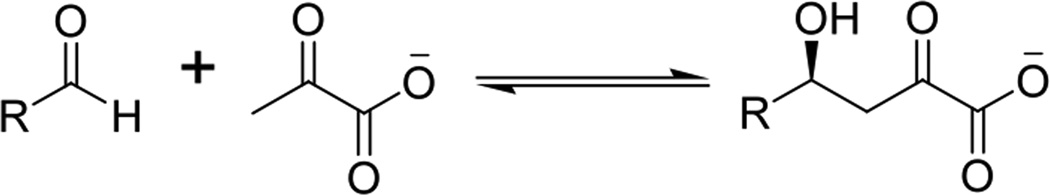Abstract
The use of biological catalysts for industrial scale synthetic chemistry is highly attractive, given their cost effectiveness, high specificity that obviates the need for protecting group chemistry, and the environmentally benign nature of enzymatic procedures. Here we evolve the naturally occurring 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolases from Thermatoga maritima and Escherichia coli, into enzymes that recognize a non-functionalized electrophilic substrate, 2-keto-4-hydroxyoctonoate (KHO). Using an in vivo selection based on pyruvate auxotrophy, mutations were identified that lower the KM value up to 100-fold in E. coli KDPG aldolase, and that enhance the efficiency of retro-aldol cleavage of KHO by increasing the value of kcat/KM up to 25-fold in T. maritima KDPG aldolase. These data indicate that numerous mutations distal from the active site contribute to enhanced “uniform binding” of the substrates, which is the first step in the evolution of novel catalytic activity.
Keywords: biocatalysis, protein engineering, thermostable enzyme, substrate specificity, random mutagenesis
1. Introduction
Protein engineering is a rapidly growing field of study where naturally occurring enzymes are altered to improve activity for industrial and laboratory applications. The advantages of biocatalysts over traditional chemical synthesis methodologies are their exquisite chemo-, stereo- and regio-selectivity, high rates of catalysis, and reactivity under mild and chemically benign conditions.1 The former properties are highly desirable for pharmaceutical applications where the production of enantiomerically pure compounds is critical, while the latter property offers cost savings for large-scale industrial applications.
Much research has focused on the development of aldolases as synthetic reagents because of the centrality of carbon-carbon bond formation in synthetic chemistry.2–4 We have focused our efforts on evolving Escherichia coli (EC) 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase and a closely related isoform from the thermophilic bacteria Thermatoga maritima (TM). These aldolases preferentially catalyze si-stereo facial aldol addition reactions between pyruvate and a range of aldehydic electrophiles5–7 (Figure 1). The types of structures afforded by the enzyme have proven useful in the synthesis of Nikkomycins.8, 9 The substrate tolerance of KDPG aldolase from a number of species includes a variety of 3 and 4 carbon sugars such as erythrose and lactaldehyde with a preference for phosphorylated substrates.6, 7 These enzymes also accept 2-,3-, and 4-pyridine carboxaldehyde as electrophilic substrates with low efficiency,6, 7 but do not catalyze aldol addition reactions with unfunctionalized aldehydes such as butyraldehyde and valeraldehyde. Since the synthesis of many valuable natural products require carbon-carbon bond formation using unfunctionalized aldehydes (for example Syrigomycin10 and Lyngbyabellin11), and since phosphorylated compounds are inconvenient for most synthetic schemes,12 we sought to develop enzymes with improved catalytic activity towards long chain acyl substrates.
Figure 1.
KDPG aldolase-catalyzed reversible aldol reaction.
To broaden the substrate tolerance of the KDPG aldolases, we employed directed evolution to selectively enhance the ability of the enzyme to catalyze aldol addition with 2-keto-4-hydroxyoctonoate (KHO), an unnatural electrophile that represents a first step towards aldol synthesis with long chain acyl aldehydes. As previously reported, our selection uses a pyruvate kinase (pykA and pykF) deficient cell line that is unable to grow on minimal media supplemented with ribose as the sole carbon source13, 14(Figure S1). Addition of exogenous pyruvate rescues the auxotrophic phenotype. Furthermore, we have previously demonstrated that cleavage of an unnatural substrate, 2-keto-4-hydroxy-4-(2’-pyridyl)butyrate (KHPB), catalyzed by a KDPG aldolase mutant can produce enough pyruvate to rescue cell growth, validating our selection approach.14
To examine the hypothesis that higher protein stability would allow for sampling of a wider range of sequence space while maintaining a properly folded structure, we carried out directed evolution experiments on both the T. maritima and E. coli KDPG aldolase enzymes. The E. coli and T. maritima KDPG aldolases have 34% identity (pairwise alignment at http://blast.ncbi.nlm.nih.gov); the sequence differences afford the TM KDPG aldolase both thermostability and minor differences in substrate specificity.6 Using random mutagenesis and selection we identified KDPG aldolase mutants that improve the efficiency for cleavage (kcat/KM) of KHO by up to 25-fold. The majority of mutations identified are located far from the active site, suggesting that subtle long-range remodeling of the active site is the mechanism of adaptation, consistent with Albery and Knowles’ model of “uniform binding”.15
