Abstract
The genome of Epstein-Barr virus (EBV), a gammaherpesvirus with potent B-cell growth-transforming ability, contains multiple copies of a 3-kb BamHI W repeat sequence; each repeat carries (i) a promoter (Wp) that initiates transformation by driving EBNA-LP and EBNA2 expression and (ii) the W1W2 exons encoding the functionally active repeat domain of EBNA-LP. The W repeat copy number of a virus therefore influences two potential determinants of its transforming ability: the number of available Wp copies and the maximum size of the encoded EBNA-LP. Here, using recombinant EBVs, we show that optimal B-cell transformation requires a minimum of 5 W repeats (5W); the levels of transforming ability fall progressively with viruses carrying 4, 3, and 2 W repeats, as do the levels of Wp-initiated transcripts expressed early postinfection (p.i.), while viruses with 1 copy of the wild-type W repeat (1W) and 0W are completely nontransforming. We therefore suggest that genetic analyses of EBV transforming function should ensure that wild-type and mutant strains have equal numbers (ideally at least 5) of W copies if the analysis is not to be compromised. Attempts to enhance the transforming function of low-W-copy-number viruses, via the activity of helper EBV strains or by gene repair, suggested that the critical defect is not related to EBNA-LP size but to the failure to achieve sufficiently strong coexpression of EBNA-LP and EBNA2 early postinfection. We further show by the results of ex vivo assays that EBV strains in the blood of infected individuals typically have a mean of 5 to 8 W copies, consistent with the view that evolution has selected for viruses with an optimal transforming function.
INTRODUCTION
Epstein-Barr virus (EBV), a B-lymphotropic herpesvirus of the Lymphocryptovirus (LCV) genus, is widespread in human populations. Successful infection of the naïve host is postulated to depend upon the ability of this orally transmitted agent to drive the clonal expansion of newly infected B cells through transient activation of EBV growth-transforming latent genes (30). Thereafter, growth-transforming gene expression is suppressed, allowing the virus to persist as an antigenically silent infection, mainly within resting memory B cells (43) but with occasional reactivation to growth-transforming infections that are extinguished by host T-cell surveillance (16). Consistent with the importance of the transforming function to LCV biology, the greater complexity of Old World (compared to New World) LCV genomes is marked by greater elaboration of the latent gene subset (31, 49).
Here we examine one basic feature of all LCV genomes, the presence of a major internal repeat containing tandemly arranged copies of a sequence of similar size, exon content, and genomic position (31, 32, 49), referred to in EBV as the 3-kb BamHI W repeat (4). Other than one study of a small number of EBV-positive Burkitt's lymphoma-derived cell lines (1), there is surprisingly little information on the number of W repeats in wild-type EBV strains. However, it is known that two characteristics of the W repeat sequence are highly relevant to the transforming function. First, each repeat contains a copy of Wp, the first viral promoter to be activated following the infection of a resting B cell (51); each Wp copy is therefore potentially active, though to what extent Wp-initiated transcript levels depend upon Wp copy numbers is not known. Second, each W repeat also contains two exons, W1 and W2, which together encode the 66-amino-acid (aa) repeat domain of EBNA-LP (7, 35, 41, 48). This is one of the two virus-coded EBV nuclear antigens (EBNAs) first expressed from Wp-initiated transcripts in newly infected B cells, the other being the transcriptional activator EBNA2. Thereafter, EBNA2, with EBNA-LP as a coactivator, induces full latent gene expression by switching on both the pan-EBNA promoter Cp (which eclipses Wp and generates EBNA1, -2, -3A, -3B, -3C, and EBNA-LP mRNAs) and the LMP promoters (generating the LMP1 and -2 mRNAs) (21). Multiple isoforms of EBNA-LP with different numbers of W1W2-encoded repeat domains are produced from this early burst of Wp activity (13). Note that the number of W repeats determines not only the maximum size of the EBNA-LP protein that can be made but possibly also the function of the protein, since certain EBNA-LP coactivating properties have been assigned to the W1W2-encoded repeat domain (27, 28). The early burst of Wp activity has also been shown to result in expression of the viral bcl2 homologue BHRF1 (19), which appears to act (possibly together with another homologue, BALF1) to protect recently infected B cells from apoptosis (2). Note that the so-called BHRF1 microRNAs (miRNAs) also have the potential to influence B-cell transformation (12, 36), but they are unlikely to be Wp products, since their expression does not peak until later, after Wp activity has waned (3).
The present work was prompted by our earlier finding, made using bacterial artificial chromosome (BAC) technology to target mutations in Wp, that recombination readily produced EBV genomes with different W copy numbers and, importantly, that viruses with only 2 copies of the wild-type W repeat (2W) were markedly impaired in transformation compared to the 11W parental virus (45). This introduced a complication for the genetic analysis of EBV's transforming activity by BAC mutagenesis, since unrecognized differences in W copy numbers between viruses with engineered mutations elsewhere in the genome could seriously confound the interpretability of results. Here we address three questions. (i) What is the true relationship between Wp copy number and transforming function? (ii) Is impaired transformation by viruses with suboptimal Wp copy numbers primarily attributable to an effect upon peak Wp activity early postinfection or to the inability of the virus to encode an EBNA-LP species with a sufficient number of W1W2 repeat domains? (iii) What is the W copy number in wild-type virus strains in the blood of infected individuals?
MATERIALS AND METHODS
Recombinant viruses.
Recombinant EBV genomes containing defined numbers of BamHI W repeats (nW viruses) were created from the B95.8 strain-derived EBV bacterial artificial chromosome (BAC) construct 2089, as previously described (9, 45). To generate the 2WrLP virus, a shuttle vector was first generated by cloning EBNA-LP cDNA as an EcoRI fragment from pW3.1LP (41) into pIRES2-EGFP (Invitrogen) to generate the pLP-IRES–EGFP plasmid. A SmaI-PciI fragment from pCP16-hygR, containing tetracycline and hygromycin resistance cassettes, was then blunt end ligated into the AflII site of pLP-IRES–EGFP. To generate the 2WrE2 virus, a shuttle vector was first constructed by replacing the EBNA-LP cDNA in the LP shuttle vector with an AvrII-BglII fragment containing the β-globin intron and EBNA2 cDNA from pSG5-EBNA2 (50). The ability of the two shuttle vector constructs to express EBNA-LP or EBNA2 protein was confirmed by Western blotting of transiently transfected DG75 cells (an EBV-negative Burkitt's lymphoma line). Homologous recombination was then used to introduce EBNA-LP or EBNA2 cDNA into the 2W BAC genome (45), with the hygromycin resistance gene and the cytomegalovirus (CMV) promoter region acting as the homologous flanking regions. The BACs were then transfected into HEK293 cells, and clones were selected that could be induced to produce high virus titers. The EBNA2-knockout (EBNA2-KO) (45) and LMP1-KO (10) viruses, both derived from the original B95.8 2089 control, were kindly provided by W. Hammerschmidt. In all cases, virus production was induced by transfection with BZLF1 and BALF4 expression vectors, supernatants were harvested and purified by density gradient centrifugation (Optiprep; Axis Shield), and virus titration was carried out using real-time quantitative PCR (QPCR) as previously described (37).
QPCR to calculate number of W repeats.
