures, such as 4-O-acetyl sialic acids or large glycans that are sterically hindered from accessing the active site, that are not on the current array, since no resistant ligands were observed on the array. It appears that the N-activating changes in HN that occur on the glycan array or on RBC surfaces do not occur, or occur to a lesser degree, when ligands are located on a soluble substrate.
Biological implications of surface-induced N activation.
At the cell surface, whether at entry as the virus balances fusion against receptor binding or at exit as the virus must ensure progeny release, N is unlikely to benefit from the same conditions (low pH and low salt concentration) that favor N activity in vitro. A conformational change that increases N might permit sufficient receptor cleavage activity for fusion regulation or progeny release but prevent activity from being great enough to alter the course of viral infection. In intracellular compartments, low-pH conditions may permit sufficient N activity in the absence of the activating conformational change to allow any biological functions of N (for example, the removal of sialic acid from other HN molecules) to take place. The way in which receptor or substrate glycans are presented to HN may therefore be important for modulating HN action at different stages of the viral life cycle.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Oklahoma Center for the Advancement of Science and Technology (OCAST), grant HR09-001. Glycan array analysis was performed by the Consortium for Functional Glycomics Core H, funded by NIGMS, grant GM62116.
We thank Shelly Gulati for technical assistance, Yi Lasanajak for execution of glycan array experiments, Ken Jackson and the OUHSC Proteomics Core Facility for amino acid analysis, and Sheryl Christofferson at the Oklahoma Medical Research Foundation Sequencing Facility for DNA sequencing.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Ah-Tye C., Schwartz S., Huberman K., Carlin E., Moscona A. 1999. Virus-receptor interactions of human parainfluenza viruses types 1, 2 and 3. Microb. Pathog. 27:329–336 [DOI] [PubMed] [Google Scholar]
- 2. Alvarez R. A., Blixt O. 2006. Identification of ligand specificities for glycan-binding proteins using glycan arrays. Methods Enzymol. 415:292–310 [DOI] [PubMed] [Google Scholar]
- 3. Alymova I. V., et al. 16 August 2011. Receptor-binding specificity of the human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Glycobiology. doi:10.1093/glycob/cwr112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amonsen M., Smith D. F., Cummings R. D., Air G. M. 2007. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with α2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 81:8341–8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bousse T., Takimoto T. 2006. Mutation at residue 523 creates a second receptor binding site on human parainfluenza virus type 1 hemagglutinin-neuraminidase protein. J. Virol. 80:9009–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burlak C., Twining L. M., Rees M. A. 2005. Terminal sialic acid residues on human glycophorin A are recognized by porcine kupffer cells. Transplantation 80:344–352 [DOI] [PubMed] [Google Scholar]
- 7. Cole G. A., et al. 1994. Analysis of the primary T-cell response to Sendai virus infection in C57BL/6 mice: CD4+ T-cell recognition is directed predominantly to the hemagglutinin-neuraminidase glycoprotein. J. Virol. 68:6863–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crennell S., Takimoto T., Portner A., Taylor G. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068–1074 [DOI] [PubMed] [Google Scholar]
- 10. Ferreira L., Villar E., Munoz-Barroso I. 2004. Gangliosides and N-glycoproteins function as Newcastle disease virus receptors. Int. J. Biochem. Cell Biol. 36:2344–2356 [DOI] [PubMed] [Google Scholar]
- 11. Green E. D., Adelt G., Baenziger J. U., Wilson S., Van Halbeek H. 1988. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J. Biol. Chem. 263:18253–18268 [PubMed] [Google Scholar]
- 12. Greengard O., Poltoratskaia N., Leikina E., Zimmerberg J., Moscona A. 2000. The anti-influenza virus agent 4-GU-DANA (zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J. Virol. 74:11108–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henrickson K. J., Savatski L. L. 1997. Antigenic structure, function, and evolution of the hemagglutinin-neuraminidase protein of human parainfluenza virus type 1. J. Infect. Dis. 176:867–875 [DOI] [PubMed] [Google Scholar]
- 14. Henrickson K. J., Savatski L. L. 1992. Genetic variation and evolution of human parainfluenza virus type 1 hemagglutinin neuraminidase: analysis of 12 clinical isolates. J. Infect. Dis. 166:995–1005 [DOI] [PubMed] [Google Scholar]
- 15. Henrickson K. J., Savatski L. L. 1996. Two distinct human parainfluenza virus type 1 genotypes detected during the 1991 Milwaukee epidemic. J. Clin. Microbiol. 34:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hermentin P., et al. 1992. The mapping by high-pH anion-exchange chromatography with pulsed amperometric detection and capillary electrophoresis of the carbohydrate moieties of human plasma alpha 1-acid glycoprotein. Anal. Biochem. 206:419–429 [DOI] [PubMed] [Google Scholar]