2. Results
According to the Haldane relationship,16 the efficiency for catalysis (kcat/KM) of the forward and reverse reactions is related by the overall equilibrium constant for the reaction, Keq. Thus changes in the efficiency of retro-aldol cleavage for a given substrate also reflect the efficiency of the enzyme to perform aldol addition reactions. Similarly, since Keq is unchanged by mutation of the enzyme, enhancements in the efficiency of retro-aldol cleavage reflect a comparable improvement in the synthetic direction. So the steady-state kinetic parameters for retro-aldol cleavage of KHO catalyzed by wild-type KDPG aldolase were measured to provide a baseline for evaluation of improvements in catalytic efficiency made during mutagenesis. The kinetic parameters for kcat/KM for the cleavage of KHO catalyzed by the T. maritima and E. coli KDPG aldolases are 2.2 ± 0.3 M−1s−1 (34 °C) and 14 ± 4M−1s−1 (25 °C), respectively. Compared to the cleavage of the natural substrate, KDPG, the catalytic efficiency for cleaving KHO is decreased by 104–106-fold, indicating that KHO is a very poor substrate for these enzymes, presumably due in part to unfavorable interactions between the hydrophobic chain of KHO and the hydrophilic phosphate binding site of the enzyme.9 Furthermore, the initial velocity has a nearly linear dependence on the concentration of KHO up to 50 mM for both enzymes, indicating that the value of KM is significantly larger than 50 mM. (Concentrations of KHO above 50 mM lead to noticeable precipitation of the coupling enzyme.) A fit of the Michaelis-Menten equation to these data provided estimated values for kcat of 0.4 ± 0.05 s−1 (TM) and 2.0 ± 0.6s−1 (EC), and KM of 200 ± 30 mM (TM) and 150 ± 50 mM (EC). These values for KM are a lower limit and could reflect the solubility of the substrate. Although KM frequently includes multiple terms and does not simply reflect binding affinity, the high value of this parameter coupled with the low turnover number suggests that wild-type KDPG aldolase binds KHO in a productive conformation with low affinity. Furthermore, the low value of kcat suggests that KHO bound to KDPG aldolase is not positioned to maximize catalytic efficiency. With such low catalytic activity, overexpression of either T. maritima or E. coli KDPG aldolase is not sufficient to rescue growth of the PB25 cell line on ribose/KHO media.14
To identify mutants of KDPG aldolase with enhanced catalytic activity towards retro-aldol cleavage of KHO, we prepared libraries of T. maritima and E. coli KDPG aldolases by random mutagenesis and selected for cells that showed growth on KHO. Libraries were generated by error prone PCR, allowing for rapid incorporation of one to five amino acid changes over the 0.6 kilobase gene, resulting in an overall library complexity of 1014. Although it is not currently feasible to exhaustively screen such a large library using in vivo selection methodologies, literature examples suggest that meaningful improvements in catalytic activity can be obtained by screening libraries of 103–105 clones.17, 18
2.1. Directed evolution and selection of T. maritima KDPG aldolase
Libraries of T. maritima aldolase were generated by error-prone PCR, using a commercially available low fidelity polymerase (Mutazyme II, Stratagene). Compared to mutagenesis methods using Taq DNA polymerase with Mn2+, Mutazyme II provides a more even distribution of mutations at all nucleotides.19 Plasmids that confer growth of PB25 cells on ribose were identified by plating on ribose/KHO minimal media at 34 °C (optimal growth temperature for PB25 cells). Colonies that grew within 120 hours of plating were isolated. Based on transformation controls, the approximate size of the total number of variants screened in this selection was 4×107.
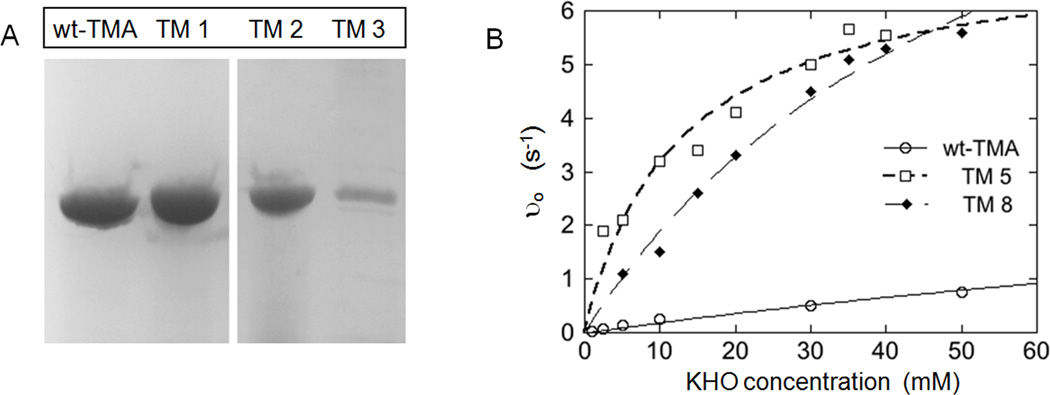
Plasmid DNA from colonies recovered in the first round of selection was isolated and the mutant eda genes were sequenced (shown in Table 1, TM 1–3). These plasmids were retransformed into PB25 cells and subjected again to the selection conditions to confirm that the growth phenotype was dependent on the plasmid encoded eda gene. Only plasmids that engendered growth were analyzed further. These isolated mutants contain 2 to 9 altered nucleotides in the KDPG gene leading to 2–7 amino acid changes (Table 1). Unexpectedly, one selected plasmid contained a TM eda gene with a frameshift mutation (TM 3) in the 2ndcodon of the gene. Expression of nearly full-length protein is still observed for this mutant since the third codon is a redundant start. However, a Western blot of whole cell lysates of each first round mutant indicated that the selected mutations decrease expression of the mutants up to 10-fold as compared to the wild-type enzyme (Figure 2A, Table 1).
Table 1.
