Abstract
Various feline APOBEC3 (fA3) proteins exhibit broad antiviral activities against a wide range of viruses, such as feline immunodeficiency virus (FIV), feline foamy virus (FFV), and feline leukemia virus (FeLV), as well as those of other species. This activity can be counteracted by the FIV Vif protein, but the mechanism by which FIV Vif suppresses fA3s is unknown. In the present study, we demonstrated that FIV Vif could act via a proteasome-dependent pathway to overcome fA3s. FIV Vif interacted with feline cellular proteins Cullin5 (Cul5), ElonginB, and ElonginC to form an E3 complex to induce degradation of fA3s. Both the dominant-negative Cul5 mutant and a C-terminal hydrophilic replacement ElonginC mutant potently disrupted the FIV Vif activity against fA3s. Furthermore, we identified a BC-box motif in FIV Vif that was essential for the recruitment of E3 ubiquitin ligase and also required for FIV Vif-mediated degradation of fA3s. Moreover, despite the lack of either a Cul5-box or a HCCH zinc-binding motif, FIV Vif specifically selected Cul5. Therefore, FIV Vif may interact with Cul5 via a novel mechanism. These finding imply that SOCS proteins may possess distinct mechanisms to bind Cul5 during formation of the Elongin-Cullin-SOCS box complex.
INTRODUCTION
The mammalian APOBEC3 (A3; apolipoprotein B mRNA-editing catalytic polypeptide 3) proteins are members of a family of natural host restriction factors (43), which exhibit broad antiviral activities against a wide range of retroviruses (2, 15, 59), endogenous retrovirus (11), and hepatitis B virus (50, 55). Among the human A3A to H proteins, A3G is the most well known since it was the first described to be part of the innate host defense against human immunodeficiency virus type 1 (HIV-1) (13). In the interaction of A3G and HIV-1, A3G is packaged into HIV-1 virions and induces dC to dU modifications of newly synthesized minus-strand viral DNA (4, 14, 24, 49). Ultimately, such modifications can result in the mutation of the TGG tryptophan codon to a TAG stop codon, thereby altering protein expression and affecting subsequent stages of the viral life cycle (57). Some of the other A3 proteins also have similar functions to various degrees of potency (3, 7, 8, 27) and sometimes with cross-species activity (20, 23, 25).
Vif (for viral infectivity factor) is a regulatory protein that is present in all lentiviruses except equine infectious anemia virus (EIAV) and is required for viral replication and pathogenicity in vivo (12, 26, 48). The primary function of HIV-1 Vif is to neutralize the antiviral function of APOBEC3 proteins (43) by triggering their degradation through polyubiquitination and proteasome activity (33, 35, 44). HIV-1 Vif acts as an adaptor protein that bridges A3 proteins to a Cullin5 (Cul5)-based E3 ubiquitin ligase complex, which includes Cul5, ElonginB, and ElonginC (45, 58, 60). The H-x(5)-C-x(17–18)-C-x(3–5)-H motif (also called the HCCH zinc finger) (30, 36, 54) and the PPLPx4L motif (also known as the Cul5-box) in the C-terminal region of HIV-1 Vif are responsible for Cul5 binding (45, 60), while another C-terminal SLQ(Y/F)LA motif (BC-box) interacts with ElonginC to help recruit HIV-1 Vif into the Cul5 ubiquitin E3 ligase complex (34, 58, 60). However, the Vif Cullin-box binds Cul5 with an affinity of ∼10-fold lower than the HCCH motif (1, 52). The PPLP motif is thought to play multiple roles in the function of HIV-1 Vif, including its dimerization (56) and its binding to A3G (51) and ElonginB (1). Moreover, HIV-1 Vif has developed rather complicated mechanisms to bind A3 proteins. There are at least 10 motifs known to regulate the interaction between Vif and A3s (5, 6, 9, 10, 17, 40, 42, 51). Although most of the interaction domains are located in the N terminus of Vif, part of its C-terminal region is also critical for Vif-mediated neutralization of A3s (6). In addition, the mechanisms of degradation of human A3C and African green monkey (agm) A3G induced by simian immunodeficiency virus (SIVagm) Vif are the same as that for HIV-1 Vif (62).
According to the nonprimate A3 nomenclature (22), there are five feline A3 (fA3) proteins: one fA3Z3 protein, three highly similar fA3Z2 proteins (a to c) and one fA3Z2-Z3 protein that is expressed by readthrough alternative splicing (37). Although the fA3Z2 (a to c) proteins target Bet-deficient feline foamy virus (FFV), they do not inhibit any other feline retrovirus (37). Infectivity of Bet-deficient FFV is reduced not only by genomic deamination but also by an A3-induced reduction of particle release (28). The infectivity of Vif-deficient feline immunodeficiency virus (FIV) and feline leukemia virus (FeLV) is reduced by fA3Z3 and fA3Z2-Z3 (28, 37) by induction of G→A hypermutation of the viral cDNA (37, 47). The antiretroviral activities of the fA3Z2s are inhibited by the foamy virus accessory Bet protein (28), while the mechanism of FeLV against fA3s is unknown (38). It is believed that FIV Vif makes a fundamental contribution to overcoming the restrictions imposed by fA3Z3 and fA3Z2-Z3 (23, 47, 63). FIV Vif colocalizes with the feline A3 proteins, targets them for proteasome degradation, and rescues the infectivity of the virus (47). The specific interactions between FIV Vif and the fA3 proteins are still unclear, and the precise mechanism of FIV Vif induced degradation of fA3s remains to be elucidated.
In the present study, we detected the expression of feline ElonginB, ElonginC, and Cul5 (fElonginB, fElonginC, and fCul5) in the feline CrFK kidney cell line. The coding sequence of ElonginC is highly conserved but that of ElonginB slightly differs among mammals. We showed evidence that FIV Vif mediated the formation of the Cul5-ElonginB-ElonginC E3 (Cul5-EB-EC E3) complex, which marks fA3s for proteasomal degradation. A dominant-negative mutant of Cul5 and a C-terminal hydrophilic replacement ElonginC mutant both inhibited FIV Vif-induced degradation of fA3s. These findings demonstrate that the FIV Vif-induced degradation of fA3s indeed required Cul5 and ElonginC. In addition, we identified an HIV/SIV Vif-like BC-box motif in FIV Vif, which is important for the interaction with ElonginC and recruitment of the E3 ligase. Moreover, FIV Vif specifically bound to Cul5 despite the fact that it has neither a Cul5-box which is used by cellular substrate receptors to recruit Cul5, nor an HCCH zinc-binding motif which is used by primate lentivirus Vif to recruit Cul5. Therefore, FIV Vif may utilize a novel mechanism for binding to Cul5, implying that the SOCS proteins which form Elongin-Cullin-SOCS box (ECS) complexes possess multiple ways to interact with Cul5.
MATERIALS AND METHODS
Cell cultures and transfections.