Multiplex PCRs were carried out on nW BAC DNA by the use of primers and a FAM (6-carboxyfluorescein)-labeled probe specific for the single-copy EBV DNA polymerase gene (Pol) (18) and primers and a VIC-labeled probe specific for the multiple-copy BamHI W repeat. The BamHI W repeat assay consisted of forward primer 5′-AGGCTTAGTATACATGCTTCTTGCTTT (B95.8 coordinates 14860 to 14896), reverse primer 5′-CCCTGGCTGATGCAACTTG (B95.8 coordinates 14958 to 14940), and probe 5′-GCAGCCTAATCCCACCCAGACTAGCC (B95.8 coordinates 14937 to 14912). Dilutions of known numbers of the 1W BAC genome (containing a single copy each of Wp and Pol) were amplified as standards for the W and Pol assays. Assays carried out on BAC genomes containing different numbers of W repeats provided a standard curve relating the W PCR signal per genome (i.e., per Pol copy) to the number of W repeats. The number of W repeats in recombinant virus preparations was then calculated using this standard curve. In other experiments, the same assay was applied to DNA from the freshly isolated peripheral blood mononuclear cells (PBMCs) of patients with infectious mononucleosis (IM) or healthy carriers and to multiple EBV-infected B-cell lines independently derived from such donors by limiting dilution seeding of peripheral blood mononuclear cells and spontaneous transformation in vitro. Written informed consent was obtained for all blood donors, and this work was approved by the University of Birmingham and South Birmingham Research Ethics Committees.
B-cell infection.
B cells were positively selected from buffy coats or apheresis cones (National Health Service Blood and Transplant [NHSBT], Birmingham, United Kingdom) by the use of CD19 Dynabeads (Invitrogen) followed by detachment performed with CD19 Detachabead (Invitrogen) according to the manufacturer's protocol. The B cells were incubated overnight with recombinant virus preparations at known multiplicities of infection (MOIs). The delivery of the recombinant virus to the B cells was assessed by binding and internalization assays as previously described (38). EBV promoter usage and gene transcription were detected using real-time quantitative reverse transcription-PCR (QRT-PCR) assays (6). Antigen expression in cells was detected by immunofluorescence (IF) cell staining with PE2 (EBNA2) and JF186 (EBNA-LP) monoclonal antibodies and an Alexa Fluor 594 (Invitrogen) conjugated anti-mouse IgG secondary antibody. Protein expression was analyzed by Western blotting with PE2 (EBNA2), JF186 (EBNA-LP), 5B11 (BHRF1), 1H4 (EBNA1), T2.78 (EBNA3A), E3CA10 (EBNA3C), and CS1–4 (LMP1) antibodies or with a β-actin antibody used as a loading control (20). Six-week transformation assays were carried out using 96-well plates with fixed cell numbers and various virus doses as previously described (38).
RESULTS
Generation and validation of recombinant viruses.
Starting with the B95.8-derived BAC 2089 construct known to contain 11 repeats (here called 11W) (Fig. 1A), we generated a series of recombinant EBV BACs containing different numbers of W repeats (nW BACs). These were obtained by recombination in Escherichia coli and selection of BAC clones with wild-type BamHI restriction digests but EcoRI-HindIII digests (cutting outside the repeat region), indicating the presence of 1 to 11 W repeats; see, for example, the 2W BAC illustrated in Fig. 1B. We also obtained a 0W BAC whose precise structure was determined by sequencing using primers complementary to the unique regions in the flanking BamHI C and BamHI Y fragments; this revealed a deletion between nucleotides 13215 and 47007 of the published B95.8 sequence. Therefore, as confirmed by the BamHI digest analyses, the 0W BAC lacked a complete W repeat but contained a residual 1,850 bp of the 3′-end sequence that is naturally located in the BamHI Y fragment but does not include the Wp promoter.
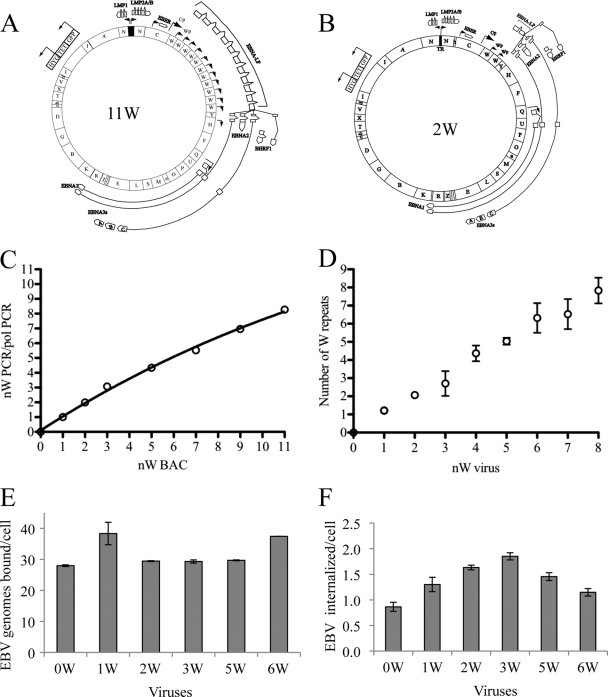
Fig. 1.
(A) Map of the B95.8 EBV strain-derived 11W BAC genome. The positions of the BamHI fragments (indicated by letters) are shown on the inner circle, promoters are represented as solid black arrows, and the spliced latent gene transcript exons, represented as boxes and connecting lines, are shown in their relative positions around the genome. (B) Map of the 11W-derived 2W BAC genome; the genome differs from that represented by panel A only in the number of BamHI W repeats and thus contains only 2 Wp promoters and produces a shorter EBNA-LP transcript. (C) A standard curve relating W QPCR signal per genome to the number of W repeats. nW and Pol PCRs (minimum of 4 replicates) were performed on a panel of BACs with known numbers of W repeats (as determined by EcoRI-HindIII restriction digestion); the ratio of mean nW PCR quantity to mean Pol PCR quantity is plotted against the number of W repeats. (D) Measurement of W repeat numbers in virus preparations. Virus preparations (1 to 4 per nW virus) were subjected to nW and Pol QPCR amplification, the nW/Pol ratio was calculated, and the numbers of W repeats were determined from the standard curve shown in panel C. (E) A representative binding assay performed on 1 × 106 B cells incubated with the 0W to 6W viruses at an MOI of 50. Data shown represent the means of the results of triplicate QPCR assays. (F) A representative nW virus internalization assay was performed on 1 × 106 B cells infected at an MOI of 50. Data shown represent the means of the results of triplicate QPCR assays.
These BACs were then transfected into HEK293 cells, recombinant EBV-containing clones selected with hygromycin and screened for virus production after induction of the lytic cycle as previously described (38). Density gradient-purified virus preparations derived from 2 different HEK293 clones were made for each nW virus, and all subsequent experiments were conducted using these duplicate virus preparations. No consistent difference in the virus titers obtained for the different nW recombinants was observed, indicating that the W repeat number did not have a significant effect on virus production (data not shown). We designed a real-time QPCR assay to check that these virus preparations did indeed carry the same number of W repeats as the parental BAC plasmid. For this purpose, we first used BAC plasmids with different W repeat numbers to construct a standard curve relating W QPCR signal per genome (i.e., per Pol gene copy number) to W repeat number (Fig. 1C). Thereafter, the different virus preparations were assayed in the same way. As shown in Fig. 1D, there was a good correlation between the expected and observed numbers of W copies in virus preparations.