T. maritima KDPG aldolase mutants selected for enhanced catalysis of KHO cleavage.
| Name | Mutations | Relative Expression Level |
|---|---|---|
| TM 1 | G34R/V172A/R190I | ++++ |
| TM 2 | K9T/G83E | +++ |
| TM 3 | *Deletion of first two AA, L16P/V31A/S82C/G83V/F99L/W167R | ++ |
| TM 4† | V136M | ++ |
| TM 5† | A30V | ++ |
| TM 6‡ | I52F/V135M/T185A | + |
| TM 7‡ | P109S | + |
| TM 8‡ | N150I | + |
100%;
50%;
10%;
5% expression relative to wild-type TMA in TMEDA-pUC plasmid
These proteins were selected in a pUC plasmid where the expression of wild-type TMA is 10% of the expression in the TMEDA-pUC plasmid.
These proteins were selected in a pUC plasmid where the expression of wild-type TMA is ~2% of the expression in the TMEDA-pUC plasmid.
Signifies a frameshift mutation where expression of the gene is initiated at the third amino acid.
Figure 2.
(A) Western blot of His-tagged KDPG aldolase mutants after the first round of selection probing with an anti-His antibody and (B) dependence of the initial velocity for pyruvate formation on the concentration of KHO catalyzed by wild-type T. maritima KDPG aldolase (open circle) and mutants TM 5 (open square) and TM 8 (filled diamond), measured using a lactate dehydrogenase coupled assay at 34 °C.
To examine whether the selection procedure identified KDPG aldolases with increased catalytic efficiency, we analyzed the kinetic parameters for KHO cleavage catalyzed by these proteins. Each KDPG aldolase mutant was expressed in PB25 cells grown on rich media and purified using a C-terminal 6×His-tag. Proteins of approximately 24 kDa, as determined by SDS-PAGE analysis, were obtained for all TM mutants. The degree of fitness towards cleavage of the KHO substrate was analyzed by measuring the steady-state kinetic parameters at the selection growth temperature, 34 °C. Values of kcat/KM for KHO cleavage increased for all three mutants by 2 to 4-fold (Table 2).
Table 2.
Kinetic parameters for KHO cleavage catalyzed by selected TM KDPG aldolase mutants.*
| KDPG aldolase |
kcat (s−1) |
KM (mM) |
kcat/KM (M−1s−1) |
relative kcat/KM |
|---|---|---|---|---|
| wt-TMA | 0.40 ± 0.05 | 200 ± 30 | 2.2 ± 0.3 | 1 |
| TM 1 | 0.07 ± 0.01 | 9 ± 2 | 9.3 ± 1.8 | 4 |
| TM 2 | 0.5 ± 0.1 | 90 ± 20 | 5.5 ± 1.0 | 2.5 |
| TM 3 | 0.14 ± 0.10 | 20 ± 14 | 7 ± 4 | 3 |
| TM 4 | 0.36 ± 0.02 | 23 ± 4 | 16 ± 2 | 7 |
| TM 5 | 0.75 ± 0.10 | 14 ± 5 | 54 ± 10 | 25 |
| TM 6 | 0.13 ± 0.02 | 8 ± 3 | 17 ± 6 | 8 |
| TM 8 | 1.2 ± 0.3 | 49 ± 19 | 25 ± 5 | 11 |
Initial velocity for pyruvate formation was measured using a lactate dehydrogenase coupled assay at 34°C. Steady state kinetic parameters were determined by fitting of the Michaelis-Menten equation to the initial velocity as a function of the substrate concentration.
The most improved mutants, TM 3 and TM 1, increase the value of kcat/KM by 3- and 4-fold and decreases the value of KM by at least 10 and 22-fold, respectively, compared to the corresponding parameters for KHO cleavage catalyzed by wild-type enzyme. Interestingly, kcat decreases up to 6-fold for these mutants, implying that the selective pressure in vivo is for increased catalytic efficiency (kcat/KM) rather than turnover (kcat). This result suggests that the in vivo concentration of KHO under selective conditions is low, a conclusion consistent with the nature of KHO, a charged molecule that cannot easily pass through a lipid bilayer by simple diffusion. Based on these data we presume that the cellular concentration of KHO is in the µM to nM range under the selection conditions.
2.2 Selection of T. maritima KDPG aldolase mutants under increased stringency
To both increase the selection pressure and to mitigate any cellular strain created by overexpression of KDPG aldolase, we constructed plasmids that confer decreased KDPG aldolase expression by making mutations in the RNA polymerase binding region of the lac promoter (between −10 and −35) that have previously been shown to significantly decrease gene expression.20 A TMEDA-pUC plasmid containing an A to C mutation at the −35 RNA polymerase binding region decreases TM KDPG aldolase expression about 10-fold, while a single guanosine nucleotide insertion into the 18 bp region between the −10 and −35 region of the promoter decreases protein expression ~50-fold. Using these plasmids, libraries of TM KDPG aldolase genes were generated by error-prone PCR amplification. A library of 1×107 plasmids was selected for conferring growth on ribose/KHO minimal media in PB25 cells and a total of 5 plasmids that allow for growth within 120 hours were identified (Table 1, TM 4–8). In this case, expression of the selected mutant proteins is comparable to the reduced level observed for the wild-type enzyme in these plasmids.
