Abstract
Long interspersed nuclear element 1 (LINE-1; L1) retrotransposons are the most common retroelements in mammalian genomes. Unlike individual families of endogenous retroviruses (ERVs), they have remained active throughout the mammalian radiation and are responsible for most of the retroelement movement and much genome rearrangement within mammals. They can be viewed as occupying a substantial niche within mammalian genomes. Our previous demonstration that L1s and B1 short interspersed nuclear elements (SINEs) are inactive in a group of South American rodents led us to ask if other elements have amplified to fill the empty niche. We identified a novel and highly active family of ERVs (mysTR). To determine whether loss of L1 activity was correlated with expansion of mysTR, we examined mysTR activity in four South American rodent species that have lost L1 and B1 activity and four sister species with active L1s. The copy number of recent mysTR insertions was extremely high, with an average of 4,200 copies per genome. High copy numbers exist in both L1-active and L1-extinct species, so the mysTR expansion appears to have preceded the loss of both SINE and L1 activity rather than to have filled an empty niche created by their loss. It may be coincidental that two unusual genomic events—loss of L1 activity and massive expansion of an ERV family—occur in the same group of mammals. Alternatively, it is possible that this large ERV expansion set the stage for L1 extinction.
INTRODUCTION
The sheer mass of transposable elements (TEs) is perhaps indicative of their enormous influence on the genomes they inhabit. TEs have been identified in the DNA of every eukaryote studied and appear to have colonized the mammalian genome throughout the radiation of mammals. These repetitive sequences comprise a major proportion of mammalian genomes, ranging from about 34% of the dog and panda genomes to 40% of the mouse genome, 46% of the human genome, 49% of the horse genome, and 52% of the genome of the short-tailed opossum (27, 28, 49, 50, 57, 83). In mammals, most elements move via an RNA intermediate by means of retrotransposition. LINE-1 (long interspersed nuclear element 1; L1) constitutes the bulk of autonomous retroelements in placental mammals and marsupials, followed by endogenous retroviruses (ERVs) (10, 17, 27, 50, 57). L1s are believed to also provide enzymes in trans for retrotransposition of the nonautonomous short interspersed nuclear elements (SINEs), small retroelements contributing more than 10% to the genomic mass of some mammals. L1s have been identified and retain the ability to retrotranspose autonomously in every mammalian order examined, except monotremes, where LINE-2 elements dominate the genome (84). There are only two known instances of L1 extinction—loss of copies of L1 that can transpose autonomously. Both occurred in the ancestors of speciose groups and thus appear to affect most species of sigmodontine rodents and all species of megabats (15, 16, 32).
The possibility that L1s evolved to provide a function for the host has been doggedly examined to offer insight into why they are more abundant and long lived than other retroelements despite the apparent cost to their hosts. The observation that L1s are at a higher density on the X chromosome than the autosomes led to the suggestion that L1s serve as the “way stations” that transmit the inactivation signal along the X chromosome to be inactivated (26, 53). Alternative roles for L1s in X chromosome inactivation (XCI) have been suggested, including demarcation of the borders of inactivity and escape (2) or in recruitment of DNA methylase and formation of heterochromatin on the inactive X (37). A recent in situ study validates some of these ideas and strongly suggests L1s facilitate compartmentalization of genes to be inactivated during random XCI (18), while another implicates them in imprinted XCI (62). Involvement of L1s in double-strand DNA break repair has been proposed by several groups (40, 51, 56, 73, 81). One study suggested that reverse transcriptase (RT) activity from L1s in particular and to a lesser extent ERVs is necessary for early embryonic development in the mouse (9). A recent in silico analysis of the genomes of six mammals, chicken, zebrafish, and five invertebrates suggests the spacing of both L1s and SINEs facilitates the formation of stems with long loops within the large introns of animals, perhaps aiding the localization of intron splice sites (74).
Examination of lineages that lack active L1s is one way to test predictions of these various functional hypotheses. For example, if L1s serve as “way stations” to transmit Xist transcript, the inactivation signal, along the X chromosome to be inactivated, one might expect an increased rate of evolution of the Xist gene in species that lack active L1s, either due to selective pressure on Xist to track the deteriorating L1 sequences or to recognize another signal to serve the function of a “way station.” However, no increase in the rate of Xist evolution was observed in Oryzomys palustris, which lacks active L1s, compared to three other rodents that retain L1 activity (13). From a different perspective, if L1s accumulate on the X chromosome because they are necessary to facilitate XCI, a species with XO females, not needing XCI, would not be expected to accumulate L1s on the single X or even have a need for L1 activity. Nevertheless, L1s in such a species, the spiny rat Tokudaia osimensis, were shown to be recently active and at high density on the lone X (71). In contrast, if L1s serve a function in double-strand DNA break repair, one might expect that species that lack active L1s might be more sensitive to agents that lead to double-stranded breaks. In fact, karyotypic variation among some sigmodontine rodents that lack active L1s is quite exceptional (4, 25, 45, 60, 61, 75), lending some support for this hypothesis.
Independent of any putative host functions, L1s have been shown to impact both gene expression and genomic evolution (19, 30, 33). L1s inserted into transcribed regions of genes cause premature termination and reduce transcription rates (34, 63), are involved in 3′ transduction and rearrangement of adjacent DNA (31, 70), and serve as sites for ectopic recombination (12, 35, 87). Although L1 retrotransposition in somatic cells has historically been thought rare, two studies have demonstrated amplification in neuronal progenitor cells, leading to speculation these insertions could increase somatic mosaicism (21, 58). Several recent analyses of L1s in multiple human genomes have revealed that their activities and contributions to individual genetic and structural variation have been grossly underestimated (6, 24, 39, 41). Whether L1s serve a function in the host or are just very successful parasites, loss of their activity could have a profound impact on the dynamic interactions within the host genome.