Experiments were then carried out to confirm that all the nW recombinant viruses were equally capable of binding to and entering resting B cells. Binding and internalization assays were carried out as previously described (38) with resting B cells exposed to recombinant virus at a multiplicity of infection (MOI) of 50. Figures 1E and F, respectively, show binding and internalization data from one typical experiment. The values obtained in the two assays were similar to those seen in earlier work performed with wild-type EBV preparations (38). In any one experiment, there were slight differences between individual nW viruses in levels of binding (in the range of 28 to 38 genomes bound/cell) and of internalization (1 or 2 genomes internalized/cell). However, no reproducible differences were observed in assays performed on successive sets of virus preparations, from which we infer that all viruses enter cells with roughly equal levels of efficiency. Indeed, this was further confirmed by a fluorescent in situ hybridization (FISH) analysis detecting individual internalized genomes in cell nuclei (45).
Analysis of transcription and antigen expression by the 0W to 6W viruses.
Resting B cells infected with each of the nW viruses at an MOI of 50 as described above were then harvested at days 1 to 3 postinfection and subjected to QRT-PCR for determination of levels of Wp- and Cp-initiated transcripts and EBNA2 and latent BHRF1 transcripts (6). The results of a representative experiment are shown in Fig. 2A. As expected, no transcripts of any type were detected with the 0W virus, while the 1W virus either was undetectable for all transcripts or, in some experiments, gave trace signals in the Wp transcript assay only. The 2W virus gave very low overall levels of transcription, with Wp transcripts failing to show the usual early peak and Cp transcripts remaining undetectable until day 2, whereas the 3W virus (and 4W virus; data not shown) did show an early peak in Wp activity at day 1 postinfection, followed by an increasing switch to Cp. However, the overall levels of these transcripts were not as high as those from viruses 5W and 6W, which were transcriptionally very similar to each other. Generally, the EBNA2 and latent BHRF1 transcripts followed a pattern similar to that shown by Wp transcripts. Increasing BamHI W repeat copy numbers beyond 5 did not lead to any further enhancement in total transcript levels (data not shown), suggesting that the 5W virus had reached an optimal level.
Fig. 2.
(A) EBV latent gene transcription in primary B cells infected with the 0W to 6W viruses. Wp, BHRF1, EBNA2, and Cp transcripts were assayed by QRT-PCR at days 1, 2, and 3 in B cells exposed to the indicated viruses at an MOI of 50. Transcripts were normalized against GAPDH (glyceraldehyde-3-phosphate dehydrogenase); values are expressed relative to those of a standard LCL (value = 1). (B) EBV latent antigen expression in B cells infected with viruses 2W to 6W. Immunofluorescence staining was performed for EBNA-LP and EBNA2 (red) in primary B cells exposed to virus at an MOI of 50 at 3 days p.i. DAPI (blue) staining shows all nuclei in the field. The percentages of cells positive for EBNA-LP and EBNA2 are shown in the corners of the images.
In parallel experiments, immunofluorescent cell staining was carried out at 3 days postinfection to determine the expression levels of the EBNA2 and EBNA-LP proteins in infected cultures. Representative EBV antigen stains (red) and DAPI (4′,6-diamidino-2-phenylindole) counterstains (blue) are shown in Fig. 2B, with the percentages of antigen-positive cells indicated. The results were consistent with the transcription data, with 2W-infected cultures containing low numbers of antigen-expressing cells and 3W cultures containing intermediate numbers compared to those seen in 5W and 6W infection. Again, viruses with 6 or more W repeats showed no further enhancement of antigen expression at that time (data not shown).
B-cell transforming ability of the 0W to 6W viruses.
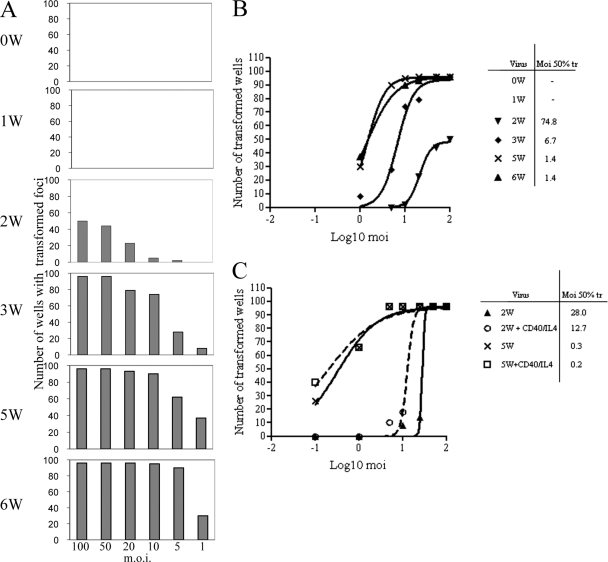
To determine the efficiency of B-cell transformation for each of the nW viruses, 106 B cells were infected at each of 6 different MOIs and plated in 96-well round-bottomed plates at 104 cells/well. The cultures were fed weekly and then scored for the number of wells showing outgrowth of transformed foci after 6 weeks. The results of one representative assay for the 0W to 6W viruses are shown in Fig. 3A as histograms denoting the incidence of transformation at different virus doses. No transformation was detected for virus 0W or 1W in this or any other assay even at the highest virus dose, whereas the yield of transformed wells increased progressively with the number of W repeats up to the maximum shown by the 5W and 6W viruses. Plotting the data from these experiments as log MOIs against the numbers of wells showing transformation allowed calculation of the MOI required for 50% transformation for each virus (Fig. 3B). This type of transformation assay was repeated several times and gave a consistent pattern of results, even though absolute transformation efficiencies can differ between individual experiments. Thus, the transformation efficiency of the 2W virus was around 100-fold lower than the transformation efficiencies of the 5W and 6W viruses, whereas the 3W and 4W viruses gave intermediate efficiency values typically between 20- and 5-fold lower than the values determined for the 5W and 6W viruses.
Fig. 3.
Transformation assays performed with nW viruses. (A) Histograms representing the numbers of 96 replicate wells, each seeded at 104 cells/well, that transformed after infection of cells with the 0W to 6W viruses at the indicated MOI; the data represent the results from one representative assay scored at 6 weeks p.i. (B) Calculation of the transformation efficiency of the 0W to 6W viruses. The MOI required for transformation of 50% of the wells (Moi 50% tr) was determined by plotting the number of transformed wells as shown in panel A against the log10 MOI. (C) The effect of a mitogenic signal on transformation. Transformation assays were carried out as described for panel A with the 2W and 5W viruses with or without the addition of soluble CD40 ligand (50 ng/ml) and IL-4 (50 ng/ml). The numbers of wells showing transformation at 6 weeks p.i. were plotted against the log10 MOI to determine the MOI required for transformation of 50% of the wells.
The results so far indicate that the impaired transforming ability of viruses with suboptimal W copy numbers was associated with reduced transcriptional activity in the first few days postinfection of resting B cells, i.e., the period in which EBNA2 and EBNA-LP cooperate to drive the cells into the cycle through the activation of key cellular genes (29, 39). We therefore asked whether a mitogenic stimulus delivered to B cells immediately postinfection might, by initiating cell cycle entry in infected cells, substitute for those early functions of EBNA2 and EBNA-LP and restore full transforming function to viruses with low W numbers. Figure 3C shows data from an experiment in which B cells infected with the 2W or 5W virus were cultured in 96-well plates in the presence or absence of soluble CD40 ligand and interleukin-4 (IL-4) for 1 week postinfection, the cytokines then being gradually diluted by the weekly feeding of the cultures. The mitogenic signal clearly had only a small effect on the transforming ability of the 2W virus, causing at most a 2-fold reduction in the MOI required for 50% transformation, and did not significantly affect transformation by the 5W virus. We suggest that the small improvement in transformation by the 2W virus simply reflects the fact that CD40L/IL-4 stimulation increases overall B-cell survival during the first week in culture.