Mutant KDPG aldolase genes were subcloned into a pET expression plasmid and the protein was expressed in XL1-Blue cells grown in rich media with IPTG induction. Mutant TM 7 precipitated during purification so kinetic data could not be obtained for this mutant. The kinetic parameters for catalysis of KHO cleavage for the other four mutants (Table 2) demonstrate that the value of kcat/KM increases up to 25-fold, from a combination of improvements in KM (4 to 25-fold), as well as changes in the turnover number (0.3 to 3-fold). The activity of TM 5 showed the greatest improvement (Figure 2B), due primarily to a >14-fold decrease in the value of KM for KHO, suggesting that new, beneficial protein-substrate contacts occur in this mutant. TM 8 shows a 3-fold increase in kcat in addition to a 4-fold decrease in KM (Figure 2B) making it one of the most promising mutants for biocatalysis, where maximal turnover under saturating conditions is critical.
2.3. Directed evolution and selection of E. coli KDPG aldolase
For comparison, we constructed a library of E. coli KDPG aldolase variants and selected for enhanced cleavage of KHO. Mutagenesis was limited to two to three mutations per KDPG aldolase gene and the library was incorporated into a pUC plasmid that contains a deletion at the −35 RNA polymerase binding region (TTTACA to TTACA) to decrease protein expression and enhance the selection stringency. Plasmids were selected for conferring growth of PB25 cells within 120 hours on minimal ribose/KHO plates at 34 °C. Based on transformation controls, the approximate size of the library screened in each case was ~107 members. Plasmids from recovered colonies were isolated and sequenced, and used to template subsequent rounds of error-prone PCR without extensive kinetic analysis. After five sequential rounds of mutagenesis and selection, the mutant aldolase proteins from the final round of selection were purified and assayed for catalysis of cleavage of KHO.
Unfortunately, 2 of the 3 selected mutant proteins lost substantial activity during purification and did not yield reproducible kinetic results. The remaining mutant, EC 11, also showed limited stability. This mutant contained substitutions both proximal and distal to the active site that improved the value of KM for KHO cleavage by 100-fold to 1.5 mM. However, the turnover number also decreased by a similar amount resulting in a mutant with a kcat/KM value comparable to that of wild-type enzyme (Table 3). These measurements may underestimate the value of kcat due to the instability of the protein.
Table 3.
Kinetic parameters for KHO cleavage catalyzed by selected EC KDPG aldolase mutants.*
| Mutant | Mutations |
kcat (s−1) |
KM (mM) |
kcat/KM (M−1s−1) |
|---|---|---|---|---|
| wt-ECA | none | 2 ± 0.6 | 150 ± 50 | 14 ± 4 |
| EC 9 | V17L/E51K/P94L/G124M/L173M | -- | -- | > 0.1 |
| EC 10 | V17L/V23I/E51K/P94L/F134I/L173M/D190E | -- | -- | > 0.2 |
| EC 11 | V17L/A53T/P94L/V118L/A137T/L173M | 0.01 ± 0.005 | 1.5 ± 0.4 | 10 ± 3 |
Initial velocity for pyruvate formation was measured using a lactate dehydrogenase coupled assay at 25 °C. Steady state kinetic parameters were determined by fitting of the Michaelis-Menten equation to the initial velocity as a function of the substrate concentration.
3. Discussion
Directed evolution is a powerful tool for altering the catalytic properties of enzymes where diversity libraries are created by error prone PCR or DNA shuffling methods and then subjected to iterative rounds of screening for the desired property.21, 22 More recently, semi-rational methods of library generation have helped minimize library complexity by focusing mutations on active site positions or mutational hotspots, and have yielded significant success.23 Here we used random whole gene mutagenesis of T. maritima and E. coli KDPG aldolase to identify mutations both proximal and distal to the active site which significantly alter the efficiency for catalysis of KHO cleavage. Selections in both isoforms clearly demonstrate that improvements in KM and kcat/KM can be achieved with a minimal number of mutations at sites that would not have been predicted a priori based on protein structural data.
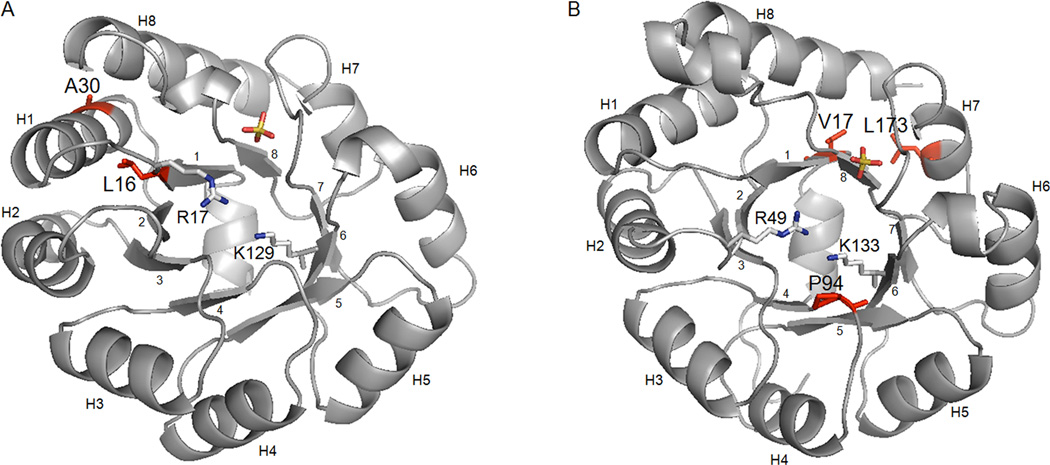
The KDPG aldolases are TIM barrel proteins that catalyze the retro-aldol cleavage of deoxy-sugars to pyruvate and the corresponding aldehyde via a Schiff base-dependent mechanism. The active site is located at the top of the pore created by the barrel structure and contains the important catalytic residues K129, E40, and R17 (Figure 3) (K133, E45 and R49 in the E. coli protein).24, 25 Lysine 129 forms a Schiff base with the substrate, while E40 serves as an acid/base catalyst, and R49 forms critical charge-charge interactions with the carboxylate of the pyruvyl- substrate.25, 26 Part of the electrophile binding pocket is created by serine 179, (equivalent to S184 in the E. coli protein), a position that is important in E. coli KDPG aldolase for recognition of phosphorylated substrates.9 The rest of the phosphate binding pocket consists of the amide nitrogen of G157, G158, and V159 (Figure 3). Since KHO is not phosphorylated, it is likely to make poor contacts with the residues that line this pocket. Surprisingly, we did not select any mutants with alterations in these residues that enhance catalytic activity for cleavage of KHO.