ERVs and L1 elements, while sharing similar environments and modes of reproduction, differ in many ways. L1s, unlike ERVs, lack long terminal repeats (LTRs). L1s have maintained activity throughout the mammalian radiation, whereas most ERV families remain active in the host genome for relatively shorter periods of time. ERV families appear to have arisen by horizontal transfer into mammalian genomes multiple times during the evolution of mammals, while L1s appear to be transmitted only vertically. In well-studied species, the copy number of closely related ERVs (i.e., recently transposed), is typically very low, at levels of less than 100 per genome (5, 8, 17, 20, 29, 57, 82). Higher copy numbers of up to 1,000 recently inserted IAP elements per genome are seen in the mouse and hamster (46, 47). L1 elements have copy numbers in the range of hundreds of thousands per genome (10, 27, 28, 49, 50, 57, 82, 83), but most of these represent fragments of ancient insertions. The copy numbers of full-length, potentially active elements vary widely from species to species—e.g., numbering approximately 100 in humans (11, 68) and 3,000 in mice (22)—although activity levels of individual retrotransposition-competent elements are also widely variable (11, 48, 72). Even though our understanding of L1s and ERVs has increased tremendously, it is still not known if activities in one group of these retroelements affect activities in the other group.
Following our identification of cessation of L1 activity in a group of sigmodontine rodents (16, 32), we hypothesized that another repetitive sequence might be amplified to higher levels in these rodents, either to fill the niche left by the loss of L1 activity or to replace lost host functions. To test this possibility, we used a phylogenetically based hybridization method which can allow identification of any type of recently amplified repetitive sequences without prior knowledge of their mode of replication (54, 85, 86). Comparison of Oryzomys palustris, a species with no L1 activity, and Sigmodon hispidus, a closely related sigmodontine retaining L1 activity, led to identification of a new family of endogenous retroviruses which we called “mysTR” (14): i.e., “mys” from the Greek for “mouse” and “TR” for “transposable.” Further analysis indicated an exceedingly high copy number of about 1,000 mysTR elements per genome in S. hispidus and an even larger number of about 10,000 copies in O. palustris. This is consistent with the hypothesis that endogenous retroviruses have been more active in L1-inactive than L1-active muroid rodents. However, examination of an L1-active outgroup, Peromyscus maniculatus, revealed an intermediate copy number of about 4,500 mysTR elements per genome, leaving open the question of whether there was any connection between endogenous retrovirus activity and L1 inactivity.
The current study examines mysTR elements from four L1-active and four L1-inactive species to ask three questions. Are copy numbers of mysTR elements indeed correlated with loss of L1 activity? Has this ERV expansion spread among the sigmodontine rodents and their relatives? How are mysTR elements within these species related to each other?
MATERIALS AND METHODS
Species examined and genomic DNA extraction.
Tissue samples from Nyctomys sumichrasti (MSB45815), Neacomys spinosus (MSB68475), Calomys callosus (MSB:NK37800), and Akodon boliviensis (MSB55219) were provided by the Museum of Southwestern Biology at the University of New Mexico. Reithrodontomys fulvescens (TK21614), Sigmodon hispidus (TK72547), and Peromyscus maniculatus (TK25418) tissues were obtained from The Museum of Texas Tech University. Oryzomys palustris (KE02) was provided by Kent Edmonds (Indiana University, New Albany, IN). Genomic DNA was isolated by the methods of Longmire et al. (52).
Degenerate PCR, isolation, and sequencing.
Previously described degenerate PCR was used to amplify beta- and beta-like retroviruses from the genomic DNA of each species (14). The primers were designed to the conserved pro/pol domains used by Herniou et al. (38) with modifications for 5′ clamps and additional degeneracies. The forward (protease) primer is 5′-ACGAATTGCTCGAGAGKIHTIITNGAYCANGG-3′ and the reverse (reverse transcriptase) primer is 5′-TGGATCGCTGCGGTARNADRTCRTCCATRTA-3′ (14). The ∼920-bp PCR products were isolated via gel purification (QIAEX, Qiagen, Inc., Valencia, CA) and ligated into pGEM-T Easy vectors (Promega Corporation, Madison, WI). At least 16 elements were sequenced from each species using BigDye Terminator cycle sequencing chemistry and a 3730 DNA analyzer (Applied Biosystems, Foster City, CA).
Sequence analysis.
Sequences with large deletions were excluded from the data analysis for this project. The nucleotide data sets were aligned with ClustalW as implemented in Lasergene MegAlign (DNA*, Madison, WI) and subsequently adjusted manually. Sequences determined to be recombinants using GENECONV (69) were removed from the data set. The best fit model of sequence evolution was determined with DT_ModSel (55), and the appropriate model was implemented in MrBayes (66) and either PAUP*v4b10 (79) or GARLI (88). Bootstrap nodal support was estimated using 100 bootstrap replicates, and GARLI consensus trees were compiled with the SumTrees script implemented in DendroPy (78). Bayesian analyses consisted of four or eight runs carried out for 10 million generations with every 1,000th being saved. The burn-in was removed, all runs were combined, and the majority rule consensus tree was used to determine the posterior probabilities for the analyses.
Dot blot construction and Southern hybridization.
To determine mysTR copy numbers of recently inserted elements, quantitative dot blot hybridizations were carried out. Genomic DNA was quantified with an LS30 luminescence spectrometer as previously described (32) and applied to charged nylon filters using standard procedures (1). DNA probes were picked on the basis of their phylogenetic relationship to other sequences. mysTR clones RfulM8, RfulM23, PmanM13, PmanM23, NsumM11, NsumM13, ShisM2, AbolM10, CcalM2, CcalM33, NspiM16, NspiM17, and OpalM27 were used both as standards and as DNA probes on the dot blots. In order to reduce hybridization to older elements, hybridization was done at a stringency such that only relatively closely related elements gave strong signal. The blots were probed for 24 h at 58°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.3% SDS, 40 μg/ml denatured salmon sperm DNA, and 10× Denhardt's solution and subsequently washed at 60°C in 2× SSCP and 0.1% SDS as previously described (14).
