Abstract
In response to environmental stress and viral infection, mammalian cells form foci containing translationally silenced mRNPs termed stress granules (SGs). As aggregates of stalled initiation complexes, SGs are defined by the presence of translation initiation machinery in addition to mRNA binding proteins. Here, we report that cells infected with poliovirus (PV) can form SGs early that contain T-cell-restricted intracellular antigen 1 (TIA1), translation initiation factors, RNA binding proteins, and mRNA. However, this response is blocked as infection progresses, and a type of pseudo-stress granule remains at late times postinfection and contains TIA but lacks translation initiation factors, mRNA binding proteins, and most polyadenylated mRNA. This result was observed using multiple stressors, including viral infection, oxidative stress, heat shock, and endoplasmic reticulum stress. Multiple proteins required for efficient viral internal ribosome entry site-dependent translation are localized to SGs under stress conditions, providing a potential rationale for the evolution and maintenance of the SG inhibition phenotype. Further, the expression of a noncleavable form of the RasGAP-SH3 domain binding protein in PV-infected cells enables SGs whose constituents are consistent with the presence of stalled 48S translation preinitiation complexes to persist throughout infection. These results indicate that in poliovirus-infected cells, the functions of TIA self-aggregation and aggregation of stalled translation initiation complexes into stress granules are severed, leading to novel foci that contain TIA1 but lack other stress granule-defining components.
INTRODUCTION
Poliovirus (PV), a member of the Picornaviridae family and a prototypical enterovirus, contains an ∼7.4-kb positive-sense, single-stranded RNA genome. Upon introduction to the cytoplasm, poliovirus RNA begins translating via an internal ribosome entry site (IRES)-mediated cap-independent mechanism to produce viral proteins required for replication of the viral genome and modulation of the intracellular environment for maximal efficiency of viral replication (reviewed in reference 47). The poliovirus open reading frame encodes a single polyprotein, which is then cleaved to intermediates and functional peptides via the internally encoded 2A and 3C proteases (reviewed in reference 47). These proteases also play a crucial role in the modulation of the intracellular environment by targeting cleavage of specific cellular proteins to inhibit cellular processes such as transcription (10, 11, 57), nuclear import (18, 43), translation (reviewed in reference 32), and protein transport and export (9, 14, 28).
Stress granules (SGs) are large cytoplasmic mRNP aggregates induced by the inhibition of translation initiation resulting from the presence of environmental stress stimuli and are thought to represent sites of mRNA storage and triage (5, 22, 24). The canonical pathway to inhibit translation during stress consists of activation of the α subunit of eukaryotic initiation factor 2 (eIF2α) kinases HRI, PERK, PKR, and GCN2 (23, 25, 35), but eIF2-independent mechanisms exist, such as inhibition of the RNA helicase eIF4A (12, 26, 34) or inhibition of translation during viral infection (34, 55). Following the initial reduction in translation, the stalled 48S preinitiation complexes consisting of small ribosome subunits, mRNA, and eIF4E, eIF4G, eIF4A, eIF4B, and eIF3 are aggregated into SG foci through an unknown mechanism that involves dynein and kinesin motor proteins and movement along microtubules (8, 20, 33, 54). It has been proposed that certain other RNA binding proteins are crucial in mediating SG focus formation, principally via an ability to self-oligomerize in response to stress stimuli. These proteins are present as markers in SGs, and overexpression of several of them, including T-cell-restricted intracellular antigen 1 (TIA1), the homologous TIA1-related protein TIAR, and RasGAP-SH3 domain binding protein (G3BP) (25, 52), results in SG formation in unstressed cells. Further, truncated forms of TIA1 and G3BP function as dominant-negative inhibitors of SG formation and are proposed to be critical effectors of SG formation (17, 52).
Numerous viral systems have been shown to interact with the SG pathway with different effects. Some viral systems induce SG formation as part of the mechanism to inhibit host translation (respiratory syncytial virus [RSV], reovirus, and coronaviruses) (31, 46, 48, 49). Other viruses induce SG formation but inhibit their formation as infection progresses (mammalian orthoreovirus [MRV], poliovirus [PV], Semliki Forest virus [SFV], and hepatitis C virus [HCV]) (36, 45, 55). Still, other viruses induce phosphorylation of eIF2α but inhibit the formation of SGs in response (rotavirus and cardioviruses) (3, 38). In most instances, the virus-stress granule relationship appears to be antagonistic; viruses may initially produce SGs in cells and then progressively inhibit the capacity of cells to form SGs. In previous work, we showed that poliovirus induced SGs early during infection but inhibited SG formation later via cleavage of G3BP by the viral 3C protease (55). We also showed that expression of a cleavage-resistant mutant of G3BP, G3BPQ326E, rescued the formation of SGs in poliovirus-infected cells and that this rescue inhibited the production of progeny virions approximately 8-fold. A subsequent study by Piotrowska et al. reported contrasting findings: that poliovirus-infected cells maintain the formation of stable SGs throughout infection that contain TIA1 and polyadenylated mRNA but not cleaved eIF4GI (44).
In light of the differing conclusions, we further investigated the interaction of poliovirus with the SG pathway and found that cells infected with poliovirus can initially form TIA1-positive stress granules that contain all tested translation initiation factors, RNA binding proteins, and mRNA. However, by mid-phase of infection, this response is blocked and pseudo-stress granules that contain TIA1 and lack all other SG components remain. In addition, cleaved eIF4GI and viral IRES transactivating factors (ITAFs) are localized to SGs under stress conditions, providing a potential rationale for the evolution and maintenance of the SG inhibition phenotype. Finally, we show that the expression of a noncleavable form of G3BP in PV-infected cells preserves SGs throughout infection that contain many components of stalled 48S translation preinitiation complexes. Together, these results indicate that in poliovirus-infected cells, the functions of TIA1 aggregation and condensation of translationally stalled mRNPs become unlinked, leading to novel TIA-positive foci that lack most stress granule-defining components.
MATERIALS AND METHODS
Cell culture and viral infection.
HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) plus penicillin, streptomycin, and glutamine at 37°C in 5% CO2. Poliovirus type I, Mahoney strain, was grown as previously described (21). For infection, HeLa cells were seeded on glass coverslips in 12-well plates at a concentration of 3 × 105 cells per well and left overnight at 37°C before being infected at a multiplicity of infection (MOI) of 20 or 30 for the indicated times in DMEM containing 2% FBS. Some infections also contained 2 mM guanidine hydrochloride as indicated. To induce the formation of stress granules, cells were treated with 0.5 mM sodium arsenite added to DMEM (Sigma) for 30 min or 10 μM thapsigargin (Calbiochem) for 90 min or heat shocked at 46°C for 40 min prior to fixation and processing for immunofluorescence.