Figure 3.
Schematic diagram of KHO binding to TM KDPG aldolase.
In the T. maritima selections, many (32 %) of the observed mutations are located 12–16 Å from the active site lysine, on solvent exposed α-helices and loops. Mutations on solvent exposed surfaces include K9T, S82C, G83V, F99L, V172A, and T185A. The proteins containing these mutations also contain additional mutations buried within the protein, and these surface mutations may constitute neutral drift. Although, we did not test these mutations individually, it is possible that some may improve protein stability.27 Several mutations (A30V, V31A, I52F, and W167R) are located in buried portions of the helices and may therefore communicate structural changes to the neighboring β-sheets that line the active site. The most remarkable of these mutants is TM 5, in which a single A30V mutation results in a 25-fold improvement in the value of kcat/KM. A30 is located on α-helix 1 with the side chain of this residue directed towards the interface with helix 8 (Figure 4A). Helix 1 attaches to the loop containing the pyruvate-binding residue R17, while helix 8 is attached to the loop containing the phosphate binding site residue S179 (equivalent position to EC S184). Substituting alanine with valine at this site may increase the distance between these two helices and alter the position of the residues on the adjoining loops to enhance catalysis of KHO cleavage (Figure 4A).
Figure 4.
Buried mutations in T. maritima and E. coli KDPG aldolases. A) Structure of T. maritima KDPG aldolase (PDB:1WA325). Mutant TM 5 contains a single mutation A30V. The side chain of A30 (in red) points between the helices 1 and 8. TM 3 contained numerous mutations but only L16P is buried. The side chain of L16 is shown in red. Helices are labeled as H1-8. The side chains of K129, R17 and a bound sulfate are shown for reference. B) Structure of E. coli KDPG aldolase (PDB:1EUA24). All final round EC mutants possess V17L, L173M, and P94V (in red). The side chains of V17 and L173 point towards each other at the interface between β-sheets 1 and 8, and α-helix 8. P94 lines the pyruvate binding pocket. Residues K133, R49 and a bound sulfate are shown for reference.
Several recovered mutations are near the active site (L16P, P109S, and N150I) and may directly affect catalytic residues and/or directly contact the substrate. Of note is mutation L16P (TM 3), located at the C-terminal end of β-sheet 1 (Figure 4A) adjacent to R17, the arginine that interacts with the carboxylate moiety of KHO. Another interesting mutation is P109S (TM 7), which was identified as a single mutation in the selection. Proline 109 is located on the inward face of β-sheet 5, and causes a kink in the sheet adjacent to the active site lysine 129. P109 is located 4 Å away from the catalytic lysine; mutation at this site may reposition or altering the mobility of the K129 side chain. Unfortunately this mutation also significantly diminishes enzyme stability, complicating analysis of catalytic properties. Additional rounds of mutagenesis might identify secondary mutations that enhance protein stability.
In contrast to the generally robust mutant proteins identified from the TM KDPG aldolase selections, recovered E. coli KDPG aldolase mutants showed marked instability. As a result the observed kcat and kcat/KM values reported for these mutants represent a lower limit for these parameters; however, the KM value is independent of the active protein concentration and should reflect a genuine characteristic of the isolated proteins. The selected E. coli KDPG aldolase mutations primarily lower the value of KM for KHO, likely reflecting enhancements in the binding affinity for this substrate. The most active mutant after five rounds of mutagenesis (EC 11) improves the value of KM by 100-fold; however, the value of kcat decreases a maximum of 200-fold, resulting in a mutant with a kcat/KM comparable to that of the wild-type enzyme. (Mutants EC 9 and 10 also showed lower values for KM in the order of 10- to 20-fold but are not reported due to rapid inactivation of the proteins in vitro.) Three mutations are common to each of the selected EC KDPG aldolase mutants: V17L, P94V, and L173M. (Figure 4B) Residue P94 lines the pyruvate binding cavity and is located ~4.6 Å from the catalytic lysine (K133). The conversion of proline 94 to valine likely alters the structure of the substrate binding pocket, enhancing interactions with KHO. V17L and L173M are located adjacent to each other, with their side chains pointing towards each other in the buried interface between β-sheets 1 and 8, and α-helix 8. Similar to the A30V mutation observed in T. maritima aldolase, the V17L and L173M pair likely reposition helices 1 and 8, thereby affecting substrate recognition. The surprising correlation in the location of mutations observed in the two related aldolases may imply that the interface between helices 1 and 8 is important for defining the structure of the substrate binding pocket of KDPG aldolases. The function of the additional mutations in EC 11 (A53T, V118L, and A137T) is unclear, although they may alter protein stability.