The human 293T cells (CRL-11268; American Type Culture Collection [ATCC], Manassas, VA), MAGI-CCR5 cells (catalog no. 3522; National Institutes of Health [NIH] AIDS Research and Reference Reagent Program, Germantown, MD), and feline CrFK cells (CCL-94; ATCC) were maintained in Dulbecco high-glucose modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 0.29 mg of l-glutamine/ml, and 100 U of penicillin-streptomycin/ml at 37°C and 5% CO2. Plasmid transfections into 293T cells and CrFK cells were performed with Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA).
RT-PCR.
The expression of feline ElonginB, ElonginC, and Cullin5 in CrFK cells were evaluated by reverse transcription-PCR (RT-PCR). Total RNA was isolated from the CrFK cells using TRIzol (Invitrogen), and the genes were amplified by RT-PCR using the following primers: fElonginB, forward (5′-ATGAACGTGTTCCTTATGATCAGT-3′) and reverse (5′-TCACTGCACCACTTGTTTGTTG-3′); fElonginC, forward (5′-ATGGATGGAGAAGAGAAAACCTATGGTGGC-3′) and reverse (5′-TTACTAACTGAACTACAGGTATTAAATACTGAAAA-3′); and fCul5, forward (5′-GGAACATGGCGACGTCTAATCTGTAAAAG-3′) and reverse (5′-GTTACCGAGTGTGCAACATGAAGGG-3′). The products were inserted into the cloning vector pGEM-T easy vector (Promega, Madison, WI) to generate fEB-T-easy, fEC-T-easy, and fCul5-T-easy.
Plasmids.
VR-FIV Vif-HA is a plasmid expressing the codon-optimized vif of FIV-34TF10. It was generated by adding an hemagglutinin (HA) tag to the C terminus of the codon-optimized vif gene and then inserted into the VR1012 vector (16) at the XbaI and BamHI restriction sites. To construct VR-FIV Vif-myc, FIV Vif-myc fragments were amplified from VR-FIV Vif-HA by PCR using forward (5′-TCTAGAACCATGAGCGAAGA-3′) and reverse (5′-GGATCCTCAAAGATCTTCTTCTGATATGAGTTTTTGTTCCAGCTCGCCGCTCCACAGCA-3′) primers. These PCR products were also inserted into VR1012. The FIV Vif mutants were constructed by site-directed mutagenesis using VR-FIV Vif-myc as a template and the following primers: FIV Vif (TLQ-AAA), forward (5′-GGAACAGCCCCCCCAAGGCAGCTGCTCGGCTGGCCATGCTGG-3′) and reverse (5′-CCAGCATGGCCAGCCGAGCAGCTGCCTTGGGGGGGCTGTTCC-3′); and FIV Vif (C184 187S), forward (5′-CGACCCCAAGAAGTGGAGCGGCGACAGCTGGAACCTGATGTG-3′) and reverse (5′-CACATCAGGTTCCAGCTGTCGCCGCTCCACTTCTTGGGGTCG-3′).
VR-fA3Z2-Z3-HA was generated by cloning the fA3Z2-Z3 gene with a 3′ fused HA tag and inserting it into the VR1012 vector at the XbaI and BglII restriction sites. The fA3Z2-Z3 transcript is encoded by exons 1 to 3 of fA3Z2a, the complete coding sequence of exon 4 of fA3Z2b, and exons 2 to 5 of fA3Z3 (37). To construct VR-fA3Z3-HA, the gene fragments were amplified from VR-fA3Z2-Z3-HA by PCR with forward (5′-TCTAGAATGAATCCACTACAGGAAG-3′) and reverse (5′-AGATCTTCACGCGTAATCTGGGACGTCG-3′) primers. To construct VR-fA3Z2a-HA, the gene fragments were first amplified from VR-fA3Z2-Z3-HA by PCR with fA3Z2a-HA (mut) forward (5′-TCTAGAATGGAGCCCTGGCGCCCC-3′) and reverse (5′-AGATCTTCACGCGTAATCTGGGACGTCGTAAGGGTACCTAAGGATTTCTTGAAGCTCTGCAGCCAGGA-3′) primers and were then point mutated twice with the following primers: R172Q, forward (5′-GAATGCGCTTTCAGAGACGGAATCTGTTA-3′) and reverse (5′-TAACAGATTCCGTCTCTGAAAGCGCATTC-3′); and G179D, forward (5′-ATCTGTTAAAAGATTATGATTTCCTG-3′) and reverse (5′-CAGGAAATCATAATCTTTTAACAGAT-3′). These PCR products were likewise inserted into VR1012.
To generate VR-fElonginC-HA, fElonginC-HA fragments were amplified from fEC-T-easy by PCR with forward (5′-TCTAGAATGGATGGAGAAGAGAAAAC-3′) and reverse (5′-AGATCTTCACGCGTAATCTGGGACGTCGTAAGGGTAACAATCTAGGAAGTTC-3′) primers. These PCR products were likewise inserted into VR1012. The fECΔ2 (A100S L103S) mutant was constructed by using forward (5′-CAATTGCACCTGAAATTTCACTGGAATCTCTGATGGCTGCGAACTTCTTA-3′) and reverse (5′-TAAGAAGTTCGCAGCCATCAGAGATTCCAGTGAAATTTCAGGTGCAATTG-3′) primers.
To generate VR-fElonginB-flag and VR-fCul5-myc, fElonginB-flag and fCul5-myc fragments were amplified from fEB-T-easy and fCul5-T-easy by PCR with the following primers sets, respectively: (i) forward, 5′-GCTCTAGAGCCACCATGAACGTGTTCCTTATG-3′; and reverse, 5′-GTCGGATCCTCACTTGTCGTCATCGTCTTTGTAGTCCTGCACCACTTGTTTGTT-3′; and (ii) forward, 5′-GCTCTAGAGCACCATGGCGACGTCTAATCTGTTAAAGAATAAAG-3′; and reverse, 5′-CTGGATCCTTAAAGATCTTCTTCTGATATGAGTTTTTGTTCTGCCATATATATGAAAG-3′. These PCR products were likewise inserted into VR1012.
The expression vectors PC-hA3G-HA, VR-HIV Vif-myc, VR-hEC-HA, VR-hEB-HA, VR-hCul5-HA, VR-hCul5ΔNedd8-HA, VR-hCul1 K720R-HA, and pNL4-3ΔVif were described previously (31, 39, 58, 61). The fA3Z2-Z3 gene with a 3′ fused HA tag and the codon-optimized vif gene with a 3′ fused HA tag were commercially synthesized (Shinegene, Shanghai, China).
Antibodies.
The following antibodies were used for the present study: anti-HA mouse monoclonal antibody (MAb; Covance, Emeryville, CA), anti-myc mouse MAb (Millipore, Billerica, MA), anti-tubulin mouse MAb (Covance), anti-Cul5 rabbit polyclonal antibody (pAb; Santa Cruz Biotechnology, Santa Cruz, CA), anti-ElonginB rabbit pAb (Santa Cruz Biotechnology), and anti-ElonginC mouse MAb (Santa Cruz Biotechnology).
Immunoprecipitation.