EBNA2 and LMP1 knockout helper viruses.
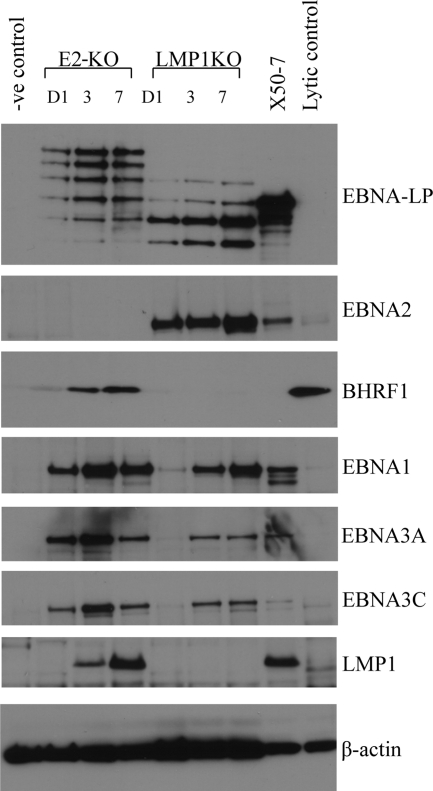
Further attempts to restore the transforming function of the 0W, 1W, and 2W viruses were then made using EBNA2-KO and LMP1-KO recombinant EBVs as helpers, i.e., virus strains which are themselves transformation incompetent but can infect B cells and provide complementary expression of some latent proteins. As a preliminary procedure, we analyzed viral antigen expression by Western blotting of B cells at 1, 3, and 7 days after exposure to these viruses; the data are shown in Fig. 4.
Fig. 4.
EBV latent gene expression in B cells infected with EBNA2 KO (E2-KO) and LMP1-KO helper viruses at an MOI of 50. Western blot assays were carried out to detect EBNA-LP, EBNA2, BHRF1, EBNA1, EBNA3A, EBNA3C, and LMP1 expression at days 1, 3, and 7 p.i. Primary uninfected B cells served as a negative control (-ve), the EBV-transformed X50-7 cell line was used as a positive control for EBNA-LP, EBNA-2, EBNA1, EBNA3A, EBNA3C, and LMP1 expression, and Akata-BL cells induced into lytic replication by surface IgG cross-linking (60% cells in the lytic cycle) were used as a positive control for BHRF1 expression (lytic control). β-Actin blotting served as a loading control.
We had anticipated that the EBNA2-KO virus would express the Wp-driven gene products EBNA-LP and latent BHRF1 but not the other latent proteins, since we saw very little Cp activity and no blast transformation or cell cycle entry in EBNA2-KO-infected cultures (45). However, we found that the EBNA2-KO virus expressed not only EBNA-LP and BHRF1 but also all of the other EBNAs (the EBNA1 and EBNA3 proteins). Moreover, the other EBNAs were readily detectable by 1 day postinfection, even earlier than they appeared in wild-type virus infections, presumably because, as under other circumstances (20), their expression from the EBNA2-deleted genome was being driven by Wp rather than Cp. Even more unexpectedly, the LMP1 protein was expressed by day 3 in EBNA2-KO-infected cells, a result which we investigated further and found to be coincident with transcription from the EBNA2-independent LMP1 terminal repeat promoter (34) (data not shown). By contrast, the LMP1-KO virus behaved more as expected, expressing EBNA-LP and EBNA2 from an active Wp by day 1 (albeit with only trace BHRF1 expression), followed by EBNA1 and EBNA3 proteins at the more usual time of day 3, once Cp had become active. As previously reported (10), the virus induced transient proliferation of infected B cells over the first 7 to 10 days, but, in the absence of the LMP1 protein, this proliferation was not sustained. Note that both the EBNA2-KO and LMP1-KO viruses encode EBNA-LP as a ladder of proteins with different numbers of W1W2-coded (66-aa) repeat domains. In each case, the size of the largest EBNA-LP product corresponded to that predicted from the numbers of W repeats in the EBNA2-KO and LMP1-KO genomes, which we determined to be 8 and 6 repeats, respectively (data not shown).
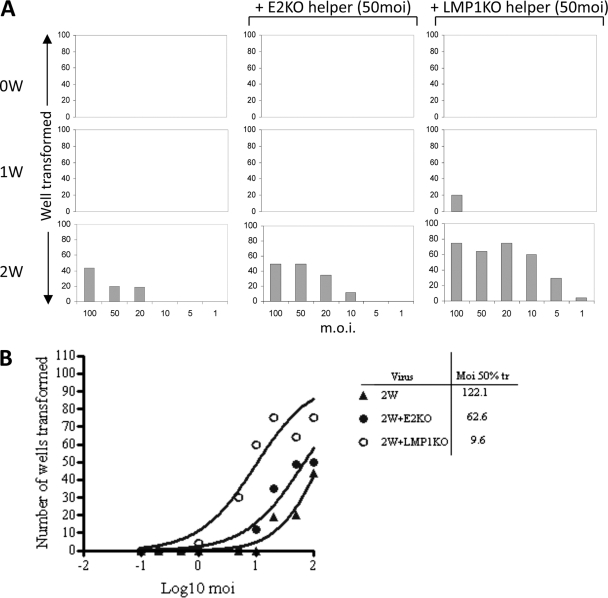
We next examined whether the transformation-defective viruses described above could act as helpers for the 0W, 1W, and 2W viruses in transformation assays. The assays again used the nW viruses at 6 different MOIs but with the addition of a standard dose (MOI of 50) of either EBNA2-KO or LMP1-KO virus. Figure 5 shows the results from one representative assay, with the number of transformed wells plotted against the MOI of nW virus. As can be seen, the EBNA2-KO virus slightly enhanced B-cell transformation by the 2W virus, with foci now being detectable in some wells at a 2W virus input MOI of 10 (rather than 20) in this particular example. Over several such assays, coinfection with the EBNA2-KO helper virus resulted in a 2- to 6-fold reduction in the 2W MOI required for 50% transformation. As a further confirmation of this small helper effect, when cell lines from the assay wells were established, PCR analysis detected the continued presence of both 2W (EBNA2-positive) and EBNA2-KO genomes in the transformed cells (data not shown). Note that coinfection with the EBNA2-KO helper never resulted in transformation by the 0W and 1W viruses even at their highest doses. In the same assays, by comparison, the LMP1-KO virus had a much more marked helper effect. Thus, coinfection with LMP1-KO resulted in a reduction of over 10- to 20-fold in the MOI of 2W virus required for 50% transformation (Fig. 5B). Moreover, in the presence of the LMP1-KO helper, transformation by the 1W virus became detectable for the first time, albeit in cells infected at the highest MOI. This was a reproducible finding in several experiments, with 20 of 96 wells showing transformed foci in the experiment illustrated in Fig. 5A. Again, PCR analysis revealed the presence of both LMP1-positive and LMP1-KO genomes in cell lines expanded from the 1W/LMP1-KO and 2W/LPM1-KO coinfections (data not shown).