- 17. Hetherington S. V., Watson A. S., Scroggs R. A., Portner A. 1994. Human parainfluenza virus type 1 evolution combines cocirculation of strains and development of geographically restricted lineages. J. Infect. Dis. 169:248–252 [DOI] [PubMed] [Google Scholar]
- 18. Huberman K., Peluso R. W., Moscona A. 1995. Hemagglutinin-neuraminidase of human parainfluenza 3: role of the neuraminidase in the viral life cycle. Virology 214:294–300 [DOI] [PubMed] [Google Scholar]
- 19. Iorio R. M., et al. 2001. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 75:1918–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawrence M. C., et al. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335:1343–1357 [DOI] [PubMed] [Google Scholar]
- 21. Lyn D., Mazanec M. B., Nedrud J. G., Portner A. 1991. Location of amino acid residues important for the structure and biological function of the haemagglutinin-neuraminidase glycoprotein of Sendai virus by analysis of escape mutants. J. Gen. Virol. 72:817–824 [DOI] [PubMed] [Google Scholar]
- 22. Mahon P. J., Deng R., Mirza A. M., Iorio R. M. 1995. Cooperative neuraminidase activity in a paramyxovirus. Virology 213:241–244 [DOI] [PubMed] [Google Scholar]
- 23. Mahon P. J., Mirza A. M., Musich T. A., Iorio R. M. 2008. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J. Virol. 82:10386–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Merz D. C., Prehm P., Scheid A., Choppin P. W. 1981. Inhibition of the neuraminidase of paramyxoviruses by halide ions: a possible means of modulating the two activities of the HN protein. Virology 112:296–305 [DOI] [PubMed] [Google Scholar]
- 25. Moscona A., Peluso R. W. 1992. Fusion properties of cells infected with human parainfluenza virus type 3: receptor requirements for viral spread and virus-mediated membrane fusion. J. Virol. 66:6280–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moscona A., Peluso R. W. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 65:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palermo L. M., et al. 2009. Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol. 83:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Porotto M., Fornabaio M., Kellogg G. E., Moscona A. 2007. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J. Virol. 81:3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Porotto M., Murrell M., Greengard O., Doctor L., Moscona A. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Portner A., Scroggs R. A., Marx P. S., Kingsbury D. W. 1975. A temperature-sensitive mutant of Sendai virus with an altered hemagglutinin-neuraminidase polypeptide: consequences for virus assembly and cytopathology. Virology 67:179–187 [DOI] [PubMed] [Google Scholar]
- 31. Potier M., Mameli L., Belisle M., Dallaire L., Melancon S. B. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287–296 [DOI] [PubMed] [Google Scholar]
- 32. Ryan C., et al. 2006. Structural analysis of a designed inhibitor complexed with the hemagglutinin-neuraminidase of Newcastle disease virus. Glycoconj. J. 23:135–141 [DOI] [PubMed] [Google Scholar]
- 33. Schonfeld D. L., Ravelli R. B., Mueller U., Skerra A. 2008. The 1.8-A crystal structure of alpha1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin. J. Mol. Biol. 384:393–405 [DOI] [PubMed] [Google Scholar]
- 34. Smith G. W., Hightower L. E. 1983. Biological consequences of neuraminidase deficiency in Newcastle disease virus. J. Virol. 47:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith G. W., Hightower L. E. 1982. Revertant analysis of a temperature-sensitive mutant of Newcastle disease virus with defective glycoproteins: implication of the fusion glycoprotein in cell killing and isolation of a neuraminidase-deficient hemagglutinating virus. J. Virol. 42:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spiro R. G. 1960. Studies on fetuin, a glycoprotein of fetal serum. I. Isolation, chemical composition, and physiochemical properties. J. Biol. Chem. 235:2860–2869 [PubMed] [Google Scholar]
- 37. Spiro R. G., Bhoyroo V. D. 1974. Structure of the O-glycosidically linked carbohydrate units of fetuin. J. Biol. Chem. 249:5704–5717 [PubMed] [Google Scholar]
- 38. Spriggs M. K., Murphy B. R., Prince G. A., Olmsted R. A., Collins P. L. 1987. Expression of the F and HN glycoproteins of human parainfluenza virus type 3 by recombinant vaccinia viruses: contributions of the individual proteins to host immunity. J. Virol. 61:3416–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Z., Iorio R. M. 1999. Amino acid substitutions in a conserved region in the stalk of the Newcastle disease virus HN glycoprotein spike impair its neuraminidase activity in the globular domain. J. Gen. Virol. 80:749–753 [DOI] [PubMed] [Google Scholar]
- 41. Warren L. 1959. The thiobarbituric acid assay of sialic acids. J. Biol. Chem. 234:1971–1975 [PubMed] [Google Scholar]
- 42. Yamashita K., Ohkura T., Ideo H., Ohno K., Kanai M. 1993. Electrospray ionization-mass spectrometric analysis of serum transferrin isoforms in patients with carbohydrate-deficient glycoprotein syndrome. J. Biochem. 114:766–769 [DOI] [PubMed] [Google Scholar]
- 43. Yuan P., et al. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803–815 [DOI] [PubMed] [Google Scholar]
- 44. Zaitsev V., et al. 2004. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 78:3733–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.