Nucleotide sequence accession number.
Previously published mysTR sequences used in the present study have the following GenBank accession numbers: DQ139724-66 and DQ139770. Non-mysTR elements and mysTR elements isolated in the course of this study were deposited in GenBank and assigned accession no. HM160000 to -91. Accession numbers for the retrovirus sequences used as references are as follows: AKV murine leukemia virus (MuLV), J01998; avian leukosis virus (ALV), AY350569; bovine syncytial virus (BSV), U94514; equine infectious anemia virus (EIAV), AF016316; feline leukemia virus (FLV), AF052723; Homo sapiens endogenous virus (HERV-K10), DQ821442; human foamy virus (HFV), Y07725; human T-cell lymphotropic virus type I (HTLV), J02029; Mus musculus intracisternal A-particle (IAP), M17551; Jaagsiekte sheep retrovirus (JSRV), M80216; mouse mammary tumor virus (MMTV), AF033807; Mason-Pfizer monkey virus (MPMV), AF033815; Mus musculus type D-like endogenous retrovirus MusD1 (MusD), AF246632; Python molurus endogenous retrovirus (PyERV), AF500296; simian T-lymphotropic virus (PTLV), FJ957880; Rattus norvegicus endogenous retrovirus (RnERV), BX883042; Rous sarcoma virus (RSV), J02342; squirrel monkey retrovirus H (SMRV), M23385; Trichosurus vulpecula retrovirus (TvERV), AF224725; walleye dermal sarcoma virus (WDSV), AF033822; and walleye epidermal hyperplasia virus (WEHV), AF014792.
RESULTS
Isolation of mysTR endogenous retroviruses from L1-active and L1-inactive species.
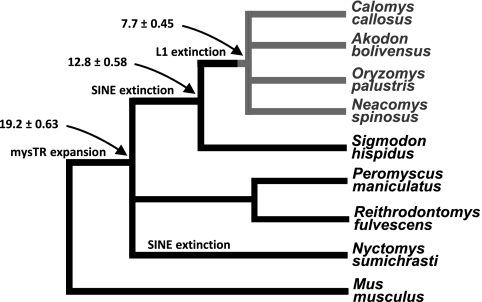
To characterize the expansion of mysTR within the L1-inactive South American rodents, we chose four L1-inactive rodent species and four L1-active sister species from which to isolate mysTR elements (Fig. 1). All eight species are in family Cricetidae, which includes New World rats and mice, voles, and hamsters. The four L1-inactive species, Akodon boliviensis, Calomys callosus, Neacomys spinosus, and Oryzomys palustris, cover a broad phylogenetic range within the Sigmodontinae, a large cricetid subfamily in which L1 activity has ceased (32). In fact, Sigmodontinae is the second largest subfamily of rodents in the muroid superfamily, with 377 species and 74 genera in eight tribes. The L1-inactive species are in three different sigmodontine tribes: O. palustris and N. spinosus in the Oryzomyini, A. boliviensis in the Akodontini, and C. callosus in the Phyllotini. The species with active L1s include Sigmodon hispidus from the basal tribe of Sigmodontinae, Sigmodontini; one species from the cricetid subfamily Tylomyinae, Nyctomys sumichrasti; and two different genera from the cricetid subfamily Neotominae, tribe Reithrodontomyini, Reithrodontomys fulvescens and Peromyscus maniculatus (59).
Fig. 1.
Phylogeny of rodents included in the analysis as per the consensus of data from Steppan et al. (77) and Smith and Patton (76). The numbers and arrows indicate estimated times of divergence in million years according to Steppan et al. (77). The color gray delineates Sigmodontinae affected by the L1 extinction event ∼8.8 million years ago (32). Basal species S. hispidus either diverged before L1 extinction occurred or regained L1 activity through mutation or recombination, but has no B1 SINE activity. The ERV mysTR expanded after the ancestor of Mus diverged.
A wide range of ERVs were isolated from the above species by use of degenerate PCR primers specific for a 920-bp region in betaretroviruses covering the 3′ portion of the protease gene and the 5′ portion of the reverse transcriptase (RT) gene (14). Sequencing and subsequent analyses yielded a total of 114 mysTR elements (Table 1). In addition to the mysTR sequences, 16 other ERVs were identified in four of the species. Phylogenetic analysis of RT domains 1 to 4 (42) of the non-mysTR elements determined their closest relatives to be betaretroviruses.
Table 1.
Types and numbers of elements isolated with betaretrovirus reverse transcriptase degenerate primers from rodents with and without L1 activitya
| Species | No. of elements |
|||
|---|---|---|---|---|
| mysTR (no. with ORF) | IAP | β6 | β | |
| L1 inactive | ||||
| Akodon boliviensis | 12 (6) | 1 | ||
| Calomys callosus | 13 (5) | |||
| Neacomys spinosus | 14 (7) | |||
| Oryzomys palustris | 9 (3) | 2 | 5 | |
| L1 active | ||||
| Nyctomys sumichrasti | 17 (11) | 1 | ||
| Peromyscus maniculatus | 23 (11) | |||
| Reithrodontomys fulvescens | 17 (16) | |||
| Sigmodon hispidus | 9 (5) | 7 | ||
Phylogenetic analysis of mysTR elements.