To determine whether FIV Vif binds Cul5, ElonginB, and ElonginC, 293T cells and CrFK cells were transfected with HIV Vif-HA, FIV Vif-HA, or FIV Vif TLQ-HA expression plasmids. At 48 h posttransfection, the cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100) with a protease inhibitor cocktail (Roche, Mannheim, Germany) at 4°C for 40 min. Cell lysates were clarified by centrifugation at 12,000 × g for 10 min at 4°C. Immunoprecipitations were performed by mixing cell lysates with anti-HA antibody and protein G (Roche), followed by incubation at 4°C for 3 h. After extensive washing with lysis buffer, the beads were eluted with 0.1 M glycine-HCl buffer (pH 2.0). The eluted materials were subsequently analyzed by Western blotting.
To determine binding of FIV Vif mutants to Cul5, 293T cells were cotransfected with FIV Vif-myc, FIV Vif mutant, or HIV Vif-myc and Cul5-HA expression plasmids. The cells were incubated, and the immunoprecipitation was performed as described above, except that the cell lysates were incubated with anti-myc antibody, along with protein G. The eluted materials were likewise analyzed by Western blotting.
For coimmunoprecipitations in the presence of N,N,N′,N′-tetrakis(2-pyridylmethyl)-ethylenediamine (TPEN; Sigma, St. Louis, MO), the culture medium was replaced with fresh Dulbecco modified Eagle medium (DMEM) containing 4 or 7 μM TPEN or dimethyl sulfoxide (DMSO). After the cells were treated for 16 h, the cell lysates were immunoprecipitated as described above.
Western blotting.
Cells were collected 48 h after transfection. Cell lysates were prepared by washing cells in phosphate-buffered saline (PBS) and lysing in radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM Tris, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) for 30 min at 4°C, followed by the addition of 4× SDS sample buffer (1 M Tris [pH 6.8], with 8% SDS, 40% glycerol, 0.4 M dithiothreitol, and 0.8% bromophenol blue). The samples were boiled for 10 min, and the proteins were separated by SDS-PAGE. The proteins were transferred onto nitrocellulose membranes by semidry transfer (Bio-Rad, Hercules, CA). After blocking in 5% nonfat milk, the membranes were probed with various primary antibodies against proteins of interest. Secondary antibodies were alkaline phosphatase-conjugated anti-rabbit, anti-mouse IgG (Jackson Immunoresearch, West Grove, PA), and staining was carried out with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium solutions.
Single-round infectivity MAGI assays.
Viral infection was determined by MAGI assay as follows. MAGI-CCR-5 cells were prepared in 24-well plates 1 day before infection. The cells were at 20 to 30% confluence on the day of infection. Cells were infected by removing the medium from each well and adding dilutions of virus in a total volume of 250 μl of complete DMEM with 5 μg of DEAE-dextran per well. After 2 h of incubation at 37°C in a 5% CO2 incubator, 500 μl of complete DMEM was added to each well, and the cells were incubated for 48 h under the same conditions. The supernatants were removed, and 500 μl of fixing solution (1% formaldehyde and 0.2% glutaraldehyde in PBS) was added. After 5 min of incubation, the cells were washed twice with PBS. A staining solution (20 μl of 0.2 M potassium ferricyanide, 20 μl of 0.2 M potassium ferricyanide, 2 μl of 1 M MgCl2, and 10 μl of a 40-mg/ml concentration of X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]) was added. The cells were incubated for 2 h at 37°C in a non-CO2 incubator. Staining was stopped by removing the staining solution, and the cells were thoroughly washed twice with PBS. β-Galactosidase activity is under the control of an HIV-1 long terminal repeat promoter; in this system, positive blue dots indicate the presence of integrated virus. The positive blue dots were counted, and the viral infectivity was determined after normalizing the amount of input virus in terms of p24 antigen content.
Sequence accession numbers.
The cDNA sequences we obtained for feline elonginB, elonginC, and Cul5 have been deposited in GenBank (accession numbers JN020919, JN020920, and JN250995, respectively). The sequence of the codon-optimized vif of FIV was previously available (accession number EF989123). The accession numbers of the protein sequences used for alignment were as follows: human SOCS3, CAG46495; human SSB1, AAL57345; human WSB1, AAD20954; human Rab40C, NP_066991; HIV-1 Vif, NP_057851; HIV-2 Vif, NP_056839; SIV Vif, NP_054370; BIV Vif, AAA42768; FIV C36 Vif, AAT12495; FIV 34TF10 Vif, NP_040974; FIV cougars Vif, ABO69514; FIV Oma Vif, AAB49924; FIV pelamuma Vif, P16089; FIV PPR Vif, AAA43077; FIV San Diego Vif, P19029; and FIV TM2 Vif, P31823.
RESULTS
Cloning and characterization of feline elonginB, elonginC, and cullin5 genes and corresponding proteins.
Cul5, ElonginB, and ElonginC are cellular factors that are highly conserved among diverse mammalian and nonmammalian species, including human, chimpanzee, dog, cow, mouse, zebrafish, Caenorhabditis elegans, and even mosquito. Here, we predicted the feline elonginB and elonginC genes from the available genomic sequences, with the use of sequence BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and automated computational analysis with Genescan. We designed oligonucleotide primers based on the predicted sequences and performed RT-PCR on RNA isolated from a feline kidney cell line (CrFK) with these primers. Both feline elonginB and elonginC cDNA were detected in the CrFK cells (Fig. 1A). The amplified cDNA bands were purified and inserted into the T-easy vector for DNA sequencing. By sequence alignment analysis, feline ElonginB and human ElonginB displayed 86.6% identity at the nucleotide level and 85.6% at the amino acid level (Fig. 1D), while feline ElonginC and human ElonginC displayed 94.7% identity at the nucleotide level and 100% at the amino acid level (Fig. 1E). Although the sequences that we obtained did not completely match the genomic sequences, probably due to the inaccuracy of the shotgun assay and differences between feline individuals, the expression of the fElonginC protein could be detected in CrFK cells with the human ElonginC MAb (Fig. 1B). Overexpression of fElonginC and fElonginB was also detected with antibodies to their protein tags (Fig. 1C).
Fig. 1.
Detection of the feline elonginB, elonginC, and cullin5 gene transcripts and the fElonginC and fCul5 proteins in CrFK cells. (A) PCR amplification of elonginB, elonginC, and cullin5 cDNA by RT-PCR from total mRNA of feline CrFK cells. The β-actin cDNA was used as a control for the RNA quality of each sample. Mk indicates the DNA molecular marker, while control is the mock DNA-negative control. (B) Cell lysates of 293T and CrFK cells were analyzed by Western blotting using anti-hCul5, anti-hElonginC, and anti-hElonginB antibodies. The control is a loading control. (C) 293T cells (106) were transfected with 1 μg of VR-hCul5-myc, VR-hEB-HA, VR-hEC-HA, VR-fCul5-myc, VR-fEB-flag, or VR-fEC-HA. Cells were harvested 48 h after transfection and analyzed by Western blotting with anti-HA, anti-myc and anti-flag antibodies. The control is nontransfected cells. (D) Alignment of the amino acid sequences of the feline ElonginB and human ElonginB. (E) Alignment of the amino acid sequences of the feline ElonginC and human ElonginC. (F) Alignment of the partial amino acid sequences of the feline Cul5 and human Cul5. The arrow indicates the single amino acid difference.