DNA constructs and transfections.
pcDNA3.1-his-G3BPQ326E was previously described (55). pcDNA3.1-hisPTB was a kind gift from Thomas Cooper, pcDNA6-V5-UNR was a kind gift from Ann-Bin Shyu, and peGFP-MS2-NLS was a kind gift from Kenneth Kosik. pcDNA3.1-hisPCBP2 was cloned by insertion of a HindIII-PCBP2-EcoRV fragment into pcDNA3.1-hisPTB cut with the same enzymes. pTRE2-CVB3-Fluc-4BoxB-4MS2 was produced by PCR amplification of a 4BoxB-4MS2-containing fragment from pGEM-5Z-globin-4BoxB-4MS2 (kind gift of Niels Gehring), with primers containing Bsu36I and AgeI and insertion into pTRE2-CVB3-Fluc (56) with the same enzymes. For stress granule rescue experiments, HeLa cells were transfected with pcDNA3.1-G3BPQ326E using FugeneHD (Roche) for approximately 40 h. Transfection of plasmids expressing PTB, PCBP2, and UNR was conducted overnight with FugeneHD (Roche). HeLa Tet-On (Clontech) cells were cotransfected with FLuc reporters and peGFP-MS2-NLS overnight in the presence of 10 μg/ml tetracycline using FugeneHD (Roche).
Immunofluorescence.
After infection as described above, HeLa cells were fixed for 30 min in 4% formaldehyde in PEM {80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8, 5 mM EGTA, and 2 mM MgCl2}, permeabilized with 0.5% Triton X-100 in PEM, and blocked with 4% powdered milk in Tris-buffered saline plus Tween 20 (TBS-T). Stress granules were stained for 2 h with antibodies against endogenous TIA1 (1:100; Santa Cruz), eIF3a (1:50; Cell Signaling), eIF4E (1:100; Cell Signaling), eIF4B (1:250; Abcam), G3BP1 (1:500) (described in reference 55), PABP (1:250) (described in reference 29), YB1 (1:1,000; gift of Thomas Cooper,), and N- and C-terminal eIF4GI (1:500 and 1:250, respectively) (described in reference 7). Exogenous PTB, PCBP2, G3BP, and G3BPQ326E were imaged using anti-T7 (1:500; Bethyl), while UNR was imaged using anti-V5 monoclonal antibody (1:500; Invitrogen). Poliovirus 3Cpro was stained with anti-3Cpro rabbit polyclonal antibody (1:1,000). After incubation with primary antibody, coverslips were washed in blocking buffer before incubation with anti-goat Alexa Fluor 594, anti-rabbit Alexa Fluor 488, or anti-mouse Alexa Fluor 488 (1:1,000; Molecular Probes) secondary antibody for 30 min at room temperature. Coverslips were then washed in TBS-T and fixed for 30 min in 4% formaldehyde in PEM before counterstaining with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) and mounting for imaging in No-Fade mounting medium (Invitrogen). Images were taken on a Zeiss Axioplan II or DeltaVision deconvolution image restoration microscope in the Baylor College of Medicine Integrated Microscopy Core, and images were compiled using Adobe Photoshop. Stress granule foci in images were enumerated and analyzed with ImageJ software.
RNA in situ hybridization.
HeLa cells were plated and transfected as described above. After infection, cells were fixed in phosphate-buffered saline (PBS) plus 2% formaldehyde for 10 min and permeabilized in 100% ice-cold methanol at −20°C for 10 min. The fixed and permeabilized cells were then washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and incubated overnight with 50 ng biotinylated oligo(dT) probe in hybridization buffer (50% formamide, 2× SSC, 10% dextran sulfate, 0.2% bovine serum albumin [BSA], and 20 mM vanadyl ribonucleoside complex [NEB]) at 43°C. The following day, the cells were washed with 2× SSC and incubated with streptavidin-Alexa Fluor 647 (1:3,000; Molecular Probes) for 45 min at room temperature. The cells were then washed with 4× SSC, incubated with biotinylated rabbit anti-streptavidin polyclonal antibody (1:1,000; Abcam) and goat anti-TIA1 polyclonal antibody (1:100; Santa Cruz) in 4× SSC plus 0.1% Triton X-100 for 1 h, washed in 2× SSC, and incubated with streptavidin-Alexa Fluor 647 and rabbit anti-goat fluorescein isothiocyanate (1:1,000; Zymed) in 2× SSC plus 0.1% Triton X-100. The cells were then washed with 2× SSC, counterstained with DAPI, and mounted for imaging as described above.
Immunoblot analysis of cell lysates.
PV-infected cells were harvested and lysed in NP-40 lysis buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, and 1% NP-40). Lysates were cleared by centrifugation at 10,000 × g, and equal volumes were resolved by SDS-PAGE. Gels were transferred to nitrocellulose membranes (ISC Bioexpress) at 0.35 amps for 80 min and probed with anti-eIF4GI rabbit polyclonal antibody (1:1,000; N-terminal specific) or anti-G3BP1 rabbit polyclonal antibody (1:1,500; N-terminal specific) in 4% powdered milk in TBS-T overnight at 4°C. Proteins were detected by incubation with horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:4,000; Bio-Rad) for 2 h at room temperature and visualized with SuperSignal West Pico substrate (Pierce).
RESULTS
Virus-induced stress granules diminish during infection.