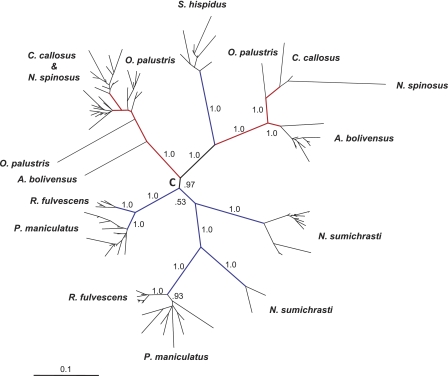
Phylogenetic analysis of mysTR elements from each species resulted in the majority of the elements grouping into one or two lineages of closely related sequences (data not shown). When the elements from all eight species were aligned and analyzed together, the majority of the sequences fell into species-specific clades (Fig. 2). Multiple lineages of mysTR are seen for all species except S. hispidus. Most mysTR elements isolated from C. callosus and N. spinosus intercalated in a lineage with two closely related, but not species-specific, sublineages and another distant lineage containing few elements.
Fig. 2.
Phylogenetic analysis of 114 mysTR elements from eight rodent species, four L1 active (blue branches) and four L1 inactive (red branches). The unrooted Bayesian tree was constructed using an average of 890 bp of the pro/pol region. Selected posterior probabilities are indicated on branches. Names of individual elements have been removed, and the species from which each clade or element originated are shown. All species except S. hispidus have two lineages of mysTR elements. “C” denotes a putative root of the tree.
In general, the terminal branch lengths are very short. Pairwise sequence distances within and between selected clades are shown in Table 2. In addition, more than half of the elements have intact open reading frames (ORFs) (Table 1). Both intact ORFs and small pairwise distances are indicative of recently transposed ERVs. The presence of intact ORFs also suggests that some of these elements may have moved autonomously. A small number of mysTR elements have no closely related neighbors and relatively long terminal branch lengths and tend to contain no intact ORFs, suggesting earlier genomic deposition.
Table 2.
Corrected pairwise distances per 100 sites of reverse transcriptase domains 1 to 4 of mysTR elements containing ORFs within and between selected clades from Fig. 2
| Speciesa | Pairwise distance/100 sitesa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| C. callosus-N. spinosus | O. palustris | A. boliviensis | S. hispidus | P. maniculatus 1 | P. maniculatus 2 | R. fulvescens 1 | R. fulvescens 2 | N. sumichrasti | |
| L1 inactive | |||||||||
| C. callosus-N. spinosus | 0.22–8.56 | ||||||||
| O. palustris | 6.18–11.16 | 2.25–5.21 | |||||||
| A. boliviensis | 27.85–31.28 | 30.62–35.6 | 0.42–4.26 | ||||||
| L1 active | |||||||||
| S. hispidus | 27.07–34.56 | 29.53–34.44 | 23.71–27.07 | 0.21–3.83 | |||||
| P. maniculatus 1 | 23.37–27.22 | 27.46–28.40 | 33.98–37.80 | 33.16–36.27 | Only 1 ORF | ||||
| P. maniculatus 2 | 17.29–25.63 | 20.44–28.41 | 27.76–35.17 | 27.58–33.91 | 21.02–26.66 | 0–6.90 | |||
| R. fulvescens 1 | 24.55–28.96 | 27.63–30.76 | 34.08–38.70 | 30.16–34.58 | 4.67–5.21 | 21.17–28.19 | 0–1.10 | ||
| R. fulvescens 2 | 16.53–22.75 | 21.65–24.42 | 27.23–31.19 | 28.22–31.92 | 22.50–25.32 | 4.22–10.06 | 22.12–24.14 | 0–2.02 | |
| N. sumichrasti | 22.86–33.86 | 26.42–30.10 | 34.20–41.13 | 29.05–33.22 | 27.98–29.73 | 16.62–21.99 | 25.48–28.48 | 19.64–24.31 | 0.22–3.00 |
Reverse transcriptase domains of mysTR elements containing ORFs within selected clades from Fig. 2 are shaded.
Figure 2 shows that clades of mysTR found in L1-active species (blue branches) are not monophyletic with respect to those found in L1-inactive species (red branches). Recently deposited elements in L1-inactive species do not appear to be more closely related to each other than elements in L1-active species, as might be the case if there has been more mysTR activity in the L1-inactive species. This observation should be considered with caution, though, because of the potential biases inherent in PCR. It is also evident that the mysTR phylogeny seen here does not follow the species phylogeny, and no matter where the root of the tree is placed, either multiple exogenous infections or lineage sorting must be inferred. Interestingly, the interdigitation of the majority of the C. callosus and N. spinosus ERVs suggests that they were deposited before the divergence of these two species and the same lineages have remained active in both species.
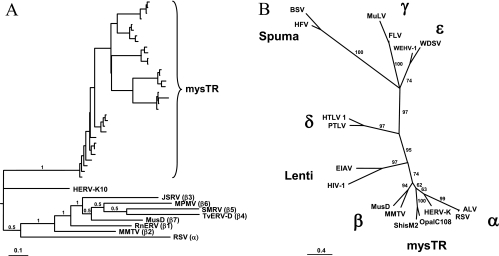
Previously isolated mysTR elements grouped with the betaretroviruses in phylogenetic analyses, but were basal to subgroups β1 to β7 (14). To determine if the elements isolated here, which appeared to be mysTR-like on initial analysis, fell into the same phylogenetic group, their RT regions were analyzed in more detail (Fig. 3). Figure 3A shows the relationships between RT regions 1 to 4 (42) of four mysTR sequences from each of the eight species under study and representatives from each of the β1 to β7 betaretrovirus groups, the human endogenous betaretrovirus HERV-K10, and the exogenous alpharetrovirus RSV. If a species contained more than one lineage of mysTR, two elements from each lineage were chosen for the analysis. All of the newly isolated mysTR elements fall into a single clade outside the β1-to-β7 grouping and, as in the previous analysis (14), equidistant from HERV-K10 and the rest of the betaretroviruses. In the event the large number of mysTR sequences in the alignment skewed the outcome of this analysis, two of the mysTR RTs were aligned with two representatives from each of the described retrovirus genera Alpharetrovirus, Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, Lentivirus, and Spuma-like virus. The resultant unrooted maximum likelihood phylogeny (Fig. 3B) also places the mysTR elements in their own clade almost equidistant between HERV-K10 and the other betaretroviruses, with the remaining retrovirus genera having approximately the same relationships as the currently accepted phylogeny (44).