The cullin5 gene consists of more than 10 exons with coding sequences that span several thousand base pairs. At present, the entire genomic sequence of Felis catus is not complete, and we could only predict the incomplete feline cullin5 gene with the available sequences (a short N-terminal sequence and a partial C-terminal sequence). We designed oligonucleotide primers based on the predicted sequences and performed RT-PCR on RNA isolated from a feline kidney cell line (CrFK) with these primers. The feline cullin5 cDNA was detected in the CrFK cells (Fig. 1A). The amplified cDNA bands were purified and inserted into the T-easy vector for DNA sequencing. By sequence alignment analysis, the fCul5 and human Cul5 (hCul5) displayed 95.5% identity at the nucleotide level and 99.9% at the amino acid level (Fig. 1F). Moreover, using a pAb against hCul5, we could detect in CrFK cells the expression of a protein of a similar size to that of hCul5 (Fig. 1B). This result suggested that the fCul5 protein is present and expressed constitutively in CrFK cells. Furthermore, Western blotting was used to confirm expression of the fCul5-myc protein with an antibody to the protein tag (Fig. 1C).
FIV Vif interacts with Cul5, ElonginB and ElonginC to induce proteasome-dependent degradation of fA3s.
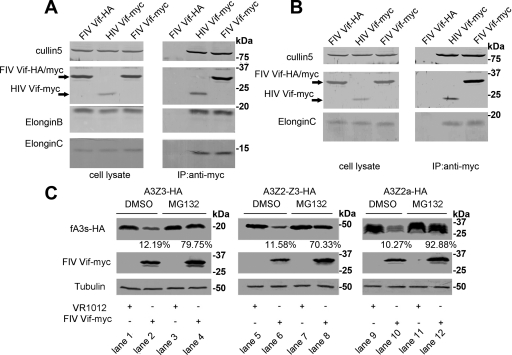
HIV-1 Vif recruits human Cul5, ElonginB and ElonginC to form an E3 complex when targeting APOBEC3G for ubiquitination, subsequently leading to its degradation (58). It is known that FIV Vif colocalizes with feline APOBEC3 proteins, targets them for degradation and rescues the infectivity of virus (47). However, the exact molecular mechanism of how FIV Vif downregulates fA3s is unclear. To explore this mechanism, we performed an immunoprecipitation assay with 293T and CrFK cells transfected with VR-HIV Vif-myc, VR-FIV Vif-myc, or VR-FIV Vif-HA. Cell lysates were precipitated with anti-myc antibody, followed by Western blot analysis with anti-HA, anti-myc, anti-hCul5, anti-hElonginB, and anti-hElonginC antibodies. The result revealed that both HIV Vif-myc and FIV Vif-myc interacted with Cul5, ElonginB, and ElonginC in 293T cells (Fig. 2A). In CrFK cells, HIV Vif-myc and FIV Vif-myc also interacted with the fCul5 protein and fElonginC (Fig. 2B). However, we did not detect the binding of fElonginB. It is likely that the differences in amino acid sequences between fElonginB and hElonginB caused the human ElonginB antibody to not recognize the feline ElonginB protein. Nevertheless, the results of the immunoprecipitation assay indicated that FIV Vif was capable of assembling with Cul5, ElonginB, and ElonginC to form an E3 complex, which is quite similar to the function of HIV Vif.
Fig. 2.
FIV Vif interacts with Cul5, ElonginB, and ElonginC and induces the degradation of fA3s in a proteasome-dependent manner. 293T cells (A) and CrFK cells (B) (2 × 106) were transfected with 4.0 μg of VR-HIV Vif-myc, VR-FIV Vif-myc, or VR-FIV Vif-HA. After 48 h, the cell lysates were immunoprecipitated with anti-myc antibody, followed by Western blot analysis with anti-HA, anti-myc, anti-hCul5, anti-hElonginB, and anti-hElonginC antibodies. (C) CrFK cells (106) were cotransfected with 100 ng of VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 400 ng of VR-FIV Vif-myc or empty plasmid (VR1012). The transfected cells were treated with the proteasome inhibitor MG132 at 10 mM (lanes 3, 4, 7, 8, 11, and 12) or DMSO (lanes 1, 2, 5, 6, 9, and 10) at 36 h after transfection. Cells were harvested 12 h later (48 h after transfection) and then analyzed by Western blotting using anti-HA, anti-myc, and anti-tubulin antibodies. The percentages of fA3s in the presence of FIV Vif with DMSO or MG132 treatment were calculated relative to that in the absence of FIV Vif (set to 100%).
To further investigate the nature of the interactions between FIV Vif and fA3s, CrFK cells were cotransfected with VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and VR-FIV Vif-myc or empty plasmid VR1012 in the presence of the proteasome inhibitor MG132 or DMSO as a negative control. We found that FIV Vif induced the degradation of fA3s (Fig. 2C, lanes 2, 6, and 10), and this degradation could be blocked by MG132 (Fig. 2C, lanes 4, 8, and 12). At the same time, FIV Vif proteins were stabilized by the addition of MG132 (Fig. 2C, lanes 4, 8, and 12), implying that their turnover was dependent on proteasome activity as with HIV Vif (19, 34). In addition, we detected fA3Z2a as triplet bands. Using the software NetNGlyc 1.0, we found that the fA3Z2a protein has two potential N-glycosylation motifs (90N and 116N), inferring that this phenomenon may be caused by glycosylation.
The activity of FIV Vif against fA3s can be disrupted by a dominant-negative Cul5 mutant or a C-terminal hydrophilic replacement ElonginC mutant.
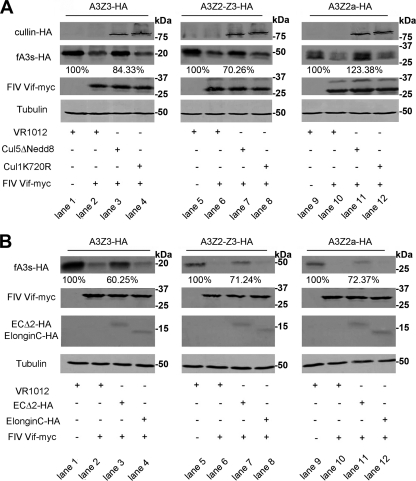
All Cullin family members are known to be modulated by the ubiquitin-like small molecule Nedd8 (18), which is critical for E3 ubiquitin ligase activity. To investigate whether the Cul5-containing complex is required for FIV Vif function against fA3s, we used a hCul5 ΔNedd8 mutant to verify if the addition of this dominant-negative hCul5 protein could inhibit the FIV Vif-induced degradation of fA3s. CrFK cells were cotransfected with VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and VR-FIV Vif-myc or empty plasmid VR1012 plus hCul5 ΔNedd8 or a control vector hCul1 K720R, which is a dominant-negative hCul1 mutant (41). We found that the degradation of the fA3s was blocked by coexpression of hCul5 ΔNedd8 (Fig. 3A, lanes 3, 7, and 11). These results suggest that Cul5 plays an important role in the FIV Vif-induced turnover of fA3s.