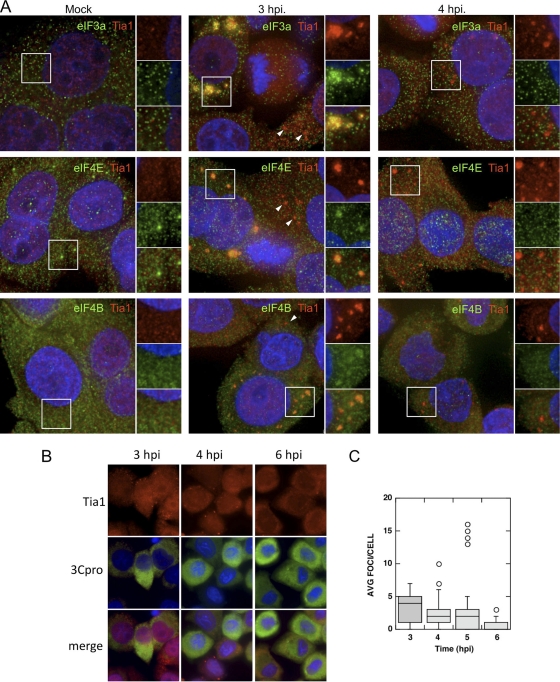
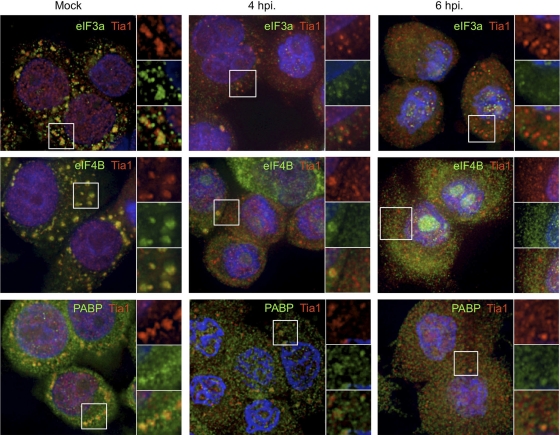
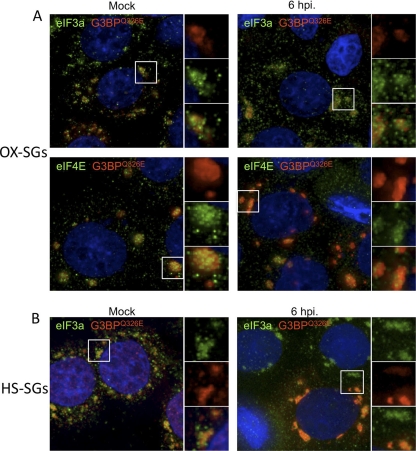
Two previous studies analyzing the status of stress granules in poliovirus-infected cells produced differing conclusions; however, both displayed a persistent formation of TIA1-positive (44, 55) or TIAR-positive (55) foci. Thus, we examined in greater detail the constituents of TIA1-containing foci during the course of infection. We first determined whether PV inhibits the formation of virus-induced SGs (V-SGs) containing translation components in HeLa cells as infection progresses. To probe V-SGs for stalled preinitiation complexes, we stained foci with TIA1 and counterstained with antibodies for three translation initiation factors, eIF3a, eIF4E, and eIF4B (Fig. 1A). None of these initiation factors are cleaved during PV infection, and both eIF3 and eIF4B function in PV IRES-mediated translation (13, 40). Analysis by immunofluorescence microscopy revealed that no significant foci containing TIA1 or initiation factors were found in mock-infected cells. Some initiation factors were found to be partly distributed into small punctae in uninfected cells, particularly eIF3 subunits and eIF4E. The significance of these microfoci are unknown; however, eIF4E enters some processing bodies in cells (larger foci boxed in Fig. 1A) (24). By 2 to 3 h postinfection (hpi), approximately 15% of infected HeLa cells mounted a V-SG response and contained numerous large foci containing TIA1 and eIF3a, eIF4E, and eIF4B. Many other cells contained TIA1 foci that did not colocalize with initiation factors (Fig. 1A, white arrowheads). However, by 4 hpi, when virus protein synthesis and RNA replication are building to a peak (56; data not shown), cells no longer contained foci with initiation factors. As we previously observed with TIAR (55), foci stained with TIA1 persisted in numerous cells. From examination of a large number of cells, we noted that at 3 to 4 hpi, the capacity of cells to form V-SGs was inversely proportional to the degree of nuclear margination and deformation (data not shown). To determine whether the presence of TIA1 foci correlated with levels of robust virus replication and gene expression, we performed similar experiments and costained with antibody and poliovirus 3Cpro. Cytoplasmic TIA1 foci were visible at 3 hpi and persisted to some degree through 6 hpi in cells that stained positive for viral 3Cpro (Fig. 1B); however, there was no correlation observed between higher intensities of 3Cpro staining and persistent TIA1-positive foci once 3Cpro staining became apparent. Quantification of TIA1-positive foci in infected cells showed that although foci were still present in numerous cells at 6 hpi, the average number of foci per cell steadily decreased during the course of infection (Fig. 1C). Together, these data indicate that viral infection initially induces aggregation of TIA1 and initiation factors into V-SGs that are disrupted by 4 hpi and that the prevalence of TIA1-containing foci decreases over the course of infection.
Fig. 1.
Virus-induced stress granules are inhibited during infection. (A) HeLa cells were infected and immunostained for TIA1 (red) and eIF3a, eIF4E, or eIF4B (green). DAPI (blue) counterstained the nuclei. White boxes indicate the locations of fields magnified in the right-hand panels. (B) Infected HeLa cells were immunostained for TIA1 (red) or PV 3Cpro (green), and nuclei were marked with DAPI (blue) at the indicated time points postinfection. (C) TIA1-positive foci were quantitated in 3Cpro-positive cells, and results are displayed with box diagrams. Horizontal lines indicate the means, boxes indicate 25th to 75th percentiles, and error bars indicate 95th percentiles. Circles indicate individual cells with statistical outliers.
TIA1-positive foci induced by oxidative stress in infected cells lack initiation factors and RNA binding proteins.