The mutants isolated from TM and EC KDPG aldolase libraries all show considerable improvements in the KM parameter with modest, if any, increases in kcat. This observation suggests that the pressure applied by this selection is for the value kcat/KM, conferring a growth advantage to the cellular host. A relatively small concentration of KHO (2.5 mM) is used in the media; the intracellular concentration is presumably much lower. Therefore, it is not surprising that selections led to improvements in the KM parameter given that the cellular concentration of KHO is likely far lower than the wild-type KM value. The subtle remodeling of the binding pocket leads to enhanced binding of the substrate and enhances reactivity with the low concentration of the available substrate. However these mutations have a similar stabilizing effect on both the ground and transition states of the rate-limiting step leaving the value of kcat unchanged. This type of enhancement, characterized as “uniform binding”,15 is proposed as a first step in the evolution of novel enzymatic activity. Alterations in numerous interactions (hydrogen bonding, hydrophobic interactions, electrostatic effects, etc.) that are often distant from the active site can lead to uniform binding enhancements, increasing the likelihood that this form of enhancement is identified when screening libraries of modest size. Once the value of KM approaches the cellular concentration of the substrate, additional enhancements in catalytic efficiency can only be achieved by “differential binding” where the transition states are stabilized relative to the substrate-bound ground states and specific catalysis of elemental steps.15
Our findings reveal significant limitations with in vivo selection methodologies to obtain enzymes suitable for biocatalytic applications: (i) when the concentration of the substrate is limiting, most improvements in catalysis are attributed to enhancements in the apparent substrate affinity rather than improving turnover at saturating substrate, and (ii) enhancements observed under in vivo conditions may not translate to the in vitro conditions used for biocatalytic transformations; and (iii) selection may lead to proteins with decreased stability. To achieve selective pressure required to increase turnover numbers, the in vivo substrate concentration must be comparable to or higher than the value of KM. For biocatalysis applications it is desirable that the binding affinity be weak and values of kcat be large. In this case, the selections must be carried out under conditions where the cellular substrate concentration is high. Unfortunately the KHO substrate is toxic to bacterial cells at concentrations above 10 mM, preventing such experiments for this application.
Our results also suggest that thermophilic, rather than the mesophilic, scaffolds may be superior for directed evolution, due to an enhanced initial stability that facilitates recovery of catalytically beneficial yet destabilizing mutations. In our hands, it was far easier to achieve an improvement in the T. maritima aldolase than the E. coli aldolase. Two rounds of mutagenesis yielded up to a 25-fold improvement in catalysis of KHO using the thermophilic aldolase; however after multiple rounds of mutagenesis, mutations in the E. coli enzyme improved the value of KM but had negligible impact on the value of kcat/KM. Moreover, the E. coli mutants proved to be difficult to handle in vitro, frustrating efforts to accurately analyze their kinetic parameters. The additional stability of the T. maritima scaffold presumably allows it to sample a greater number of mutations while still remaining properly folded, thereby making the screening effort more effective. A similar result was reported in the case of subtilisin.28
4. Conclusions
Our data demonstrate that selection using a pyruvate auxotrophic strain can generate enzymes with enhanced kinetic efficiency towards cleavage of substrates with unnatural electrophiles. In theory, this method could select for reaction of aldolases with any unnatural aldehyde within the limits of solubility, chemical stability, competing reactivity, and acute toxicity to the cell. It is clear that significant improvements in KM and kcat/KM can be achieved with a minimal number of mutations at sites that would not have been identified a priori from protein structural data. However, in vivo selections may not be optimal for generating improvements in the turnover number (kcat) of an enzyme, as high substrate concentrations may not be readily achieved inside the cell. These findings also point to one of the challenges of in vivo selections; the overall fitness of the organism is a function of careful tuning of expression levels and activity of multiple proteins.
Most of the selected mutants improve KHO catalysis by lowering the KM parameter, suggesting that the cellular steady-state concentration of KHO is low. As a result, these in vivo selections mainly identify improvements in KM until the value of KM is comparable to the cellular concentration of the substrate. Beyond this point further improvements in KM will not increase the catalytic efficiency of the enzyme or viability of the cells; additional improvements must derive from an increase in the value of the turnover number. For biocatalyst development, the value of kcat is arguably the most important kinetic parameter, so developing methods to engineer enzymes with improved kcat is desirable. The intracellular concentration of KHO is difficult to control; however the periplasmic influx of small molecules is generally unregulated29 and KHO concentrations in the periplasm should parallel those in the medium. Therefore, localization of the selected enzyme to the periplasm may circumvent issues with substrate concentration. A key advantage of this approach would be that the concentration of KHO can be directly controlled by the growth conditions, presumably allowing for the rapid engineering of aldolases with improved values of kcat.