Fig. 5.
Transformation assays with nW and helper viruses. (A) Primary B cells were exposed to viruses at each of the indicated MOIs (as described for Fig. 1) with or without the addition of the E2-KO or LMP1-KO helper virus at a constant MOI of 50. The numbers of wells showing transformation were scored at 6 weeks p.i. (B) The results of the transformation assays described for panel A were plotted as the numbers of transformed wells versus the log10 MOI to determine the MOI required for 50% transformation.
Repaired EBNA-LP and EBNA2 expression in 2W viruses.
As a complement to the helper virus approach, we turned to a second means of determining whether artificially increasing EBNA-LP or EBNA2 expression from a low-W-copy-number virus genome might improve the transforming function of the virus. This involved inserting either an EBNA-LP or EBNA2 coding sequence into the 2W virus genome under the control of a CMV immediate-early (IE) promoter rather than the native Wp.
A CMV IE promoter-driven cassette carrying a complete EBNA-LP cDNA that included 7 copies of the W1W2 exons (7LP) was generated and expression of the appropriately sized EBNA-LP protein confirmed by Western blotting of transiently transfected DG75 cells. As illustrated in Fig. 6A, the 7LP cassette was then introduced by recombination into the 2W BAC to generate a 2WrLP BAC. The 7LP cassette in the BAC was verified by sequencing, and the genome integrity of the 2WrLP BAC was confirmed by restriction enzyme digestion (data not shown). Initial assays were carried out, confirming that the viruses produced from the 2 different 2WrLP HEK293 clones (2WrLP-1 and -2) bound and were internalized with similar levels of efficiency to each other and to all the other nW viruses (data not shown).
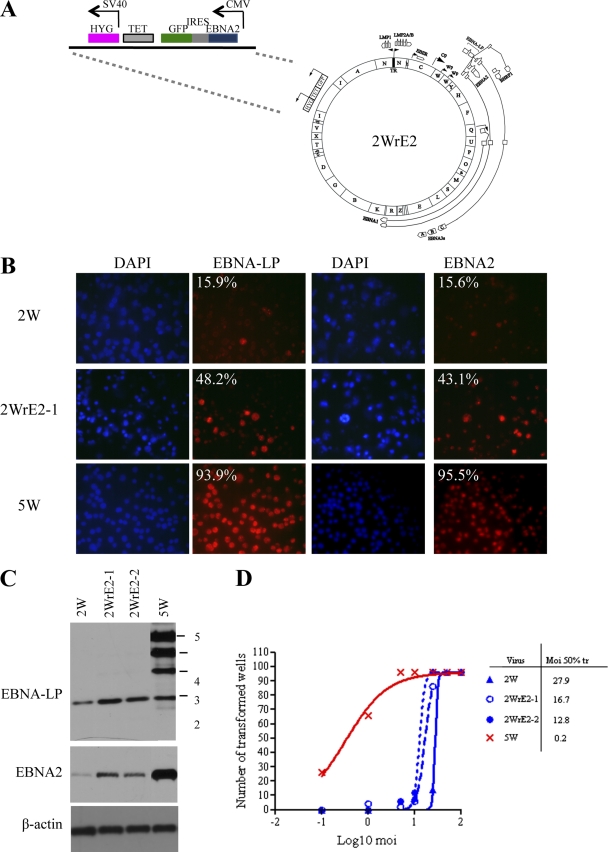
Fig. 6.
EBNA-LP repair of the 2W virus. (A) Map of the 2WrLP BAC, including details of the EBNA-LP expression cassette that was recombined into the 2W BAC genome in place of the green fluorescent protein (GFP)-tetracycline-hygromycin selection cassette. (B) EBV latent antigen expression in B cells infected with 2W, EBNA-LP-repaired 2WrLP-1, and 5W viruses. Immunofluorescence staining was performed for EBNA-LP and EBNA2 in primary B cells exposed to virus at an MOI of 50 at 3 days p.i. DAPI (blue) staining shows all nuclei in the field. The percentages of cells positive for EBNA-LP and EBNA2 (red) staining are shown in the corners of the images. (C) EBNA-LP and EBNA2 expression in 2WrLP-infected cells. Primary B cells were infected with 2WrLP virus preparations derived from 2 different 293 producer clones (2WrLP-1 and -2) or with 2W and 5W control viruses at an MOI of 50. Cell lysates at 3 days postinfection were subjected to Western blotting to determine levels of EBNA-LP and EBNA2 expression, with β-actin serving as a loading control. (D) Transformation efficiency of the EBNA-LP-repaired virus. Transformation assays were carried out with 2WrLP-1 and -2 or with 2W and 5W control viruses (as described for Fig. 1) to determine the efficiency of transformation; the results are expressed as the MOIs required for 50% transformation.
Primary B cells were therefore exposed to the 2WrLP, 2W, and 5W viruses at an MOI of 50 and, as described above, stained for EBNA-LP- and EBNA-2-expressing cells on day 3 p.i. Results of representative antigen staining (red) and DAPI counterstaining (blue) and the percentages of antigen-positive cells in these cultures are shown in Fig. 6B. Both the percentage of EBNA-LP-positive cells and the intensity of EBNA-LP staining were higher in 2WrLP-1-infected than in 2W-infected cells; interestingly, EBNA2 staining also seemed to increase, but neither antigen reached the expression levels seen with the 5W virus. Experiments performed with a second recombinant virus, 2WrLP-2, produced a similar pattern of results (data not shown). Western blots of cell lysates harvested on day 3 p.i. also showed increased levels of EBNA-LP and, to some extent, EBNA2 proteins in cells infected with the 2WrLP-1 and 2WrLP-2 viruses compared to the parental 2W virus results (Fig. 6C). Notably, however, the 2WrLP viruses do not produce a full-length 7-repeat EBNA-LP protein, the predominant bands on the blot representing LP proteins with 2, 3, and 4 repeats, whereas the 2W virus produces only a 2-repeat protein and the 5W virus predominantly produces proteins with 5, 4, and 3 repeats (Fig. 6C). Note that RT-PCR analysis performed using primers specific to the EBNA-LP expression cassette transcript confirmed that the introduced EBNA-LP cDNA was indeed expressed in the 2WrLP-infected cells within the first few days of infection (data not shown). The same viruses were then compared in transformation assays; the results of a representative experiment are shown in Fig. 6D. Both the 2WrLP-1 and 2WrLP-2 viruses transformed more efficiently than 2W but still not as well as 5W. This was a consistent observation, with improvements in transformation efficiency of 2- to 9-fold (mean, 5-fold) across several experiments.
Figure 7 shows the corresponding sets of results obtained using EBNA2 gene-inserted viruses. Again, the genome integrity of the resulting 2W-EBNA2 BAC (2WrE2) (Fig. 7A) was verified by restriction digestion, stable virus-producing 293 clones were generated, 2 clones (2WrE2-1 and 2WrE2-2) were selected to produce virus stock, and efficient binding of the viruses to B cells was confirmed (data not shown). Cells infected with the 2WrE2 viruses consistently showed increased levels of EBNA2 expression at 3 days p.i. by immunofluorescence staining and Western blotting, and RT-PCR analysis confirmed that at least some of the EBNA2 expression was derived from the expression cassette (data not shown). Interestingly, we also noticed that expression of the appropriately sized EBNA-LP from the 2W virus genome was slightly increased in 2WrE2-infected cells. Note that, despite these increases, EBNA2 and EBNA-LP expression levels were again still well below those seen in 5W virus infections (Fig. 7B and C). Finally, as illustrated in Fig. 7D (results are from the same assay as that shown for 2WrLP viruses in Fig. 6D), both 2WrE2 viruses transformed B cells slightly better than the parental 2W virus, with 2- to 8-fold (mean, 5-fold) increases in transformation efficiency consistently observed across several assays.