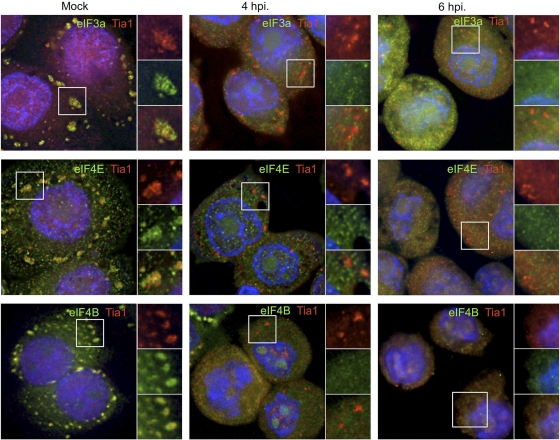
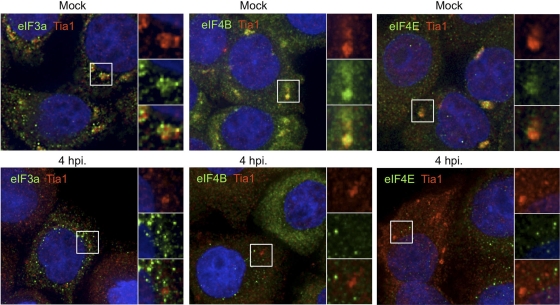
Emerging evidence suggests that SGs induced under different stress conditions can contain unique protein constituents and thus may form via overlapping but unique processes (25, 44, 50). To determine whether stress granules containing stalled preinitiation complexes could be induced by oxidative stress (Ox-SGs) in infected cells, we treated PV-infected cells with sodium arsenite at 4 and 6 hpi. As seen in Fig. 2, eIF3, eIF4E, and eIF4B colocalize extensively with TIA1 in large Ox-SGs in uninfected HeLa cells subjected to arsenite treatment. Upon infection with PV, colocalization of these factors with TIA1 in stressed cells declined rapidly after 2 hpi (data not shown), with little to no colocalization observed at 4 hpi or 6 hpi (Fig. 2). eIF4GI was previously screened with similar results (55). These data indicate that poliovirus infection inhibits the inclusion of translation initiation factors in TIA1-positive foci as infection progresses.
Fig. 2.
PV infection inhibits the inclusion of initiation factors in Ox-SGs. HeLa cells were infected for the indicated times and stressed with 0.5 mM arsenite for 30 min before fixation and processing for analysis. Ox-SGs were immunostained with TIA1 (red) and costained for the indicated translation initiation factors (green). Cell nuclei were stained with DAPI (blue). White boxes indicate fields magnified in the right-hand panels.
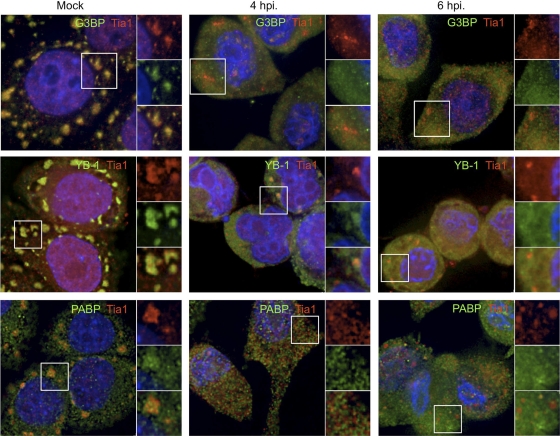
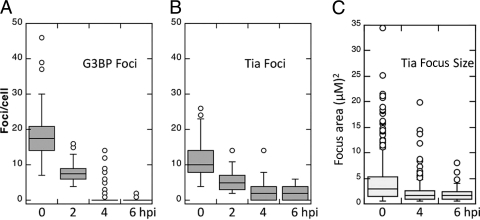
To determine whether the inhibition of localization with TIA1-positive foci was specific only to translation initiation factors, we also monitored the localization of RNA binding proteins G3BP, YB1, and PABP, which are known to enter SGs (8, 25, 55). As was observed with translation initiation factors, the concentration of these RNA binding proteins with TIA1-positive foci during infection waned nearly completely by 4 hpi and was inhibited at 6 hpi (Fig. 3). Similar results were obtained when other RNA binding proteins were assayed (data not shown). Quantification of G3BP-positive foci and TIA1-positive foci in PV-infected cells (Fig. 4) revealed that G3BP-positive foci formed more readily in response to oxidative stress and also at slightly higher numbers in uninfected cells but decreased rapidly in frequency as infection progressed, until they were essentially absent at 4 to 6 hpi. In contrast, the frequency of TIA1-positive foci decreased at a lower rate and foci remained in many cells at 6 hpi. Particle analysis of TIA1 foci revealed that a progressive decrease in the size of foci occurred as infection progressed (Fig. 4). Taken together, these data indicate that many RNA binding proteins and translation initiation factors that enter Ox-SGs are excluded from TIA1-positive foci by mid-phase of PV infection, yet TIA1-positive foci persist but also decrease in frequency and size.
Fig. 3.
PV infection inhibits the inclusion of mRNA binding proteins in Ox-SGs. HeLa cells were infected for the indicated times and stressed with 0.5 mM arsenite. SGs were visualized by immunostaining with TIA1 (red) and the indicated mRNA binding proteins (green). Cell nuclei were immunostained with DAPI (blue). Fields magnified in the right-hand panels are outlined in white.
Fig. 4.
TIA1-containing foci persist during infection but decrease in number and size. Foci positive for G3BP (A) and TIA1 (B) were counted at the indicated times postinfection. (C) TIA1-positive focus areas were calculated using ImageJ software. Lines indicate means, boxes indicate 25th to 75th percentiles, and error bars indicate 95th percentiles. Circles denote statistical outliers.
Multiple factors are absent from SGs formed by other stressors in infected cells.
Recently, a number of reports have indicated that SGs formed by different stressors may not contain equivalent mRNP constituents (25, 44, 50). To determine whether the localization of translation initiation factors and RNA binding proteins to TIA1-positive foci is inhibited under other stress conditions, we subjected PV-infected HeLa cells to heat shock or thapsigargin treatment and monitored the subcellular localization of the same factors. Heat shock treatment induced inclusion of eIF3, eIF4B, and PABP into heat shock SG (HS-SG) foci that colocalized with TIA1 (Fig. 5) in more than 80% of cells (data not shown). However, translation initiation factors were inhibited from inclusion in TIA1-positive foci in heat-shocked cells at 4 hpi and at 6 hpi except in rare instances; two of these examples are shown in Fig. 5. Treatment with thapsigargin to induce endoplasmic reticulum (ER) stress granules (ER-SGs) resulted in colocalization of initiation factors and TIA1 in a smaller subset of cells (∼60%). Cells were imaged at 2, 4, and 6 hpi, and effects on ER-SGs were observed to be very similar to effects on HS-SGs. Cells could still form ER-SGs at 2 hpi (data not shown); however, when thapsigargin-treated infected cells were imaged at 4 hpi (Fig. 6) or 6 hpi (data not shown), mobilization of initiation factors into TIA1-positive foci was not observed. Similar results were obtained with other factors (eIF4E, YB1, and PABP) for both heat shock and thapsigargin treatments (data not shown). Overall, these data indicate that during PV infection, TIA1-positive foci that lack initiation factors and RNA binding proteins form under multiple conditions of oxidative stress, heat shock, and ER stress. This suggests that poliovirus inhibits the formation of SGs at a point shared by multiple stress pathways.