Fig. 3.
Phylogenetic relationships of mysTR to known retrovirus genera, both endogenous and exogenous. Unrooted maximum likelihood trees are based on nucleotide sequences of reverse transcriptase domains 1 to 4 (42). Posterior probabilities or bootstrap values are noted on select branches. Panel A contains four mysTR elements from each species included in the study, representative elements of betaretrovirus subgroups β1 to β7 (3), the endogenous betaretrovirus HERV-K10, and RSV (an alpharetrovirus). Panel B contains two representatives each of mysTR and the retrovirus genera Alpharetrovirus (α), Betaretrovirus (β), Deltaretrovirus (δ), Epsilonretrovirus (ε), Gammaretrovirus (γ), Lentivirus, and Spuma-like virus plus HERV-K10.
Copy numbers of mysTR elements in multiple sigmodontine species.
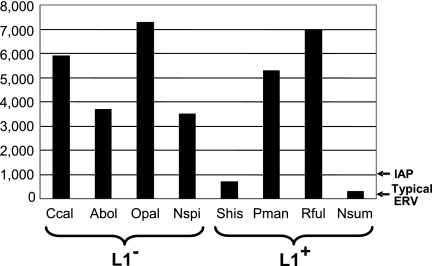
The copy numbers of relatively recently inserted msysTR elements in the L1-active and L1-inactive species were determined by dot blot hybridizations (Fig. 4). For each species, probes representative of each major lineage were hybridized at a stringency that allowed strong signals from sister elements within clades but minimized hybridization at the sequence distances which exist between separate clades in the species. Dot blots contained DNA from multiple species as controls, and in all cases, hybridization was only detectable with the species-specific probes, indicating mysTR was amplified independently and recently in each species. The copy number of the minor mysTR lineage in C. callosus was determined, but not those of the minor mysTR lineages in O. palustris, N. spinosus, and A. boliviensis, because PCR results indicated that these lineages contain few elements. Every species exhibits an extremely high number of copies of mysTR, ranging from around 300 in N. sumichrasti to just over 7,000 in O. palustris. These numbers represent a conservative estimate of recent mysTR insertion since they do not include lone LTRs or unsampled clades. However, there appears to be little correlation between mysTR copy number and L1 activity.
Fig. 4.
Copy number of recently inserted mysTR elements in eight rodent species. The pro/pol domains of selected mysTR elements were hybridized to dot blots of genomic DNA from the following L1-inactive (L1−) and L1-active (L1+) species: C. callosus (Ccal), A. boliviensis (Abol), O. palustris (Opal), N. spinosus (Nspi), S. hispidus (Shis), P. maniculatus (Pman), R. fulvescens (Rful), and N. sumichrasti (Nsum). The copy number of a typical endogenous retrovirus (ERV) is noted with an arrow to the right, as is the copy number of mouse IAP elements, one of the most numerous ERVs previously known in a mammalian genome.
DISCUSSION
The null hypothesis for mammalian retroelements is that they are genomic parasites. The dynamic mammalian genome appears to be the site of an ongoing arms race among retroelements and host cells for host factors and genomic space. A change in activity of one family of elements, particularly one that constitutes a major portion of the genome, could dramatically impact the host and the elements that reside there. Alternatively, if a group of elements serve a specific host function, major impacts would occur if the elements providing the function were lost. Our previous observation that the mysTR family of endogenous retroviruses had undergone a much greater expansion in the L1-inactive species O. palustris than in its L1-active sister species S. hispidus led to this study: the isolation and examination of mysTR elements from eight New World rodent species covering a phylogenetic range around the L1 extinction event. The species chosen for this study cover a broad phylogenetic range of L1-inactive species and as similar a range as possible of L1-active species, although there is slightly greater divergence among the L1-active species than the L1-inactive species (Fig. 1).
mysTR endogenous retroviruses are at exceedingly high levels within each species from this group of New World rodents, but there is no clear correlation between mysTR copy numbers and loss of L1 activity. An average of approximately 5,100 recently inserted mysTR elements per species is seen among the L1-inactive rodents, while a lower average of approximately 3,300 elements per species is seen among the L1-active rodents, but this difference is not significant. While the lowest mysTR copy numbers are seen in two of the L1-active species, N. sumichrasti and S. hispidus, the other two L1-active species, P. maniculatus and R. fulvescens, show copy numbers in the upper range of those seen for the four L1-inactive species.
Given the time since divergence of most of the species studied here and the lineage-specific stringency of the dot blot hybridizations, it is clear that the amplification of mysTR elements to such high copy numbers occurred relatively recently and independently in each species. The amplification of the mysTR ERV family itself is unprecedented. The copy numbers of recently inserted elements in most of the species studied here are far higher than have been found for any group of such closely related mammalian elements. ERV families that have average sequence distances between elements of less than 20% are typically found at copy numbers of 5 to 50, with a few ranging into the upper hundreds (5, 8, 20, 29). The only notable exception to this is the still active mouse and hamster IAP elements, which are found at about 1,000 closely related copies per genome (46, 47). All other known groups of ERVs numbering in the thousands show much higher sequence divergence and a preponderance of lone LTRs, the footprint of former elements excised as a result of recombination between their LTRs.