Fig. 3.
A dominant-negative Cul5 mutant and a C-terminal hydrophilic replacement ElonginC mutant can both disrupt the activity of FIV Vif against fA3s. (A) CrFK cells (106) were cotransfected with 100 ng of VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 400 ng of VR-FIV Vif-myc or VR1012, adjusted to 700 ng with 200 ng of VR-hCul5ΔNedd8, a control vector VR-hCul1K720R or VR1012. The cells were harvested 48 h after transfection and analyzed by Western blotting with anti-HA, anti-myc, and anti-tubulin antibodies. The percentages of fA3s in the presence of Cul5 ΔNedd8 were calculated relative to that in the absence of FIV Vif (set to 100%). (B) CrFK cells (106) were cotransfected with 100 ng of VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 400 ng of VR-FIV Vif-myc or VR1012, adjusted to 700 ng with 200 ng of fElonginC WT-HA, fElonginC Δ2-HA, or VR1012. The cells were harvested 48 h after transfection and analyzed by Western blotting with anti-HA, anti-myc and anti-tubulin antibodies. The percentages of fA3s in the presence of ElonginCΔ2 were calculated relative to that in the absence of FIV Vif (set to 100%).
To further strengthen the notion that ElonginC is essential for the activity of FIV Vif against fA3s, we constructed a feline ElonginC mutant (fEC Δ2-HA) in which the critical hydrophobic amino acids (A100 and L103) were replaced with hydrophilic serine (46). CrFK cells were cotransfected with VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and VR-FIV Vif-myc or empty plasmid VR1012 plus fEC Δ2-HA or a control vector fEC WT-HA. As expected, the expression of fEC Δ2-HA in CrFK cells significantly increased the amount of fA3s (Fig. 3B, lanes 3, 7, and 11). These results indicated that ElonginC was required for the degradation of fA3s by FIV Vif. In addition, the fact that ElonginC interacts with the BC-box of the cellular proteins through its hydrophobic binding pocket (46) implies that FIV Vif may possess an HIV/SIV Vif-like BC-box.
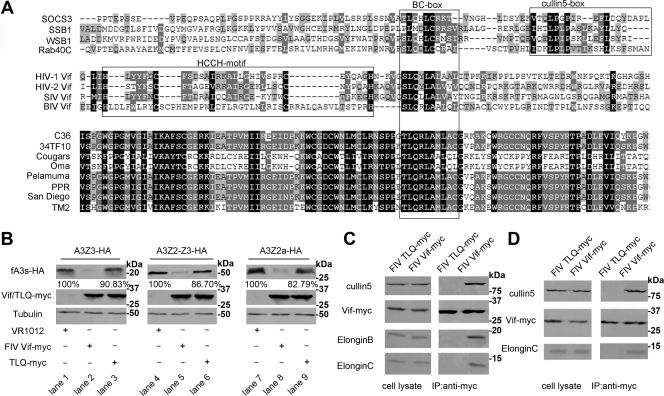
FIV Vif contains a BC-box motif that is required for its interaction with the Cul5-EB-EC E3 complex.
In SOCS-box proteins, a sequence motif with consensus sequence [STP] LXXX [CSA] XXXΦ is referred to as the BC-box, is required for binding to the Elongin BC complex (32). By analyzing FIV Vif and a variety of retroviral Vif protein sequences, we identified a BC-box motif in FIV Vif (amino acids 200 to 209). This motif was found to be highly conserved among diverse FIV Vif and SOCS box proteins (Fig. 4A). Like the divergent BC-box motif present in SIV Vif and adenovirus E4orf6 molecules (29, 30), it lacks a downstream Cul5-box (Fig. 4A). To determine whether the BC-box is required for the interaction between FIV Vif and the Cul5-EB-EC E3 complex, we introduced a TLQ-AAA mutation into the putative BC-box. CrFK cells were cotransfected with VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and VR-FIV Vif-myc or VR-FIV Vif TLQ-myc, followed by analysis of the cell lysates by Western blotting. We found that the FIV Vif TLQ mutant not only lost the ability to degrade fA3s (Fig. 4B, lanes 3, 6, and 9), but it also lost the ability to inhibit the antiviral activity of fA3Z3 and fA3Z2-Z3 (see Fig. 6), which confirmed the theory that the BC-box of FIV Vif was critical for its activity against fA3s. To further support this point, 293T and CrFK cells were transfected with VR-FIV Vif-myc or VR-FIV Vif TLQ-myc. The cell lysates were precipitated with an anti-myc antibody, followed by Western blotting with anti-myc, anti-hCul5, anti-hElonginB, and anti-hElonginC antibodies. The data showed that the interaction of the Cul5-EB-EC E3 complex with FIV Vif TLQ was significantly decreased compared to wild-type FIV Vif (Fig. 4C and D). These results indicated that the BC-box motif in FIV Vif was required for interaction with the Cul5-EB-EC E3 complex and degradation of fA3s.
Fig. 4.
The BC-box motif in FIV Vif is critical for interaction with the Cul5-EB-EC E3 complex. (A) Alignment of partial FIV Vif sequences from 34TF10 with lentiviral Vif and cellular SOCS-box proteins by vector NTI. (B) CrFK cells (106) were cotransfected with 100 ng of VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 400 ng of VR-FIV Vif-myc or VR-FIV Vif TLQ-myc or VR1012. Stabilities of fA3 proteins were analyzed by Western blotting with anti-HA, anti-myc, and anti-tubulin antibodies. The percentages of fA3s in the presence of FIV Vif TLQ mutant were calculated relative to that in the absence of FIV Vif (set to 100%). 293T cells (C) and CrFK cells (D) (2 × 106) were transfected with 4 μg of VR-FIV Vif-myc or VR-FIV Vif TLQ-myc. Cell lysates were immunoprecipitated with anti-myc antibody, followed by Western blotting with anti-myc, anti-hCul5, anti-hElonginB, or anti-hElonginC antibodies.
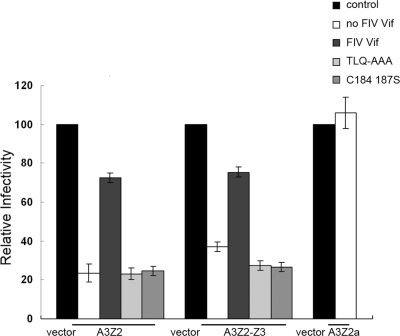
Fig. 6.
Antiretroviral properties of feline A3 proteins and their sensitivity to FIV Vif and mutants. 293T cells were cotransfected with 500 ng of pNL4-3ΔVif plus 10 ng of VR1012, VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 40 ng of VR-FIV Vif-myc, FIV Vif mutant, or VR1012. The virus infectivity was assayed by infecting MAGI cells. Cells were fixed and stained for β-galactosidase activity. Virus infectivity in the absence of fA3s was set to 100%.