Fig. 5.
Initiation factors are excluded from HS-SGs in PV-infected cells. HeLa cells infected with PV were immunostained for TIA1 (red) and additional protein factors (green) in cells subjected to heat shock prior to fixation at the indicated times postinfection. Cell nuclei were stained with DAPI (blue). Fields magnified in the right-hand panels are outlined in white.
Fig. 6.
Initiation factors are excluded from ER-SGs in PV-infected cells. HeLa cells were infected for 4 h and treated with thapsigargin, and then SGs were visualized by immunostaining for TIA1 (red) and the indicated initiation factors (green). Cell nuclei were stained with DAPI (blue), and white boxes indicate fields magnified in the right-hand panels.
Cleaved eIF4GI fragments localize to SGs.
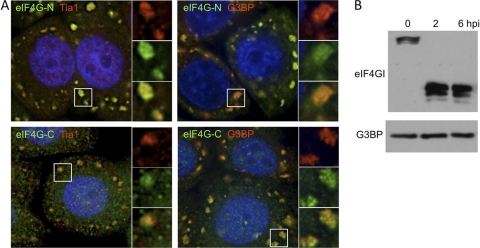
One possible mechanism to explain the inhibition of localization of some factors to SGs during PV infection is their cleavage by virus-encoded proteases as previously suggested (44). To test this, we infected HeLa cells treated with guanidine hydrochloride to inhibit viral RNA replication and therefore the normal replicative cycle (51). Upon application of arsenite treatment at 6 hpi, we observed the efficient colocalization of both the N- and C-terminal fragments of eIF4GI with both TIA1 and G3BP into large, abundant SGs (Fig. 7A). Immunoblot analysis of lysates from these cells showed that eIF4GI is completely cleaved at 6 hpi but that G3BP is not, which is consistent with the ability of these infected cells to form SGs (Fig. 7B). These results indicate that, at least in the case of eIF4GI, cleavage by a viral protease does not preclude the inclusion of either the N- or C-terminal fragment of an initiation factor in Ox-SGs.
Fig. 7.
Cleaved eIF4GI can mobilize to SGs in response to arsenite treatment. (A) HeLa cells were infected in the presence of 2 mM guanidine-HCl and were coimmunostained for TIA1 or G3BP (red), N-terminal or C-terminal eIF4GI fragment (green), and cell nuclei (blue) after arsenite treatment. White boxes indicate fields magnified in the right-hand panels. (B) Immunoblot analysis of eIF4GI in guanidine-HCl-treated, PV-infected HeLa cells.
TIA1-positive foci lacking mRNA accumulate during infection.
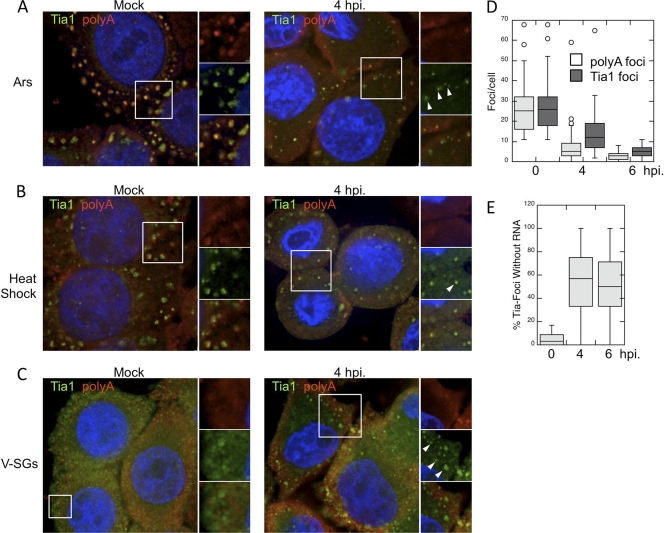
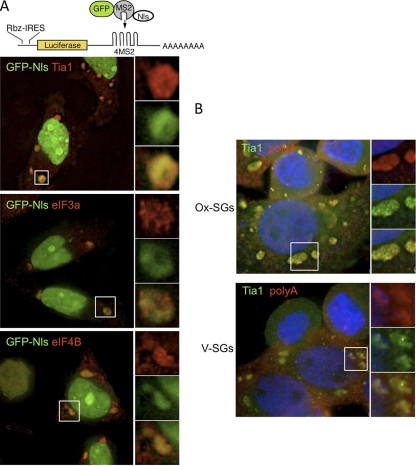
Central to the concept of stress granules is their role as storage and triage centers for mRNA under stress conditions (22, 24). After observing a lack of translation initiation factors and RNA binding proteins in TIA1-positive foci during PV infection, we wanted to determine whether PV was selectively inhibiting the inclusion of protein components or whether PV infection blocked the inclusion of entire mRNPs into TIA1-positive foci. To test this, we monitored the distribution of total polyadenylated mRNA in PV-infected cells by in situ hybridization together with TIA1 coimmunostaining. In mock-infected cells, the application of heat shock or arsenite treatment induced large SGs with TIA1 and poly(A) RNA localization. As described above, we consistently observed the formation of smaller TIA1-positive foci at 4 hpi in both arsenite-treated and heat-shocked cells (Fig. 8A and B). Surprisingly, we also observed a lack of inclusion of polyadenylated mRNA in many, but not all, foci at 4 hpi, similar to what we observed with initiation factors and RNA binding proteins (Fig. 8A to C, white arrowheads). Quantification of TIA1-positive foci and mRNA-positive foci in infected cells stressed with arsenite revealed that TIA1 and mRNA colocalized within foci extensively in uninfected cells, although occasional foci with only a TIA or poly(A) signal could be found (Fig. 8E). However, the frequency of mRNA-positive foci decreased more rapidly than that of TIA1-positive foci, and the frequency of mRNA-positive foci remained consistently lower throughout later times in infection (Fig. 8D). As infection progressed, a high percentage (average of 50 to 60%) of TIA1-positive foci was detected in PV-infected cells that lacked an mRNA signal (Fig. 8E). This indicates that during PV infection, TIA1-positive foci are forming in response to stressors that do not contain initiation factors, RNA binding proteins, or polyadenylated mRNA, and thus, the ability of TIA1 to aggregate and its ability to mediate Ox-SG or HS-SG formation become unlinked. We also examined colocalization of TIA1 and poly(A) RNA within V-SGs in the absence of other stressors (Fig. 8C). In cells infected for 4 h, TIA1 foci were present as expected, and similar to the results described above, not all TIA1 foci stained for poly(A) RNA. This indicates that TIA1 aggregation and V-SG formation also become unlinked.