It is also intriguing that the two species containing the lowest copy numbers of mysTR have inactive B1 SINEs while retaining L1 activity (65). The phylogenetic relationships of the species under study are unresolved, so although it is obvious the mysTR expansion preceded the extinction of the SINEs, we are unable to determine whether the latter was a single event in a common Sigmondontinae/Tylominae ancestor or occurred independently at least twice. In addition, it raises questions about the relationships among the elements themselves as well as how cellular defenses against one may inadvertently affect another. One idea we previously entertained to explain how B1 SINE extinction could precede that of L1 was the possibility of an arms race with a third genetic parasite, imagining another SINE family outcompeting B1s and possibly L1s for cellular factors needed for retrotransposition (65). Although we favored another hypothesis—that a gradual extinction of L1s eliminated B1 activity early on and allowed for resurrection of L1s in some groups—the mysTR expansion is exactly the type of burst characterizing the periodic cycling of retrotransposition that we imagine in the arms race scenario.
The life cycle of an ERV begins with the colonization of one or more germ cells by an exogenous retrovirus, thus creating a vertically transmitted family. Generally, if not immediately selected against, the rate of amplification of a novel ERV lineage is rapid at first and then declines over time due to suppression by a host mechanism or mutation of the ERV insertions during host cell replications (29). Decline can be postponed by trans complementation, as with IAP elements (67), or reinstatement of the ability to replicate by recombination or reinfection following purifying selection, as with HERV-K (7, 23) and IAP (64), but usually the autonomous copies will eventually mutate until nonautonomous ERVs prevail in the population. As copy numbers increase during this sometimes long process of endogenization, recombination between left and right long terminal repeats (LTRs) increases the percentage of elements present as lone LTRs. The eventual loss of all autonomous forms stops transposition, and mutational decay then leads to eventual loss of recognizable sequences. In some small number of cases, specific ERV inserts may come to supply essential host functions (43). A thorough understanding of the interplay between ERVs and the genomes they occupy will require comparison of multiple ERV families in different host genomes.
The present expansion of the mysTR family in the muroid rodents may lie in an early part of this life cycle and provides a valuable natural system for comparison to more commonly studied species to ascertain what evolutionary aspects are common and what are unique to the invasion and distribution of different ERVs into mammalian genomes. The high copy numbers of mysTR elements in conjunction with likely continuing activity in their mammalian hosts make this expansion distinctive. A few mammalian ERV expansions appear to have been initiated more recently (36, 80), but the mysTR expansion provides the opportunity for examining an amplification to unusually high copy numbers in a natural system still experiencing unprecedented endogenous activity.
Two observations suggest mysTR arose from a single infection of an exogenous retrovirus in a common ancestor of these species, followed primarily by intracellular retrotransposition and vertical transmission, rather than from multiple infections by an exogenous precursor of mysTR in multiple species. First, the sister species R. fulvescens and P. maniculatus share the same two relatively distantly related lineages (Fig. 2), suggesting that both lineages existed in a common ancestor and were transmitted vertically. Second, previously characterized full-length mysTR elements from O. palustris (14) lack a discernible envelope gene, even though they were shown to have been recently deposited, suggesting that the mysTR family has been transmitted vertically in the genome for an extended period of time. Interestingly, the tree seen in Fig. 2 is consistent with an initial exogenous mysTR infection occurring such that the root of the phylogeny is on branch C, followed by only endogenous transposition and limited lineage sorting.
The question remains open as to whether the majority of the mysTR elements have moved autonomously or nonautonomously. Among the pro/pol regions analyzed, a number of the recently deposited elements have retained single ORFs, supporting the possibility of autonomous movement. The three full-length mysTR elements previously sequenced (14) do not contain full-length ORFs, but very few changes are required to restore those reading frames. Thus, it appears many of these mysTR elements were either recently able to move autonomously or still can.
MysTR appears to form an exclusive group of class II retroviruses, falling out phylogenetically between the alpha- and betaretroviruses. Whether some intrinsic characteristic of mysTR itself, such as an aggressive reverse transcriptase, the ability to monopolize host factors or evade cell defenses, or evolution of a strong promoter is responsible for the extraordinary proliferation of the mysTR lineage is not evident here. Alternatively, some characteristic of the host species may have contributed to mysTR success.
Finally, recent insights into the role of L1s facilitating X chromosome inactivation raise questions about the long-term viability of these L1-inactive species. Although Chow et al. (18) specifically state that young elements are not essential to their XCI model, their data suggest that some genes would likely escape inactivation without the benefit of L1 transcription. The putative L1 function of facilitating heterochromatization of the inactive X, even if it does not require L1 activity, will eventually be affected by the gradual loss of L1 sequence identity as the elements decay and are not replaced. Indeed, deep extinctions of mammalian L1 activity have not yet been documented. The only other well-characterized L1 extinction, in the common ancestor of the bat family Pteropodidae (15), occurred ∼24 million years ago, resulting in the youngest megabat L1s having about 8% divergence from the last active L1s—about the same divergence as the youngest L1s in these sigmodontine rodents. However, no evidence of unusual TE activity has been found in the one megabat genome currently being assembled (unpublished data).
Several unusual genomic events have occurred in the Sigmodontinae. The rapid expansion of mysTR to unprecedented levels, the loss of L1 activity, and the preceding loss of SINE activity are all rare events among mammals. The order in which these events occurred is unexpected and intriguing. Thus, mysTR expansion in species lacking the typical genomic effects of L1 provides a unique opportunity to examine the interplay among elements and with the host.
ACKNOWLEDGMENTS
We thank The Museum of Texas Tech University, the Museum of Southwestern Biology at the University of New Mexico, and Kent Edmonds of Indiana University for the invaluable gifts of tissue samples. The Dixie Mager lab was especially generous with technical advice and the sequence files of reference retroviruses.