5. Materials and Methods
5.1. Library creation
Error prone PCR amplification of the T. maritima eda genes was conducted using the Genemorph mutagenesis kit (Stratagene). Primers containing SacI and XhoI restriction sites were used (forward 5’-GGAAACAGCTATGACCATGATTACGAATTCGAGCTCTACCATG-3’ and reverse 5’-CTCAGTGGTGGTGGTGGTGGTGCTCGACTTC -3’). The eda genes were encoded on pUC derived plasmid TMEDA-pUC as previously reported.14 Following PCR amplification, the DNA was fractionated on a 1% agarose gel and extracted using Microcon Ultrafree DA spin filters. The plasmid was simultaneously digested with SacI and XhoI in NEB Buffer 4 and BSA (New England BioLabs) and the DNA encoding the eda gene was purified on a 1% agarose gel and extracted with phenol:chloroform followed by precipitation with ethanol using standard protocols.30 The vector for ligation reactions was made by digestion of the original TMEDA-pUC plasmid, (or reduced expression vectors described below) with SacI and XhoI and purification on a gel followed by phenol:chloroform extraction. Ligations were run using a 1:1.2 molar excess of insert to vector at 125 ng/µL total DNA concentration. T4 DNA Ligase (New England BioLabs) was used with the supplied buffer and 10 mM ATP. Ligation reactions were incubated on ice overnight, allowing the reaction to slowly reach room temperature. The product DNA was extracted with phenol:chloroform and precipitated with ethanol before resuspending in water to a final concentration of 625 ng/µL. The E.coli KDPG aldolase library was constructed similarly to the TM library using the following primers: forward 5’-GAAAACAGCTATGACCATGATTACGAAT TCGAGCTCGCTCATG-3’; reverse 5’-ATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTT-3’ and the pUC-ECEDA derived plasmid.14
The PB25 strain is an E. coli K12 JM101-derived strain where the pyruvate kinase genes, pykA and pykF, have been disrupted [supE thiΔ(lac-proAB) pykA::kanr pykF::catr]. Electrocompetent PB25 cells were made according to the protocol described by Hanahanet. al31 except that 0.2% glucose was added to the growth media to enhance growth of the auxtrophic cell line and stored in 80 µL aliquots at −80 °C for up to 3 months. Electroporation of up to 1 µg of DNA was done using a BIORAD Gene Pulser in 0.1 mm cuvettes under standard E. coli transformation conditions.31 The cells were immediately resuspended in a total of 4 mL of SOC media and allowed to recover for 1 h at 34 °C and 250 rpm. The cells were pelleted and resuspended in 4 mL of M9 media twice to remove trace amounts of glucose before plating on selective minimal media.
5.2. Low expression KDPG aldolasevectors
Mutations were made in the lac promoter of the pUC plasmid using site-directed mutagenesis. The −35 region was changed from TTTACA to TTTCCA by whole plasmid PCR amplification using the following primer and its reverse complement: 5’-GCACCCCAGGCTTTCCACTTTATGC-3’.A single guanosine nucleotide was introduced in the spacer region between −10 and −35 using the following primer and its reverse complement: 5’-CACTTTATGCTGTCCGGCTCGTATGTTG-3’. A point deletion was made in the −35 region of the lac promoter (TTTACA to TTACA) using the following primer and its reverse complement; 5’-GCACCCCAGGCTTACACTTTATGC-3’. The PCR products were transformed into SmartCells (Genlantis) using the standard heat shock methods. Following overnight growth on LB agar plates containing 100 µg/mL ampicillin (LB/AMP), a single colony was grown up and the plasmid DNA was purified and sequenced.
The level of protein expression achieved by various plasmids was determined by isolating PB25 cells that were transformed with each plasmid. Ten large colonies were picked from LB/AMP and resuspended in 100 µL H2O to a final concentration of OD600 = 1.25. The cells were lysed by heating for 10 min at 95 °C after addition of SDS loading dye. The cell debris was pelleted and the supernatant was fractionated on a 12% polyacylamide gel. The proteins were transferred electrophoretically to nitrocellulose paper and blotted according to the manufacturer’s procedures using Mouse anti-His antibody (Novagen) followed by Goat anti-Mouse AP-conjugate secondary antibody (Novagen). The presence of bound antibody was visualized by staining with 5-bromo-4-chloro-3'-indolyphosphate p-toluidine and nitro-blue tetrazolium chloride to detect phosphatase activity. The blot was quantified using the Gel-Pro Analyzer software (Media Cybernetics) to determine the level of expression.
5.3. Selection conditions
Transformed cells were plated on selective M9 minimal media that contained the following: 0.2% ribose, 1 µg/mL thiamine, 40 µg/mL proline, 50 µg/mL carbenecillin, and 2.5 mM KHO. The plates were further supplemented with 1 µM FeCl3, 50 µM ZnSO4, 0.1 µM CaCl2, 40 µg/mL each adenosine, guanosine, thymidine, cytosine and uracil, and 1 µg/mL each of D-biotin, cyanocobalamin, folic acid, niacinamide, D-pantothenate, pyridoxal hydrochloride and riboflavin. These supplements improve the growth of the PB25 strain on minimal plates without negating the pyruvate auxotroph phenotype. The library size was estimated by counting the number of recovered colonies on LB plates containing carbenicillin. Selection plates were grown at 34 °C.
As controls, plasmids containing the wild-type aldolase genes were transformed into PB25 cells and plated on selective media alongside the libraries. No significant growth was seen on this plate within 120 hr. All colonies that grew in the first 120 hours on selection plates were picked and restreaked on selective media and then grown in 5 mL LB liquid cultures containing carbenicillin to recover the plasmid DNA. The plasmid DNA was isolated using the MoBio UltraClean DNA spin kit and was sequenced at the University of Michigan DNA sequencing core. The plasmids were retransformed and only plasmids that reproduced the growth phenotype on selection plates are reported.