Fig. 8.
Loss of polyadenylated mRNA from TIA1-positive foci in PV-infected cells. TIA1-positive foci (green) induced by arsenite treatment (A), heat shock (B), or PV infection (C) were costained with an oligo(dT) probe for polyadenylated mRNA (red) at 4 hpi. Cell nuclei were stained with DAPI (blue). (D) Quantification of TIA1- and poly(A)-positive foci in HeLa cells at the indicated times postinfection. (E) Colocalization of TIA1- and poly(A)-positive foci was quantified, and data are shown as the percentage of TIA1-positive foci that lacked poly(A) staining. In box plots showing TIA1 focus quantification (D, E), lines indicate means, boxes represent 25th to 75th percentiles, error bars indicate 95th percentiles, and circles denote individual cells with statistical outliers.
IRES transactivating factors localize to SGs.
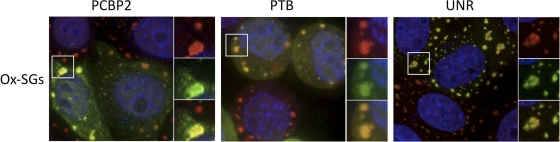
In order to efficiently recruit ribosomes for translation initiation, picornavirus IRES elements require critical cellular RNA binding proteins to bind to the IRES and recruit initiation factors and ribosomes (reviewed in reference 15). In the case of PV, these IRES transactivating factors (ITAFs) include cellular polypyrimidine tract binding protein (PTB), poly(rC) binding protein 2 (PCBP2), upstream of N-ras (UNR), and La autoantigen (2, 4, 19, 37). One possible explanation for the evolution and maintenance of SG inhibition by PV is that these required RNA binding proteins, along with a large portion of the cellular translation apparatus, may be sequestered in SGs and become inaccessible for use by viral mRNA. Considering the observed localization of many other RNA binding proteins to SGs, including PCBP2 (16), we investigated the localization of exogenously expressed PTB, PCBP2, and UNR in cells subjected to arsenite treatment. Figure 9 shows that all three ITAFs strongly localize to Ox-SGs, indicating that factors essential to viral RNA translation, including initiation factors and ITAFs, are sequestered in SGs under stress conditions.
Fig. 9.
PV IRES transactivating factors (ITAFs) localize to SGs. HeLa cells expressing the indicated ITAFs were stressed with 0.5 mM sodium arsenite and immunostained for exogenously expressed ITAFs (green), TIA1 (red), and nuclei (blue). Fields magnified in the right-hand panels are outlined in white.
Rescue of SG formation restores localization of initiation factors and RNA binding proteins to SGs.
In a previous study, we identified the cleavage of G3BP as a mechanism by which PV inhibits the formation of SGs. We also showed that the expression of a cleavage-resistant mutant of G3BP, G3BPQ326E, rescues SG formation late in infection, as measured by eIF4GI and TIAR colocalization in PV-infected, arsenite-stressed cells (55). To further extend that study, we expressed G3BPQ326E and tested the distribution of additional translation initiation factors in response to multiple stressors in infected cells. Ectopic expression of G3BP is known to induce the formation of very large SGs in a subset of uninfected unstressed cells (24, 52; data not shown). Figure 10A shows that under arsenite treatment, PV-infected cells expressing G3BPQ326E display extensive localization of eIF3 and eIF4E with G3BPQ326E in large Ox-SG foci at 6 hpi, similar to that observed in uninfected cells. Parallel experiments revealed Ox-SGs rescued under the same conditions contained other initiation factors and RNA binding proteins (eIF4B, PABP, and YB1) (data not shown). We also tested the ability of G3BPQ326E to rescue stress granule formation in response to heat shock and found extensive colocalization of G3BPQ326E and eIF3 in large foci in both uninfected and infected cells at 6 hpi (Fig. 10B). Other initiation factors and RNA binding proteins (eIF4B, eIF4E, PABP, and YB1) were also tested with heat shock, and similar results were observed (data not shown). Together, these data indicate that the rescue of SGs by the expression of G3BPQ326E also rescues the localization of translation initiation factors to both Ox-SGs and HS-SGs in poliovirus-infected cells.
Fig. 10.
Initiation factors localize to SGs containing cleavage-resistant G3BP. HeLa cells expressing G3BPQ326E were infected for 6 hpi and subjected to arsenite treatment (A) or heat shock (B). Rescued SGs were immunostained for G3BPQ326E (red) and translation initiation factors (green), and cell nuclei were stained with DAPI (blue). White boxes indicate fields magnified in the right-hand panels.
In light of the rescue of initiation factor localization to SGs in G3BPQ326E-expressing cells during infection, we also monitored the localization of IRES-containing reporter RNA and total polyadenylated mRNA in SGs in poliovirus-infected cells at 6 hpi. The cytoplasmic distribution of firefly luciferase reporter mRNA bearing the CVB3 IRES and MS2 binding loops was monitored using a previously described system that tethers a coexpressed green fluorescent protein (GFP)-MS2-coat protein fusion that also contains three nuclear localization signals (24). As expected, distribution of the GFP signal was skewed toward the nucleus in unstressed cells and stressed cells not expressing the reporter RNA (data not shown). In stressed cells expressing the fusion protein, reporter RNA, and G3BPQ326E, most of the GFP signal was still retained in the nucleus; however, the GFP signal entered cytoplasmic SG foci in PV-infected cells (Fig. 11A). The mRNA-tethered GFP fusion protein colocalized with TIA1, eIF3a, and eIF4B in infected cells. This indicates that the expression of G3BPQ326E rescues the localization of mRNA to reconstituted Ox-SGs even when the mRNA contains an enteroviral IRES element. Additionally, staining for total polyadenylated mRNA in cells expressing G3BPQ326E and subjected to poliovirus infection for 6 h showed a similar relocalization of mRNA to SGs formed in response to oxidative stress and viral infection (Fig. 11B). Taken together, these results indicate that the expression of noncleavable G3BP results in the rescue of the SG response and the localization of initiation factors and mRNA. Further, the data indicate that mRNA containing virus IRES elements is capable of entering SGs in virus-infected cells.
Fig. 11.