Fig. 7.
EBNA-2 repair of the 2W virus. (A) Map of the 2WrE2 BAC, including details of the EBNA2 expression cassette that was recombined into the 2W BAC genome in place of the GFP-tetracycline-hygromycin selection cassette. (B) EBV latent antigen expression in B cells infected with 2W, EBNA2-repaired 2WrE2-1, and 5W viruses. Immunofluorescence staining was performed for EBNA-LP and EBNA2 in primary B cells exposed to virus at an MOI of 50 at 3 days p.i. DAPI (blue) staining shows all nuclei in the field. The percentages of cells positive for EBNA-LP and EBNA2 (red) staining are shown in the corners of the images. (C) EBNA-LP and EBNA2 expression in 2WrE2-infected cells. Primary B cells were infected with 2WrE2 virus preparations derived from 2 different 293 producer clones (2WrE2-1 and -2) or 2W and 5W control viruses at an MOI of 50. Cell lysates at 3 days postinfection were subjected to Western blotting to determine levels of EBNA-LP and EBNA2 expression, with β-actin serving as a loading control. (D) Transformation efficiency of the EBNA2-repaired virus. Transformation assays were carried out with 2WrE2-1 and -2 or with 2W and 5W control viruses (as described for Fig. 1) to determine the efficiency of transformation, expressed as the MOI required for 50% transformation.
BamHI W repeat copy numbers in EBV strains in vivo and in vitro.
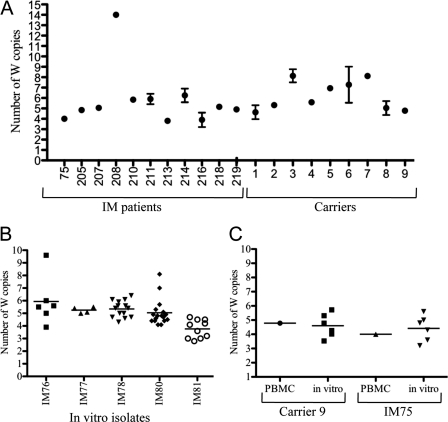
The relationship between W copy number and the EBV growth-transforming function, shown above for in vitro studies, prompted us to examine the W copy content of EBV strains circulating in the wild, an issue that has received very little attention to date. Figure 8A shows the results obtained when the newly established W copy assay (Fig. 1C and D) was applied to DNA samples from freshly isolated peripheral blood mononuclear cell samples from 11 patients with infectious mononucleosis (IM) and from 9 long-term virus carriers. The majority (16/20) gave W repeat copy numbers between 5 and 8, three IM patients gave a value of 4, and one IM patient gave a value of 14, an unusually high number. Note that, for each individual, the ex vivo assay gives a mean value representing all the EBV genomes present within latently infected B cells in the blood at the time of the assay. We were therefore interested to look at the range of W repeat numbers present in viruses rescued from the blood by spontaneous transformation in vitro. Figure 8B shows results determined with multiple B-cell lines independently established from each of 5 different IM patients bled in the acute phase of infection. In each case, the W repeat numbers in these cells were distributed around a mean lying between 4 and 6; in three cases, the distribution around the mean was reasonably tight, but for two patients (IM76 and IM81), their in vitro EBV isolates included occasional outliers with high W repeat copy numbers. Figure 8C shows data from one IM patient and one long-term carrier where we were able to assay the original ex vivo sample and multiple in vitro lines established from the same bleed. For both donors, W repeat numbers in the in vitro isolates were distributed fairly tightly around a mean that was similar to the value determined for the original ex vivo sample.
Fig. 8.
The numbers of W repeats in naturally occurring virus strains. (A) The numbers of W repeats in ex vivo samples. A PBMC sample from each of 11 IM patients and 9 healthy virus carriers was subjected to nW and Pol QPCR (3 to 4 replicates) in order to calculate the number of W repeats in resident EBV genomes. The means and standard errors are shown. (B) The number of W repeats in in vitro isolates. For 5 IM donors, multiple independent in vitro spontaneously transformed cell lines, derived from a single bleed, were subjected to nW and Pol QPCR (in duplicate) in order to calculate the number of W repeats in the resident virus strain. The mean QPCR result for each cell line is shown as an individual point and the mean value for all the cell lines from each donor as a horizontal line. (C) Comparison of the number of W repeats in ex vivo PBMCs and in vitro isolates from the same bleed of one IM patient and one virus carrier. For each sample, the mean of the results of nW and Pol PCRs (carried out in duplicate) is shown as an individual point and the mean value for all the cell lines from each donor as a horizontal line.
DISCUSSION
The major internal repeat is a common feature of all LCV genomes studied so far, with multiple tandemly arranged copies of a sequence of similar size, exon content, and genomic position being found not just throughout Old World primate LCVs but even in CalHV-3, the distantly related New World primate LCV (31, 32). From the study of EBV, it is known that each W repeat contains both the W promoter that initiates B-cell growth-transforming infections and two exons, W1 and W2, that together encode the repeat domain of EBNA-LP, a protein that is highly expressed from Wp early in the transformation process. Despite the likely importance of the W repeat to virus biology, very little is known about the influence the repeat number has upon the transforming function of the virus or about the numbers of repeats present in naturally circulating EBV strains.
The starting point for this work was our observation that, during the manipulation of the B95.8 EBV strain BAC in recA-positive E. coli, W repeat copies can be lost from the genome, thereby providing a series of products with different numbers of W repeats (45); thereafter, the BACs can be maintained in recA-deficient hosts without further alteration. For the present work, we developed a QPCR-based quantitative assay to determine W repeat numbers that confirmed that the repeat number was also faithfully maintained both in the EBV BAC carried as an episome in stably transfected 293 cell clones and in the virus preparations produced from those cell clones following induction of the EBV lytic cycle. Furthermore, the nW virus preparations were all capable of binding to and entering B cells with efficiencies similar to those shown by wild-type EBV. Thus, we believe that differences in the progress of infection in resting B cells can reasonably be ascribed to differences in W repeat numbers.
Under conditions of high MOIs, where the EBV genome accesses the nucleus of most if not all resting B cells, we noted a striking, nonlinear relationship between the number of W repeats and the level of Wp activity, which typically peaks at 1 day postinfection. Wp transcripts were never detected in 0W and rarely in 1W virus infections. However, such transcripts were reproducibly detectable at a low level with the 2W virus and levels then increased stepwise with extra W copies, reaching a maximum shown by the 5W virus that was >10-fold greater than the level seen with 2W; thereafter, increasing W copy numbers above 5 had no further effect. Interestingly, EBNA2 and latent BHRF1 transcript levels showed a similar relation to the W copy number, which is consistent with the transcripts being products of Wp activity at early times postinfection (19, 51). These differences between viruses with respect to Wp transcription were closely mirrored at the protein level when, on day 3 p.i., the percentage of B cells with detectable EBNA2 and EBNA-LP expression was determined by immunofluorescence staining. By contrast, in long-term outgrowth assays, the impact of suboptimal W copy numbers was much more marked, with the 2W viruses now 100-fold less efficient than the 5W optimum. This is not surprising, given that transformation is a more demanding process than acute infection, with culling and clonal selection involved in lymphoblastoid cell line (LCL) outgrowth (24, 26, 33, 42); furthermore, transformation assay endpoints reflect viral activity under dose-limiting conditions rather than at the high MOIs used in the short-term infection experiments. Importantly, however, even using this more demanding readout, we never detected any enhancement of transforming function when the W copy number was increased above 5.