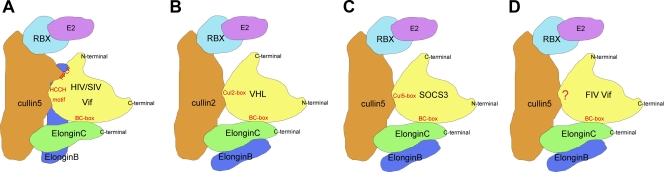
FIV Vif binds with Cul5 through a novel mechanism.
Cellular proteins assemble with Cul2/Cul5-EB-EC E3 complexes through a BC-box and a downstream Cul-box (21). Some cellular proteins such as von Hippel-Lindau tumor suppressor (VHL) use a Cul2-box to bind with Cul2 (see Fig. 7B), while others such as SOCS3 use a Cul5-box to bind with Cul5 (Fig. 7C). Primate lentiviral (HIV/SIV) Vif use a zinc-binding HCCH motif to interact with Cul5 (30, 54) (Fig. 7A). However, through in-depth sequence analysis, we found that FIV Vif contains neither a Cul5-box nor an HCCH motif (Fig. 7A). To determine whether zinc is required for the interaction between FIV Vif and Cul5, we used the membrane-permeable zinc chelator TPEN. TPEN is known to inhibit the binding of HIV Vif with Cul5 (53). Here, 293T cells were cotransfected with VR-HIV Vif-myc or VR-FIV Vif-myc and VR-hCul5-HA and treated with increasing amounts of TPEN or control DMSO. HIV Vif was set as a positive control. The presence of TPEN disrupted the association of HIV Vif with Cul5 but had no effect on the interaction between FIV Vif and Cul5 (Fig. 5A). In parallel, these data suggested that zinc chelation by 7 μM TPEN did not disrupt the degradation of fA3s (Fig. 5B, lanes 8, 12, and 16), while the HIV Vif-induced degradation of hA3G was inhibited (Fig. 5B, lane 4). This indicated that zinc was not an essential cofactor for FIV Vif function. At the same time, HIV Vif proteins also were stabilized by the addition of TPEN (Fig. 5B, lane 4), implying that HIV Vif was degraded together with hA3G, or the degradation process of HIV Vif was related to the zinc finger (34).
Fig. 7.
Schematic representation of multiple classes of the Cul5-EB-EC E3 complex. HIV/SIV Vif select Cul5 based on the presence of the HCCH motif (A), whereas cellular substrate receptors (VHL or SOCS3) utilize a Cul2/Cul5 box to interact with Cul2/Cul5 (B and C). FIV Vif uses a motif that is yet to be determined (D).
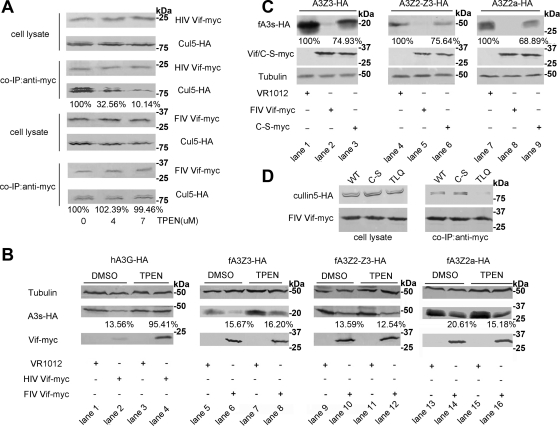
Fig. 5.
FIV Vif selects Cul5 via a novel mechanism. (A) 293T cells (106) were cotransfected with 1 μg of VR-HIV Vif-myc or VR-FIV Vif-myc and 1 μg of VR-Cul5-HA. Cells were treated with increasing concentrations of TPEN (0, 4, and 7 μM) or control DMSO 12 h before harvesting. The cell lysates were coimmunoprecipitated and analyzed by Western blotting with anti-HA and anti-myc antibodies. The percentages of Cul5 binding to Vif with different concentrations of TPEN were calculated relative to that in the absence of TPEN (set to 100%). (B) 293T cells (106) were cotransfected with 100 ng of PC-hA3G-HA, VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 400 ng of VR-HIV Vif-myc, VR-FIV Vif-myc, or VR1012. Transfected cells were treated with the membrane-permeable zinc chelator TPEN at 7 μM (lanes 3, 4, 7, 8, 11, 12, 15, and 16) or DMSO (lanes 1, 2, 5, 6, 9, 10, 13, and 14) at 36 h after transfection. Cells were harvested 12 h later (48 h after transfection) and then analyzed by Western blotting with anti-HA, anti-myc, and anti-tubulin antibodies. The percentages of APOBEC3s in the presence of Vif with DMSO or TPEN treatment were calculated relative to that in the absence of Vif (set to 100%). (C) CrFK cells (106) were cotransfected with 100 ng of VR-fA3Z3-HA, VR-fA3Z2-Z3-HA, or VR-fA3Z2a-HA and 400 ng of VR-FIV Vif-myc, VR-FIV Vif C-S-myc or VR1012. The stabilities of fA3 proteins were analyzed by Western blotting with anti-HA, anti-myc, and anti-tubulin antibodies. The percentages of fA3s in the presence of FIV Vif C-S mutant were calculated relative to that in the absence of FIV Vif (set to 100%). (D) 293T cells (106) were cotransfected with 1 μg of VR-Cul5-HA and 1 μg of VR-FIV Vif-myc, VR-FIV Vif C-S-myc, or VR-FIV Vif TLQ-myc. Cell lysates were precipitated with anti-myc antibody, followed by Western blotting with anti-myc and anti-HA antibodies.
Based on sequence analysis of variant FIV and other lentiviral Vif proteins, a putative Cul5 binding domain (KCCC) in FIV Vif, which is similar to the HCCH motif of HIV Vif, was previously reported (47). To investigate whether this putative motif functions in an analogous manner to that of the HCCH motif, we constructed a mutant of FIV Vif in which two amino acids (C184 and C187) in the C-terminal region were replaced with serine. 293T cells were cotransfected with VR-hCul5-HA and VR-FIV Vif-myc, VR-FIV Vif C184,187S-myc, or VR-FIV Vif TLQ-myc. The cell lysates were precipitated with anti-myc antibody, followed by Western blotting with anti-myc and anti-HA antibodies. The data showed that FIV Vif C184,187S maintained the ability to bind with Cul5, revealing that the KCCC motif may not be the key feature involved in the interaction of FIV Vif with Cul5 (Fig. 5D). However, the levels and the antiviral activity of fA3s were upregulated in the presence of FIV Vif C184,187S, suggesting that the C-S mutation affected the FIV Vif-induced degradation of fA3s (Fig. 5C and Fig. 6). Taken together, we propose that FIV Vif may possess a novel, but yet to be defined mechanism to bind with Cul5 (Fig. 7D).