RNA localizes to SGs in cells expressing G3BPQ326E. (A) Cartoon of reporter RNA containing a hammerhead ribozyme to remove the cap structure and four MS2 binding loops. HeLa cells expressing luciferase reporter and GFP-MS2-NLS in the presence of G3BPQ326E were treated with 0.5 mM arsenite, and reporter localization (green) with initiation factors (red) was monitored. (B) Polyadenylated mRNA (red) was monitored along with TIA1 (green) in cells expressing G3BPQ326E. Cell nuclei were stained with DAPI (blue).
DISCUSSION
Stress granules are cytoplasmic aggregates of stalled preinitiation complexes, containing polyadenylated mRNA, 40S ribosomal subunits, associated initiation factors and mRNA binding proteins, and key marker proteins, notably G3BP and TIA1 or TIAR. Here, we report that in poliovirus-infected cells, novel TIA1-positive granules that lack initiation factors and polyadenylated mRNA are present in infected cells treated with different stressors, indicating that the ability of TIA1 to form aggregates is separated from its ability to form stress granules in poliovirus-infected cells. Further, the expression of noncleavable G3BP resulted in the relocalization of initiation factors, mRNA binding proteins, and mRNA to Ox-SGs formed in response to arsenite stress late in infection (6 hpi), when control-infected cells did not mount this response. Finally, analysis of IRES transactivating factors PCBP2, PTB, and UNR as well as the eIF4GI cleavage fragments and IRES-containing RNA revealed that these crucial factors localize to stress granules, providing a potential rationale for poliovirus evolving and maintaining the function of inhibiting stress granules.
Several results reported here are consistent with previous reports of SG formation during PV infection. We previously reported that the expression of cleavage-resistant G3BP could rescue SGs in PV-infected cells that contained transgenic G3BP and endogenous TIAR. In this case, those cells not ectopically expressing G3BP did not contain G3BP-stained SGs, but a subset did contain small foci that stained only with TIAR (Fig. 6) (55). We have determined that TIAR-stained pseudo-SG foci also form that are similarly devoid of initiation factors and that are indistinguishable from the TIA1 foci reported here (data not shown). We have no evidence that TIA1 and TIAR behave differently in PV infections; rather, they consistently enter the same structures, likely together. Piotrowska et al. also described foci containing both TIA1 and mRNA in PV-infected cells at 4 hpi but did not examine initiation factors other than eIF4GI, which displayed a marked decline in focus formation at 4 hpi (44). Our results indicate that TIA1- and mRNA-containing foci still exist at 4 hpi, though many TIA foci are becoming devoid of mRNA at this time point. We also note that at 4 hpi, the capacity to inhibit SG formation on an individual cell basis is inversely proportional to the degree of nuclear chromatin margination and nuclear deformation observed. Those few cells displaying low cytopathic effect (CPE) retain some residual capacity to form SGs; however, those displaying marked CPE do not mount this response.
Stress granules are defined as aggregates composed of stalled initiation complexes induced by the inhibition of translation initiation by various forms of environmental stress (5). Virus infection produces cell stress on multiple levels that could signal SG formation. PV shuts off host translation principally from cleavage of eIF4GI and PABP but also induces significant levels of eIF2α phosphorylation (41, 56). As sites of storage and triage of stalled 48S preinitiation complexes (22, 24), stress granules contain eIF2, eIF4GI, eIF4A, eIF4E, eIF4B, 40S ribosomal subunits, and polyadenylated mRNA (6, 23, 27, 34). Although the precise mechanism by which stress granules aggregate is still unknown, it is known that several proteins induce the spontaneous formation of stress granules upon overexpression, and evidence suggests that G3BP1 and TIA1 (17, 52), as well as other factors such as metastatic lymph node 51, HDAC-6, ataxin 2, and Grb7 (1, 30, 39, 53), play important roles in the stress granule formation process. Both TIA1 and G3BP contain self-oligomerization domains proposed to be instrumental in the process of SG formation, as evidenced by the dominant-negative effect on SG formation exerted by either oligomerization domain when overexpressed (17, 52). In this report, we show that in poliovirus-infected cells, the ability of TIA1 to aggregate is separated from its ability to form stress granules, as evidenced by the continued presence of TIA1-positive foci that lack any other tested marker, including mRNA (Fig. 1, 2, 3, 5, 6, and 8). This indicates that, as previously indicated (55), poliovirus actively inhibits the inclusion of translation initiation factors and mRNA binding proteins in stress granules. Also in agreement with previous data (55), we found that the rescue of stress granules by the expression of cleavage-resistant G3BP results in the relocalization of initiation factors, RNA binding proteins, and mRNA in poliovirus-infected cells (Fig. 10 and 11). This implicates the cleavage of G3BP in the release of these factors from stress granules or, conversely, the inhibition of their inclusion in response to stress. Together with the finding that PV infection unlinks TIA1 aggregation and SG formation under multiple stress conditions, our data indicate that by cleaving G3BP, PV inhibits canonical SG formation caused by the activation of several different pathways. This suggests that although SG compositions can vary under different stress conditions (25, 44, 50), central effectors to SG formation are common between SG pathways, with G3BP being one of these primary effectors. Further biochemical analysis of TIA1 and TIAR is required under these conditions, as the mechanism behind the unlinking of TIA1-mediated SG formation and TIA1 self-oligomerization is unknown.
Enteroviruses require RNA binding proteins termed IRES transactivating factors (ITAFs) to bind to their IRES in order to efficiently mediate ribosome recruitment (for reviews, see references 15 and 42). In the case of poliovirus, the IRES requires UNR (4), PTB (19), PCBP2 (2), and La autoantigen (37). PCBP2 and PTB have been shown to localize to stress granules (16, 49), and here we show that PCBP2, PTB, and UNR localize to stress granules in response to stress (Fig. 9). In addition, the poliovirus IRES requires the C-terminal cleavage fragment of eIF4GI produced during infection, which localized to stress granules in response to stress (Fig. 7). During PV infections of HeLa and 293 cells, virus protein synthesis reached peak levels between 4 and 5 hpi (56), after most SGs were blocked or dispersed. Together, these data suggest that cellular proteins required for viral replication, both ITAFs and translation initiation factors, localize to stress granules, possibly reducing the local concentration in the cytoplasm and inhibiting viral translation and therefore viral replication early during infection. While circumstantial, the possibility exists that stress granules can play an antiviral role, at least in the case of enteroviruses. We also report evidence that RNA containing an enterovirus IRES can enter SGs. Further investigation is required to determine whether this IRES mRNA is actually translationally silenced in the context of infection.