5.4. Kinetics
2-keto-4-hydroxyoctonoate, synthesized and purified as described by Griffiths et al,14 was used as the substrate. Aldolase activity was measured using a lactate dehydrogenase (LDH) coupled assay where the production of pyruvate was measured by the decrease in NADH absorbance at 340 nm.7 All assays were done in quartz microcuvettes (70 µL) using a CARY 100 Bio UV/Vis spectrometer fitted with a Peltier temperature controller. The reactions contained the following final concentrations: HEPES (100 mM, pH 7.5), NADH (250 µM), LDH (0.023 Units/µL), 100 µM to 50 mM KHO. Each reaction was initiated with addition of 1 µM enzyme. Kinetic parameters for the TM aldolase variants were measured at the selection temperature (34 °C), while the EC aldolase variants were measured at 25 °C to minimize thermal denaturation of unstable mutants. Each measurement was made 3 times and averaged, and the Michaelis-Menten equation was fit to the data using the curve-fitting program KaleidaGraph (Synergy Software).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [grant number R01 GM61596]. M.C. received support from the Chemical Biology Interface training program [GM08597]. M.J.W. was supported by the Biological Chemistry training program [T32 GM008558].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Faber K. Biotransformations in Organic Chemistry. Berlin: Springer; 2004. [Google Scholar]
- 2.Fessner WD, Helaine V. Curr Opin Biotechnol. 2001;12:574. doi: 10.1016/s0958-1669(01)00265-8. [DOI] [PubMed] [Google Scholar]
- 3.Bolt A, Berry A, Nelson A. Arch Biochem Biophys. 2008;474:318. doi: 10.1016/j.abb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clapes P, Fessner WD, Sprenger GA, Samland AK. Curr Opin Chem Biol. 2010;14:154. doi: 10.1016/j.cbpa.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 5.Henderson DP, Cotterill IC, Shelton MC, Toone EJ. J Org Chem. 1998;63:906. doi: 10.1021/jo971549s. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths JS, Wymer NJ, Njolito E, Niranjanakumari S, Fierke CA, Toone EJ. Bioorg Med Chem. 2002;10:545. doi: 10.1016/s0968-0896(01)00307-8. [DOI] [PubMed] [Google Scholar]
- 7.Shelton MC, Cotterill IC, Novak ST, Poonawala RM, Sudarshan S, Toone EJ. J Am Chem Soc. 1996;118:2117. [Google Scholar]
- 8.Henderson DP, Shelton MC, Cotterill IC, Toone EJ. J Org Chem. 1997;62:7910. doi: 10.1021/jo971549s. [DOI] [PubMed] [Google Scholar]
- 9.Cheriyan M, Toone EJ, Fierke CA. Protein Science. 2007;16:2368. doi: 10.1110/ps.073042907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stock SD, Hama H, Radding JA, Young DA, Takemoto JY. Antimicro Agents & Chemo. 2000;44:1174. doi: 10.1128/aac.44.5.1174-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luesch H, Yoshida WY, Moore RE, Paul VJ. J Nat Prod. 2000;63:1437. doi: 10.1021/np000104n. [DOI] [PubMed] [Google Scholar]
- 12.Royer SF, Haslett L, Crennell SJ, Hough DW, Danson MJ, Bull SD. J Am Chem Soc. 2010;132:11753. doi: 10.1021/ja104412a. [DOI] [PubMed] [Google Scholar]
- 13.Ponce E, Flores N, Martinez A, Valle F, Bolivar F. J Bacteriol. 1995;177:5719. doi: 10.1128/jb.177.19.5719-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths JS, Cheriyan M, Corbell JB, Pocivavsek L, Fierke CA, Toone EJ. Bioorg Med Chem. 2004;12:4067. doi: 10.1016/j.bmc.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 15.Albery WJ, Knowles JR. Biochemistry. 1976;15:5631. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 16.Haldane JBS. Enzymes. London: Longmans; 1930. [Google Scholar]
- 17.Fong S, Machajewski TD, Mak CC, Wong CH. Chem Biol. 2000;7:873. doi: 10.1016/s1074-5521(00)00035-1. [DOI] [PubMed] [Google Scholar]
- 18.Ran N, Frost JW. J Am Chem Soc. 2007;129:6130. doi: 10.1021/ja067330p. [DOI] [PubMed] [Google Scholar]
- 19.Zhang VQ, Hogrefe HH. Methods Mol Biol. 2010;634:399. doi: 10.1007/978-1-60761-652-8_28. [DOI] [PubMed] [Google Scholar]
- 20.Piette J, Decuyper-Debergh D, Gamper H. Proc Natl Acad Sci USA. 1985;82:7355. doi: 10.1073/pnas.82.21.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reetz MT. Angew Chem Int Ed Engl. 2011;50:138. doi: 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- 22.Jackel C, Hilvert D. Curr Opin Biotechnol. 2010;21:753. doi: 10.1016/j.copbio.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Lutz S. Curr Opin Biotechnol. 2010;21:734. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allard J, Grochulski P, Sygusch J. Proc Natl Acad Sci USA. 2001;98:3679. doi: 10.1073/pnas.071380898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fullerton SWB, Griffiths JS, Merkel AB, Cheriyan M, Wymer NJ, Hutchins MJ, Fierke CA, Toone EJ, Naismith JH. Bioorg Med Chem. 2006;14:3002. doi: 10.1016/j.bmc.2005.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meloche HP, Wood WA. J Bio Chem. 1964;239:3511. [PubMed] [Google Scholar]
- 27.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Nature. 2003;424:805. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 28.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Proc Natl Acad Sci USA. 2006;103:5869. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sroga GE, Dordick JS. Prot Engineer. 2001;14:929. doi: 10.1093/protein/14.11.929. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 31.Hanahan D, Jessee J, Bloom FR. Method Enzymol. 1991;204:63. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.