DISCUSSION
The predominant mechanism of HIV Vif for overcoming the antiviral activity of human A3 proteins is to form an E3 ubiquitin ligase with Cul5, ElonginB, and ElonginC and target these proteins for degradation by the ubiquitin-proteasome pathway (33, 35, 44, 45, 58, 60). FIV Vif also inhibits the antiviral activity of feline A3Z3 and A3Z2-Z3 by inducing the degradation of these host restriction factors (23, 47, 63). We hypothesized that FIV Vif utilizes a similar mechanism to degrade fA3s. However, fCul5, fElonginB, and fElonginC, the feline components of the E3 complex, were not previously reported. Here, we predicted the feline elonginB, elonginC, and partial cullin5 (a short N-terminal sequence and a partial C-terminal sequence) genes within the available feline genomic sequences and amplified the corresponding cDNAs from CrFK cells by RT-PCR. We further confirmed the expression of fElonginC and fCul5 in CrFK cells by Western blotting. Moreover, analysis of the sequence alignments demonstrated that fElonginB, fElonginC, and the fCul5 were highly homologous with the human counterparts (Fig. 1D, E, and F).
In order to further understand the function of FIV Vif, we investigated the interaction between FIV Vif and cellular factors. We observed that FIV Vif specifically assembled with Cul5, ElonginB, and ElonginC in 293T cells (Fig. 2A). In CrFK cells, we detected interactions between FIV Vif, fElonginC, and fCul5 (Fig. 2B). Because the human ElonginB antibody was unable to recognize the feline ElonginB protein (Fig. 1B), we could not confirm whether fElonginB was present in the complex. However, HIV/SIV Vif, E4orf6, and other SOCS-box proteins can bind with Cul5, ElonginB, and ElonginC to form E3 complexes (21, 29, 60, 62), and we found that hElonginB and fElonginB are highly homologous (Fig. 1D). As a consequence, it is likely that FIV Vif is able to form a similar complex in CrFK cells.
We also showed that a dominant-negative Cul5 mutant and a C-terminal hydrophilic replacement ElonginC mutant could disrupt FIV Vif-induced degradation of fA3s, which proved that both Cul5 and ElonginC are required for the FIV Vif activity against fA3s. In the presence of the proteasome inhibitor MG132, the turnover of fA3s mediated by FIV Vif was reduced. Therefore, we concluded that FIV Vif assembled with Cul5, ElonginB, and ElonginC to form an E3 complex to degrade feline A3Z2a, which has low anti-FIV activity, and feline A3Z3/A3Z2-Z3 with high anti-FIV activity mediated by the proteasome. This mechanism is quite similar to that of HIV/SIV Vif. It has been theorized that the counteraction between different retroviruses and their hosts results in the distinct evolution of the viral antagonist (Vif) to resist the natural antivirus factors (A3 proteins). As a consequence, the mechanisms of biological action between FIV Vif and HIV Vif are similar, even though their primary structures are quite different.
The BC-box is a highly conserved motif among many proteins that interact with Cullin and ElonginB/C (21, 29, 30). With HIV, for example, the Vif protein binds to ElonginC with its BC-box motif, and then its PPLP motif interacts with ElonginB. These interactions, together with the interaction between Cul5 and Vif HCCH domain, are necessary for the assembly of a functional ubiquitin ligase (1). A BLAST search of the conserved domain database revealed that FIV Vif contains a C-terminal sequence motif resembling the BC-box. A previous report indicated that the FIV Vif TLQ mutant which contains a substitution in the core region of BC-box lacks the ability to inhibit the antiviral activity of A3 proteins (23). In our study, wild-type FIV Vif was coimmunoprecipitated with Cul5, ElonginB, and ElonginC, while the FIV Vif TLQ mutant was not and also could not degrade fA3s. Meanwhile, the mutation disrupted the ability of the mutant Vif protein to rescue virus infectivity. These findings confirmed that the BC-box motif in FIV Vif is essential for the recruitment of the E3 complex. On the other hand, the TLQ mutation in the BC-box not only blocked the interaction with ElonginB/C, but it also disrupted the binding of Cul5. This result may be due to the interaction between FIV Vif and ElonginB/C that is prerequisite for the binding of Cul5 (1, 52). Binding of FIV Vif to Cul5, ElonginB, and ElonginC was strongly dependent on the presence of an intact BC-box within the Vif protein. Further analysis is needed to properly address the question of whether other amino acids in this motif are critical, as well as to examine the effect of FIV Vif T200 phosphorylation on ElonginC binding and fA3 degradation (34).
Cellular substrate receptors such as SOCS3 use a Cul5-box downstream from the BC-box to select Cul5 (21), while primate lentiviral Vif proteins use a highly conserved HCCH zinc-binding motif to recruit Cul5 (30, 54). However, we did not find either a Cul5-box or a HCCH motif after a careful search of the FIV Vif sequence. In addition, the membrane-permeable zinc chelator TPEN could neither inhibit the interaction between FIV Vif and Cul5, nor affect the degradation of fA3s, even with TPEN at concentrations up to 7 μM. The putative KCCC motif of FIV Vif was found to be not critical for Cul5 binding, although the FIV Vif C184,187S mutations inhibited the degradation of fA3s and rescued the antiviral activity of fA3Z2 and fA3Z2-Z3. Thus, we propose these results may be caused by other reasons. For example, Vif may fail to dimerize which is essential for degradation of A3 proteins, or these mutations may account for the destruction of other functional domains in FIV Vif. Therefore, FIV Vif may possess a novel mechanism to bind with Cul5 that does not require zinc, implying that SOCS proteins which are able to form E3 complexes may use distinct mechanisms to bind with Cul5. Meanwhile, this finding also indicated that the selectivity of Cul5 binding is extensive. Since the adenovirus protein E4orf6 also utilizes an unknown motif to bind Cul5 (29), whether it utilizes the same mechanism as that of FIV Vif to interact with Cul5 remains to be seen.
In conclusion, our results presented here define the detailed mechanism by which the FIV Vif induces the degradation of feline A3 proteins. FIV Vif uses a conserved viral BC-box to interact with ElonginC and to recruit Cul5 and ElonginB. Although lacking a Cul5 box and an HCCH motif, FIV Vif specifically selects Cul5, suggesting that it may utilize an alternative, possibly zinc-independent interface, for Cul5 selection. Future studies to define the interactions between FIV Vif and feline A3s would shed light on the novel functional domain in this lentiviral Vif protein, as well as on the potential for cross-species transmission of FIV.
ACKNOWLEDGMENTS
We thank the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease (NIAID), for generously providing the MAGI-CCR5 cell lines and the pNL4-3Δvif plasmid. We thank Xiaofang Yu (Johns Hopkins University, Baltimore, MD) for kindly providing some important plasmids such as the human Cul5, human Cul1, and HIV-1 Vif expression plasmids. We thank Jiaxin Wu for advice and technical assistance. We also thank Phuong Thi Sarkis for editorial assistance.
This study was supported in part by grant 30570363 from the National Nature Science Foundation of China.
Footnotes
Published ahead of print on 28 September 2011.
ADDENDUM IN PROOF
We report here the cloning of feline ElonginB sequences (JN020919) that are equivalent to human ElonginB variant 1 (NM_0071108.3), which encodes the a isoform. The homology comparisons reported here are to this variant and not to variant 2 (NM_207013.2), which encodes the b isoform. There is substantial evidence in the literature that human ElonginB isoform a is involved in the formation of the E3 complexes described here.