Overall, the findings in this report indicate that the formation of SGs is a multilayered process involving multiple factors, providing an explanation for the requirement of several proteins for SG formation. Of specific interest is the cleavage of G3BP, the kinetics of which correspond to the observed loss of initiation factors from SGs during infection but not mRNA. The expression of a cleavage-resistant form of G3BP results in the inhibition of this dispersal of initiation factors and mRNA from SGs despite the continued cleavage of endogenous G3BP (55). Additional studies are under way to further investigate the role of G3BP in the recruitment of initiation factors to SGs as well as the effects of this recruitment on viral replication. In addition, the novel TIA1 pseudo-stress granules reported in this study highlight the importance of utilizing multiple markers during analysis of SGs in infected cells, specifically translation initiation factors and G3BP.
ACKNOWLEDGMENTS
We are grateful to Thomas Cooper, Ann-Bin Shyu, Kenneth Kosik, and Niels Gehring for plasmids as well as to members of the Lloyd lab for helpful discussions.
This work was supported by NIH Public Health Service grant AI 50237. J.P.W. was partly supported by NIH training grant T32 AI07471.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Baguet A., et al. 2007. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J. Cell Sci. 120:2774–2784 [DOI] [PubMed] [Google Scholar]
- 2. Blyn L. B., Towner J. S., Semler B. L., Ehrenfeld E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borghese F., Michiels T. 2011. The leader protein of cardioviruses inhibits stress granule assembly. J. Virol. 85:9614–9622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boussadia O., et al. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77:3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchan J. R., Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchan J. R., Yoon J. H., Parker R. 2011. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124:228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Byrd M. P., Zamora M., Lloyd R. E. 2005. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J. Biol. Chem. 280:18610–18622 [DOI] [PubMed] [Google Scholar]
- 8. Chernov K. G., et al. 2009. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J. Biol. Chem. 284:36569–36580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choe S. S., Dodd D. A., Kirkegaard K. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337:18–29 [DOI] [PubMed] [Google Scholar]
- 10. Clark M. E., Hammerle T., Wimmer E., Dasgupta A. 1991. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 10:2941–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark M. E., Lieberman P. M., Berk A. J., Dasgupta A. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dang Y., et al. 2006. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878 [DOI] [PubMed] [Google Scholar]
- 13. de Breyne S., Yu Y., Unbehaun A., Pestova T. V., Hellen C. U. 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U. S. A. 106:9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dodd D. A., Giddings T. H., Jr., Kirkegaard K. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fitzgerald K. D., Semler B. L. 2009. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta 1789:518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujimura K., Kano F., Murata M. 2008. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 14:425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilks N., et al. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gustin K. E., Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hellen C. U. T., et al. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivanov P. A., Chudinova E. M., Nadezhdina E. S. 2003. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290:227–233 [DOI] [PubMed] [Google Scholar]
- 21. Jones C., Ehrenfeld E. 1983. The effect of poliovirus infection on the translation in vitro of VSV messenger ribonucleoprotein particles. Virology 129:415–430 [DOI] [PubMed] [Google Scholar]
- 22. Kedersha N., Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 [DOI] [PubMed] [Google Scholar]
- 23. Kedersha N., et al. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13:195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kedersha N., et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim W. J., Kim J. H., Jang S. K. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 26:5020–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., Harding H. P. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273–C284 [DOI] [PubMed] [Google Scholar]
- 28. Kondratova A. A., Neznanov N., Kondratov R. V., Gudkov A. V. 2005. Poliovirus protein 3A binds and inactivates LIS1, causing block of membrane protein trafficking and deregulation of cell division. Cell Cycle 4:1403–1410 [DOI] [PubMed] [Google Scholar]
- 29. Kuyumcu-Martinez M., et al. 2004. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J. Virol. 78:8172–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwon S., Zhang Y., Matthias P. 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21:3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindquist M. E., Lifland A. W., Utley T. J., Santangelo P. J., Crowe J. E., Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lloyd R. 2006. Translational control by viral proteinases. Virus Res. 119:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loschi M., Leishman C. C., Berardone N., Boccaccio G. L. 2009. Dynein and kinesin regulate stress-granule and P-body dynamics. J. Cell Sci. 122:3973–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mazroui R., et al. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell 17:4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McEwen E., et al. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925–16933 [DOI] [PubMed] [Google Scholar]
- 36. McInerney G. M., Kedersha N. L., Kaufman R. J., Anderson P., Liljeström P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meerovitch K., et al. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montero H., Rojas M., Arias C. F., Lopez S. 2008. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. J. Virol. 82:1496–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nonhoff U., et al. 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 18:1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ochs K., et al. 2002. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 76:2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Neill R. E., Racaniello V. R. 1989. Inhibition of translation in cells infected with a poliovirus 2Apro mutant correlates with phosphorylation of the alpha subunit of eucaryotic initiation factor 2. J. Virol. 63:5069–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pacheco A., Martinez-Salas E. 2010. Insights into the biology of IRES elements through riboproteomic approaches. J. Biomed. Biotechnol. 2010:458927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park N., Katikaneni P., Skern T., Gustin K. E. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 82:1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Piotrowska J., et al. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84:3654–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qin Q., Carroll K., Hastings C., Miller C. L. 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2alpha phosphorylation and PKR. J. Virol. 85:8798–8810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raaben M., Groot Koerkamp M. J., Rottier P. J., de Haan C. A. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 9:2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rozovics J. M., Semler B. L. 2010. Genome replication. I. The players, p. 107–125 In Ehrenfeld E., Domingo E., Roos R. P. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 48. Smith J. A., et al. 2006. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 80:2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sola I., et al. 2011. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J. Virol. 85:5136–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stoecklin G., et al. 2004. MK2-induced tristetraprolin:14-13-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tolskaya E. A., et al. 1994. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236:1310–1323 [DOI] [PubMed] [Google Scholar]
- 52. Tourriere H., et al. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Tsai N. P., Ho P. C., Wei L. N. 2008. Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO J. 27:715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsai N. P., Tsui Y. C., Wei L. N. 2009. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience 159:647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. White J. P., Cardenas A. M., Marissen W. E., Lloyd R. E. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 56. White J. P., Reineke L. C., Lloyd R. E. 2011. Poliovirus switches to an eIF2-independent mode of translation during infection. J. Virol. 85:8884–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yalamanchili P., Datta U., Dasgupta A. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]