Based on these findings, we conclude that there is a clear connection between the level of Wp transcription initiated postinfection of a resting B cell and the transforming efficiency of the virus, with a 1W copy being virtually inactive in both respects and 5W copies sufficient to achieve optimal levels. The data suggest that, for the 2W, 3W, 4W, and 5W viruses, all available copies of Wp (except perhaps the most 3′ copy) are transcriptionally active early in infection. This would accord with the notion that all the cellular factors required for Wp activity are present in resting B cells (5, 22, 44) and that they can access the incoming unmethylated viral genome (17, 47). Furthermore, using multiple Wps also helps to explain how the 5W virus encodes EBNA-LPs with 5, 4, 3, and 2 repeat domains (see Fig. 6C and 7C). Interestingly, however, 5 copies represents a threshold beyond which adding more W copies has little effect on total W transcription. Why this threshold exists is unclear, but it may reflect interference by transcripts initiated at upstream Wp copies with the firing of downstream Wps, just as transcriptional interference by Cp-initiated transcripts is thought to contribute to the eventual reduction of Wp activity (52). Whatever the reason, the existence of this threshold has important implications for an increasingly active area of research, the genetic analysis of EBV transforming function performed using mutants generated by BAC cloning (11). Thus, all genetically manipulated constructs need to be checked not just for the engineered mutation in the targeted gene but also for the number of W copies. We suggest that wild-type and mutant strains should have equal numbers (ideally at least 5) of W copies to avoid compromising the analysis.
Three proteins, EBNA-LP, EBNA2, and BHRF1, are thought to be encoded by Wp-initiated transcripts in the first few days postinfection. Impaired transformation by low-W-copy-number viruses could therefore reflect suboptimal expression of one or all of these proteins, with any effect on EBNA-LP levels possibly compounded by the limited number of W1W2-encoded repeat domains in that protein. Of these possible explanations, a change in the BHRF1 expression level seemed least likely, since a complete knockout of the BHRF1 gene has relatively little effect on early events postinfection and then only at limiting virus doses (2, 36). By contrast, EBNA-LP and EBNA2 are both considered essential for the transformation process (8, 14, 25). We therefore undertook further experiments to investigate whether increasing EBNA-LP and/or EBNA2 expression in low-W-copy-number virus infections could restore transforming efficiency. In this regard, the helper virus experiments showed that the EBNA2-KO virus, which provided optimal expression of a full-length EBNA-LP (plus BHRF1 and all other latent proteins) in the absence of EBNA2, only slightly improved transformation by 2W virus. By contrast, the LMP1-KO virus, which provided optimal expression of full-length EBNA-LP and EBNA2 (plus the other EBNAs but minimal BHRF1), had a much greater helper effect, improving the transforming efficiency of the 2W virus by up to 20-fold and even revealing the transforming potential of the 1W virus. Clearly, therefore, the impaired transforming function of low-W-copy-number viruses could not be compensated simply by providing full-length EBNA-LP but was greatly improved if increased levels of both EBNA-LP and EBNA2 were supplied early postinfection.
There are, however, limitations to the helper virus approach. First, the experiments are dependent on the ability of both test and helper viruses to coinfect the same cells, and competition for binding and entry could be a complication at low MOIs of the test virus. Second, as the EBNA2-KO virus data show, antigen expression from helper viruses is not always as restricted as one might anticipate. Third, and most crucially, there is no corresponding EBNA-LP-KO virus available with which to investigate whether providing EBNA2 alone would rescue efficient transformation by low-W-copy-number viruses. For these various reasons, we took the alternative approach of repairing the 2W virus genome with either a full-length EBNA-LP or EBNA2 coding sequence under the control of the strong CMV IE promoter. Consistent results were obtained with two independent virus isolates per construct. Both LP-repaired viruses showed increased levels of EBNA-LP expression in B cells 3 days p.i. (although the levels were still well short of those shown by the 5W virus) and transformation efficiencies that were, on average, 5-fold greater than that of the parental 2W virus. Notably, this improvement occurred in the absence of a full-length EBNA-LP, since, for reasons that are not clear, the sizes of the CMV promoter-driven LP species indicated the presence of proteins with just 3 and 4 (rather than the expected 7) W1W2 repeat domains. In the same assays, the EBNA2-repaired viruses also showed enhancement of EBNA2 expression on day 3 p.i., although (as noted with the LP-repaired viruses and EBNA-LP expression) EBNA2 expression levels remained well below that achieved by the 5W virus. Importantly, EBNA2 repair was no more effective at improving transformation than was EBNA-LP repair, again giving only a 5-fold improvement. In this context, one should also note that increasing viral expression of EBNA-LP or EBNA2 via the CMV promoter-driven insert led in each case to a slight increase, detectable 3 days p.i., in the levels of the non-CMV-driven partner antigen (i.e., of EBNA2 and EBNA-LP, respectively), possibly as a result of an increased cooperative effect on EBV genome activation.
These findings, taken together with those from the helper virus work, suggest that the impaired transforming function of low-W-copy-number viruses is primarily due to the suboptimal levels of Wp transcription from the incoming virus genome and therefore to the suboptimal levels of the two key transactivators of viral and cellular gene expression, EBNA 2 and EBNA-LP. We see no clear evidence that EBNA-LP size, that is, the number of W1W2-encoded repeat domains in the protein, is a major limiting factor in low-W-copy-number virus infections. This is indeed consistent with the results of other studies examining EBNA-LP coactivating function in transient transfection assays, which demonstrated that an EBNA-LP with just 2 repeat domains could efficiently coactivate Cp and the LMP1 promoter (15, 27, 28).
What is the relevance of these findings in the broader context of EBV biology? The extent to which EBV's colonization of the naïve host depends upon B-cell transforming function remains unclear (23, 30, 43). If evolution has indeed selected for, among other things, good transforming function, then we would predict from our in vitro studies that contemporary EBV strains in the wild would have preserved adequate numbers of W repeats. Interestingly, only one attempt has been made to measure W repeat numbers in EBV strains; however, that study focused on viruses in EBV-positive Burkitt's lymphoma cell lines, a sample that may not be generally representative of wild-type stains (1). The present study, applying a new QPCR assay to ex vivo peripheral blood mononuclear cell samples, provides the first data on viruses circulating in the general population. Of 20 individuals examined during the primary or persistent stage of infection, 17 gave W repeat values of 5 or greater, while the others gave a value of 4. We point out that these are mean values for all resident virus genomes in a sample and that, given the fact that many individuals carry more than one EBV strain (40, 46), there may be a range of genomes present with different copy numbers. Preliminary evidence from experiments performed with in vitro isolates suggests that this may be the case in some individuals, but even in such cases most in vitro isolate values cluster closely around the ex vivo mean. More work is needed to fully resolve this interesting issue. However, the balance of current evidence supports the general concept that wild-type strains have been selected to carry sufficient numbers of W repeats to preserve good transforming function.