This study was supported by funding from the National Institutes of Health (GM38727 to H.A.W.). Analytical resources were provided by NIH grants from the INBRE (RR016454) and COBRE (RR016448) Programs of the National Center for Research Resources.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Ausubel F. M., et al. 1989. Current protocols in molecular biology. Green Publications/Wiley-Interscience, New York, NY [Google Scholar]
- 2. Bailey J. A., Carrel L., Chakravarti A., Eichler E. E. 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. U. S. A. 97:6634–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baillie G. J., van de Lagemaat L. N., Baust C., Mager D. L. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78:5784–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barros M. A., Reig O. A., Perez-Zapata A. 1992. Cytogenetics and karyosystematics of South American oryzomyine rodents (Cricetidae: Sigmodontinae). IV. Karyotypes of Venezuelan, Trinidadian, and Argentinian water rats of the genus Nectomys. Cytogenet. Cell Genet. 59:34–38 [DOI] [PubMed] [Google Scholar]
- 5. Baust C., et al. 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J. Virol. 77:11448–11458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beck C. R., et al. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141:1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belshaw R., et al. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U. S. A. 101:4894–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benit L., Lallemand J. B., Casella J. F., Philippe H., Heidmann T. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 73:3301–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beraldi R., Pittoggi C., Sciamanna I., Mattei E., Spadafora C. 2006. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 73:279–287 [DOI] [PubMed] [Google Scholar]
- 10. Bovine Genome Sequencing and Analysis Consortium 2009. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324:522–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brouha B., et al. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U. S. A. 100:5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burwinkel B., Kilimann M. W. 1998. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J. Mol. Biol. 277:513–517 [DOI] [PubMed] [Google Scholar]
- 13. Cantrell M. A., Carstens B. C., Wichman H. A. 2009. X chromosome inactivation and Xist evolution in a rodent lacking LINE-1 activity. PLoS One 4:e6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantrell M. A., et al. 2005. MysTR: an endogenous retrovirus family in mammals that is undergoing recent amplifications to unprecedented copy numbers. J. Virol. 79:14698–14707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cantrell M. A., Scott L., Brown C. J., Martinez A. R., Wichman H. A. 2008. Loss of LINE-1 activity in the megabats. Genetics 178:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Casavant N. C., et al. 2000. The end of the LINE? Lack of recent L1 activity in a group of South American rodents. Genetics 154:1809–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87 [DOI] [PubMed] [Google Scholar]
- 18. Chow J. C., et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141:956–969 [DOI] [PubMed] [Google Scholar]
- 19. Cordaux R., Batzer M. A. 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10:691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costas J. 2003. Molecular characterization of the recent intragenomic spread of the murine endogenous retrovirus MuERV-L. J. Mol. Evol. 56:181–186 [DOI] [PubMed] [Google Scholar]
- 21. Coufal N. G., et al. 2009. L1 retrotransposition in human neural progenitor cells. Nature 460:1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeBerardinis R. J., Goodier J. L., Ostertag E. M., Kazazian H. H., Jr 1998. Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat. Genet. 20:288–290 [DOI] [PubMed] [Google Scholar]
- 23. Dewannieux M., et al. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ewing A. D., Kazazian H. H., Jr 2010. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 20:1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fagundes V., Christoff A. U., Yonenaga-Yassuda Y. 1998. Extraordinary chromosomal polymorphism with 28 different karyotypes in the neotropical species Akodon cursor (Muridae, Sigmodontinae), one of the smallest diploid number in rodents (2n = 16, 15 and 14). Hereditas 129:263–274 [DOI] [PubMed] [Google Scholar]
- 26. Gartler S. M., Riggs A. D. 1983. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17:155–190 [DOI] [PubMed] [Google Scholar]
- 27. Gentles A. J., et al. 2007. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 17:992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibbs R. A. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521 [DOI] [PubMed] [Google Scholar]
- 29. Gifford R., Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315 [DOI] [PubMed] [Google Scholar]
- 30. Goodier J. L., Kazazian H. H., Jr 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23–35 [DOI] [PubMed] [Google Scholar]
- 31. Goodier J. L., Ostertag E. M., Kazazian H. H., Jr 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9:653–657 [DOI] [PubMed] [Google Scholar]
- 32. Grahn R. A., Rinehart T. A., Cantrell M. A., Wichman H. A. 2005. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 110:407–415 [DOI] [PubMed] [Google Scholar]
- 33. Han J. S., Boeke J. D. 2005. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays 27:775–784 [DOI] [PubMed] [Google Scholar]
- 34. Han J. S., Szak S. T., Boeke J. D. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429:268–274 [DOI] [PubMed] [Google Scholar]
- 35. Han K., et al. 2008. L1 recombination-associated deletions generate human genomic variation. Proc. Natl. Acad. Sci. U. S. A. 105:19366–19371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanger J. J., Bromham L. D., McKee J. J., O'Brien T. M., Robinson W. F. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J. Virol. 74:4264–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hansen R. S. 2003. X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum. Mol. Genet. 12:2559–2567 [DOI] [PubMed] [Google Scholar]
- 38. Herniou E., et al. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang C. R., et al. 2010. Mobile interspersed repeats are major structural variants in the human genome. Cell 141:1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hutchison C. A., III, Hardies S. C., Loeb D. D., Shehee W. R. W., Edgell M. H. 1989. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome, p. 593–617 In Berg D., Howe M. (ed.), Mobile DNA. American Society for Microbiology, Washington, DC [Google Scholar]
- 41. Iskow R. C., et al. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141:1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jacobo-Molina A., Arnold E. 1991. HIV reverse transcriptase structure-function relationships. Biochemistry 30:6351–6356 [DOI] [PubMed] [Google Scholar]