REFERENCES
- 1. Bergeron J. R., et al. 2010. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 6:e1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bieniasz P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115 [DOI] [PubMed] [Google Scholar]
- 3. Bogerd H. P., Wiegand H. L., Doehle B. P., Lueders K. K., Cullen B. R. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cen S., et al. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177–33184 [DOI] [PubMed] [Google Scholar]
- 5. Chen G., He Z., Wang T., Xu R., Yu X. F. 2009. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J. Virol. 83:8674–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dang Y., Davis R. W., York I. A., Zheng Y. H. 2010. Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J. Virol. 84:5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang Y., et al. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283:11606–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang Y., Wang X., Esselman W. J., Zheng Y. H. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dang Y., Wang X., York I. A., Zheng Y. H. 2010. Identification of a critical T(Q/D/E)x5ADx2(I/L) motif from primate lentivirus Vif proteins that regulate APOBEC3G and APOBEC3F neutralizing activity. J. Virol. 84:8561–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dang Y., Wang X., Zhou T., York I. A., Zheng Y. H. 2009. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 83:8544–8552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esnault C., et al. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433 [DOI] [PubMed] [Google Scholar]
- 12. Fisher A. G., et al. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888–893 [DOI] [PubMed] [Google Scholar]
- 13. Goila-Gaur R., Strebel K. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 15. Harris R. S., Liddament M. T. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868–877 [DOI] [PubMed] [Google Scholar]
- 16. Hartikka J., et al. 1996. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther. 7:1205–1217 [DOI] [PubMed] [Google Scholar]
- 17. He Z., Zhang W., Chen G., Xu R., Yu X. F. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000–1011 [DOI] [PubMed] [Google Scholar]
- 18. Hori T., et al. 1999. Covalent modification of all members of human Cullin family proteins by NEDD8. Oncogene 18:6829–6834 [DOI] [PubMed] [Google Scholar]
- 19. Izumi T., et al. 2009. MDM2 is a novel E3 ligase for HIV-1 Vif. Retrovirology 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jonsson S. R., et al. 2007. The restriction of zoonotic PERV transmission by human APOBEC3G. PLoS One 2:e893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamura T., et al. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. LaRue R. S., et al. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83:494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larue R. S., Lengyel J., Jonsson S. R., Andresdottir V., Harris R. S. 2010. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 84:8193–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lecossier D., Bouchonnet F., Clavel F., Hance A. J. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 25. Lee J., et al. 2011. Repression of porcine endogenous retrovirus infection by human APOBEC3 proteins. Biochem. Biophys. Res. Commun. 407:266–270 [DOI] [PubMed] [Google Scholar]
- 26. Lee T. H., et al. 1986. A new HTLV-1II/LAV protein encoded by a gene found in cytopathic retroviruses. Science 231:1546–1549 [DOI] [PubMed] [Google Scholar]
- 27. Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391 [DOI] [PubMed] [Google Scholar]
- 28. Lochelt M., et al. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo K., et al. 2007. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 21:1742–1750 [DOI] [PubMed] [Google Scholar]
- 30. Luo K., et al. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with Cullin5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lv M., et al. 2011. Polarity changes in the transmembrane domain core of HIV-1 Vpu inhibits its anti-tetherin activity. PLoS One 6:e20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahrour N., et al. 2008. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J. Biol. Chem. 283:8005–8013 [DOI] [PubMed] [Google Scholar]
- 33. Marin M., Rose K. M., Kozak S. L., Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403 [DOI] [PubMed] [Google Scholar]
- 34. Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mehle A., et al. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792–7798 [DOI] [PubMed] [Google Scholar]
- 36. Mehle A., Thomas E. R., Rajendran K. S., Gabuzda D. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259–17265 [DOI] [PubMed] [Google Scholar]
- 37. Munk C., et al. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Munk C., Hechler T., Chareza S., Lochelt M. 2010. Restriction of feline retroviruses: lessons from cat APOBEC3 cytidine deaminases and TRIM5α proteins. Vet. Immunol. Immunopathol. 134:14–24 [DOI] [PubMed] [Google Scholar]
- 39. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., Wu K. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23:1985–1997 [DOI] [PubMed] [Google Scholar]
- 40. Pery E., Rajendran K. S., Brazier A. J., Gabuzda D. 2009. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J. Virol. 83:2374–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Read M. A., et al. 2000. Nedd8 modification of cul-1 activates SCF(β(TrCP))-dependent ubiquitination of IκBα. Mol. Cell. Biol. 20:2326–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Russell R. A., Pathak V. K. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 44. Sheehy A. M., Gaddis N. C., Malim M. H. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 45. Stanley B. J., et al. 2008. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 82:8656–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stebbins C. E., Kaelin W. G., Jr., Pavletich N. P. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455–461 [DOI] [PubMed] [Google Scholar]
- 47. Stern M. A., et al. 2010. Productive replication of Vif-chimeric HIV-1 in feline cells. J. Virol. 84:7378–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Strebel K., et al. 1987. The HIV “A” (sor) gene product is essential for virus infectivity. Nature 328:728–730 [DOI] [PubMed] [Google Scholar]
- 49. Svarovskaia E. S., et al. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822–35828 [DOI] [PubMed] [Google Scholar]
- 50. Turelli P., Mangeat B., Jost S., Vianin S., Trono D. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 51. Walker R. C., Jr., et al. 2010. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J. Virol. 84:5201–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wolfe L. S., Stanley B. J., Liu C., Eliason W. K., Xiong Y. 2010. Dissection of the HIV Vif interaction with human E3 ubiquitin ligase. J. Virol. 84:7135–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiao Z., Ehrlich E., Luo K., Xiong Y., Yu X. F. 2007. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 21:217–222 [DOI] [PubMed] [Google Scholar]
- 54. Xiao Z., et al. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290–299 [DOI] [PubMed] [Google Scholar]
- 55. Xu R., et al. 2007. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology 46:1810–1820 [DOI] [PubMed] [Google Scholar]
- 56. Yang S., Sun Y., Zhang H. 2001. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J. Biol. Chem. 276:4889–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu Q., et al. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442 [DOI] [PubMed] [Google Scholar]
- 58. Yu X., et al. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060 [DOI] [PubMed] [Google Scholar]
- 59. Yu X. F. 2006. Innate cellular defenses of APOBEC3 cytidine deaminases and viral counter-defenses. Curr. Opin. HIV AIDS 1:187–193 [DOI] [PubMed] [Google Scholar]
- 60. Yu Y., Xiao Z., Ehrlich E. S., Yu X., Yu X. F. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang W., Chen G., Niewiadomska A. M., Xu R., Yu X. F. 2008. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host antiviral proteins. PLoS One 3:e3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang W., et al. 2008. Conserved and non-conserved features of HIV-1 and SIVagm Vif mediated suppression of APOBEC3 cytidine deaminases. Cell Microbiol. 10:1662–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zielonka J., et al. 2010. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J. Virol. 84:7312–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]