ACKNOWLEDGMENTS
This work was funded by Cancer Research UK (CR-UK) grant C910/A8829. K.-Y.K. was supported by a studentship from the Lady Tata Memorial Trust.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Allan G. J., Rowe D. T. 1989. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt's lymphoma and lymphoblastoid cell lines. Virology 173:489–498 [DOI] [PubMed] [Google Scholar]
- 2. Altmann M., Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amoroso R., et al. 2011. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J. Virol. 85:996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baer R., et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 [DOI] [PubMed] [Google Scholar]
- 5. Bell A., Skinner J., Kirby H., Rickinson A. 1998. Characterisation of regulatory sequences at the Epstein-Barr virus BamHI W promoter. Virology 252:149–161 [DOI] [PubMed] [Google Scholar]
- 6. Bell A. I., et al. 2006. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J. Gen. Virol. 87:2885–2890 [DOI] [PubMed] [Google Scholar]
- 7. Bodescot M., Chambraud B., Farrell P., Perricaudet M. 1984. Spliced RNA from the IR1-U2 region of Epstein-Barr virus: presence of an open reading frame for a repetitive polypeptide. EMBO J. 3:1913–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen J. I., Wang F., Mannick J., Kieff E. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. U. S. A. 86:9558–9562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delecluse H. J., Hammerschmidt W. 2000. The genetic approach to the Epstein-Barr virus: from basic virology to gene therapy. Mol. Pathol. 53:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dirmeier U., et al. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982–2989 [PubMed] [Google Scholar]
- 11. Feederle R., Bartlett E. J., Delecluse H. J. 2010. Epstein-Barr virus genetics: talking about the BAC generation. Herpesviridae 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feederle R., et al. 2011. A Viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 7:e1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finke J., et al. 1987. Monoclonal and polyclonal antibodies against Epstein-Barr virus nuclear antigen 5 (EBNA-5) detect multiple protein species in Burkitt's lymphoma and lymphoblastoid cell lines. J. Virol. 61:3870–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammerschmidt W., Sugden B. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393–397 [DOI] [PubMed] [Google Scholar]
- 15. Harada S., Kieff E. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hislop A. D., Taylor G. S., Sauce D., Rickinson A. B. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu. Rev. Immunol. 25:587–617 [DOI] [PubMed] [Google Scholar]
- 17. Hutchings I. A., et al. 2006. Methylation status of the Epstein-Barr virus (EBV) BamHI W latent cycle promoter and promoter activity: analysis with novel EBV-positive Burkitt and lymphoblastoid cell lines. J. Virol. 80:10700–10711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Junying J., et al. 2003. Absence of Epstein-Barr virus DNA in the tumor cells of European hepatocellular carcinoma. Virology 306:236–243 [DOI] [PubMed] [Google Scholar]
- 19. Kelly G. L., et al. 2009. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in Burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 5:e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly G. L., et al. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt's lymphoma cells and with increased resistance to apoptosis. J. Virol. 79:10709–10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kieff E., Rickinson A. B. 2007. Epstein-Barr virus and its replication, p. 2603–2654 In Knipe D. M., Howley P. M. (ed.), Fields virology, vol. 5 Lippincott-Williams and Wilkins Publishers, Philadelphia, PA [Google Scholar]
- 22. Kirby H., Rickinson A., Bell A. 2000. The activity of the Epstein-Barr virus BamHI W promoter in B cells is dependent on the binding of CREB/ATF factors. J. Gen. Virol. 81:1057–1066 [DOI] [PubMed] [Google Scholar]
- 23. Küppers R. 2003. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat. Rev. Immunol. 3:801–812 [DOI] [PubMed] [Google Scholar]
- 24. Lacoste S., et al. 2010. Chromosomal rearrangements after ex vivo Epstein-Barr virus (EBV) infection of human B cells. Oncogene 29:503–515 [DOI] [PubMed] [Google Scholar]
- 25. Mannick J. B., Cohen J. I., Birkenbach M., Marchini A., Kieff E. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 65:6826–6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nikitin P. A., et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nitsche F., Bell A. I., Rickinson A. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng R., Tan J., Ling P. D. 2000. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953–9963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Portal D., et al. 2011. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc. Natl. Acad. Sci. U. S. A. 108:7808–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rickinson A. B., Kieff E. 2007. Epstein-Barr virus, p. 2655–2700 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed., vol. II Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 31. Rivailler P., Cho Y. G., Wang F. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055–12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rivailler P., Jiang H., Cho Y. G., Quink C., Wang F. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ryan J. L., et al. 2006. Clonal evolution of lymphoblastoid cell lines. Lab. Invest. 86:1193–1200 [DOI] [PubMed] [Google Scholar]
- 34. Sadler R. H., Raab-Traub N. 1995. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J. Virol. 69:4577–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sample J., Hummel M., Braun D., Birkenbach M., Kieff E. 1986. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc. Natl. Acad. Sci. U. S. A. 83:5096–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seto E., et al. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 6:e1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shannon-Lowe C., et al. 2009. Features distinguishing Epstein-Barr virus infections of epithelial cells and B cells: viral genome expression, genome maintenance, and genome amplification. J. Virol. 83:7749–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shannon-Lowe C., et al. 2005. Epstein-Barr virus-induced B-cell transformation: quantitating events from virus binding to cell outgrowth. J. Gen. Virol. 86:3009–3019 [DOI] [PubMed] [Google Scholar]
- 39. Sinclair A. J., Palmero I., Peters G., Farrell P. J. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sitki-Green D., Covington M., Raab-Traub N. 2003. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J. Virol. 77:1840–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Speck S. H., Pfitzner A., Strominger J. L. 1986. An Epstein-Barr virus transcript from a latently infected, growth-transformed B-cell line encodes a highly repetitive polypeptide. Proc. Natl. Acad. Sci. U. S. A. 83:9298–9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sugden B., Mark W. 1977. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J. Virol. 23:503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thorley-Lawson D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75–82 [DOI] [PubMed] [Google Scholar]
- 44. Tierney R., Kirby H., Nagra J., Rickinson A., Bell A. 2000. The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5. J. Virol. 74:10458–10467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tierney R., et al. 2007. Epstein-Barr virus exploits BSAP/Pax5 to achieve the B-cell specificity of its growth-transforming program. J. Virol. 81:10092–10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tierney R. J., et al. 2006. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J. Infect. Dis. 193:287–297 [DOI] [PubMed] [Google Scholar]
- 47. Tierney R. J., et al. 2000. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 74:10468–10479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang F., Petti L., Braun D., Seung S., Kieff E. 1987. A bicistronic Epstein-Barr virus mRNA encodes two nuclear proteins in latently infected, growth-transformed lymphocytes. J. Virol. 61:945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang F., Rivailler P., Rao P., Cho Y. 2001. Simian homologues of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang F., Tsang S. F., Kurilla M. G., Cohen J. I., Kieff E. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Woisetschlaeger M., Yandava C. N., Furmanski L. A., Strominger J. L., Speck S. H. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 87:1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoo L. I., Mooney M., Puglielli M. T., Speck S. H. 1997. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J. Virol. 71:9134–9142 [DOI] [PMC free article] [PubMed] [Google Scholar]