- 43. Jern P., Coffin J. M. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709–732 [DOI] [PubMed] [Google Scholar]
- 44. Knipe D. M., Howley P. M. (ed.). 2007. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 45. Koop B. F., Baker R. J., Genoways H. H. 1983. Numerous chromosomal polymorphisms in a natural population of rice rats (Oryzomys, Cricetidae). Cytogenet. Cell Genet. 35:131–135 [DOI] [PubMed] [Google Scholar]
- 46. Kuff E. L., et al. 1986. Chromosome distribution of intracisternal A-particle sequences in the Syrian hamster and mouse. Chromosoma 93:213–219 [DOI] [PubMed] [Google Scholar]
- 47. Kuff E. L., Lueders K. K. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51:183–276 [DOI] [PubMed] [Google Scholar]
- 48. Lee S. H., Cho S. Y., Shannon M. F., Fan J., Rangasamy D. 2010. The impact of CpG island on defining transcriptional activation of the mouse L1 retrotransposable elements. PLoS One 5:e11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li R., et al. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lindblad-Toh K., et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819 [DOI] [PubMed] [Google Scholar]
- 51. Liu J., et al. 1997. LINE-I element insertion at the t(11;22) translocation breakpoint of a desmoplastic small round cell tumor. Genes Chromosomes Cancer 18:232–239 [DOI] [PubMed] [Google Scholar]
- 52. Longmire J. L., et al. 1988. Isolation and molecular characterization of a highly polymorphic centromeric tandem repeat in the family Falconidae. Genomics 2:14–24 [DOI] [PubMed] [Google Scholar]
- 53. Lyon M. F. 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80:133–137 [DOI] [PubMed] [Google Scholar]
- 54. Martin S. L., Wichman H. A. 1993. Molecular approaches to mammalian retrotransposon isolation. Methods Enzymol. 224:309–322 [DOI] [PubMed] [Google Scholar]
- 55. Minin V., Abdo Z., Joyce P., Sullivan J. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52:674–683 [DOI] [PubMed] [Google Scholar]
- 56. Morrish T. A., et al. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31:159–165 [DOI] [PubMed] [Google Scholar]
- 57. Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562 [DOI] [PubMed] [Google Scholar]
- 58. Muotri A. R., et al. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435:903–910 [DOI] [PubMed] [Google Scholar]
- 59. Musser G. G., Carleton M. D. 2005. Superfamily Muroidea, 3rd ed., vol. 2 The Johns Hopkins University Press, Baltimore, MD [Google Scholar]
- 60. Nachman M. W. 1992. Geographic patterns of chromosomal variation in South American marsh rats, Holochilus brasiliensis and H. vulpinus. Cytogenet. Cell Genet. 61:10–16 [DOI] [PubMed] [Google Scholar]
- 61. Nachman M. W., Myers P. 1989. Exceptional chromosomal mutations in a rodent population are not strongly underdominant. Proc. Natl. Acad. Sci. U. S. A. 86:6666–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Namekawa S. H., Payer B., Huynh K. D., Jaenisch R., Lee J. T. 2010. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol. Cell. Biol. 30:3187–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Perepelitsa-Belancio V., Deininger P. 2003. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 35:363–366 [DOI] [PubMed] [Google Scholar]
- 64. Ribet D., et al. 2008. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 18:597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rinehart T. A., Grahn R. A., Wichman H. A. 2005. SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet. Genome Res. 110:416–425 [DOI] [PubMed] [Google Scholar]
- 66. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 67. Saito E. S., Keng V. W., Takeda J., Horie K. 2008. Translation from nonautonomous type IAP retrotransposon is a critical determinant of transposition activity: implication for retrotransposon-mediated genome evolution. Genome Res. 18:859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sassaman D. M., et al. 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16:37–43 [DOI] [PubMed] [Google Scholar]
- 69. Sawyer S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526–538 [DOI] [PubMed] [Google Scholar]
- 70. Schwartz A., et al. 1998. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum. Mol. Genet. 7:1–11 [DOI] [PubMed] [Google Scholar]
- 71. Scott L. A., Kuroiwa A., Matsuda Y., Wichman H. A. 2006. X accumulation of LINE-1 retrotransposons in Tokudaia osimensis, a spiny rat with the karyotype XO. Cytogenet. Genome Res. 112:261–269 [DOI] [PubMed] [Google Scholar]
- 72. Seleme M. C., et al. 2006. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc. Natl. Acad. Sci. U. S. A. 103:6611–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sen S. K., Huang C. T., Han K., Batzer M. A. 2007. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 35:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shepard S., McCreary M., Fedorov A. 2009. The peculiarities of large intron splicing in animals. PLoS One 4:e7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Silva M. J., Yonenaga-Yassuda Y. 1998. Karyotype and chromosomal polymorphism of an undescribed Akodon from Central Brazil, a species with the lowest known diploid chromosome number in rodents. Cytogenet. Cell Genet. 81:46–50 [DOI] [PubMed] [Google Scholar]
- 76. Smith M., Patton J. 1999. Phylogenetic relationships and the radiation of Sigmodontine rodents in South America: evidence from cytochrome b. J. Mamm. Evol. 6:89–128 [Google Scholar]
- 77. Steppan S., Adkins R., Anderson J. 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 53:533–553 [DOI] [PubMed] [Google Scholar]
- 78. Sukumaran J., Holder M. T. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571 [DOI] [PubMed] [Google Scholar]
- 79. Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods) version R, 4.0 ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- 80. Tarlinton R. E., Meers J., Young P. R. 2006. Retroviral invasion of the koala genome. Nature 442:79–81 [DOI] [PubMed] [Google Scholar]
- 81. Teng S. C., Kim B., Gabriel A. 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383:641–644 [DOI] [PubMed] [Google Scholar]
- 82. Venter J. C. 2001. The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- 83. Wade C. M., et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Warren W. C., et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wichman H. A., Payne C. T., Reeder T. W. 1990. Intrageneric variation in repetitive sequences isolated by phylogenetic screening of mammalian genomes, p. 153–160 In Clegg M. T., O'Brien S. J. (ed.), Molecular evolution. Wiley-Liss, New York [Google Scholar]
- 86. Wichman H. A., Potter S. S., Pine D. S. 1985. Mys, a family of mammalian transposable elements isolated by phylogenetic screening. Nature 317:77–81 [DOI] [PubMed] [Google Scholar]
- 87. Xing J., et al. 2009. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 19:1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation University of Texas, Austin, TX [Google Scholar]