Abstract
Detecting genomic alterations that result in more aggressive prostate cancer may improve clinical treatment and our understanding of the biology underlying this common but complex disease. To this end, we undertook a genome-wide copy number alterations (CNAs) study of clinicopathological characteristics of 62 prostate tumors using the Illumina 1M SNP array. The highest overall frequencies of CNAs were on chromosomes 8q (gains), 8p (loss and copy-neutral) and 6q (copy-loss). Combined loss and copy-neutral events were associated with increasing disease grade (p=0.03), stage (p=0.01), and diagnostic PSA (p=0.01). Further evaluation of CNAs using gene ontology identified pathways involved with disease aggressiveness. The ‘regulation of apoptosis’ pathway was associated with stage of disease (p=0.004), while the ‘reproductive cellular process’ pathway was associated with diagnostic PSA (p=0.00038). Specific genes within these pathways exhibited strong associations with clinical characteristics; for example, in the apoptosis pathway BNIP3L was associated with increasing prostate tumor stage (p=0.007). These findings confirm known regions of CNAs in prostate cancer, and localize additional regions and possible genes (e.g., BNIP3L, WWOX, and GATM) that may help clarify the genetic basis of prostate cancer aggressiveness.
INTRODUCTION
Prostate cancer is characterized by the accumulation of multiple copy number alterations (CNAs) across the genome. Prior studies of CNAs in prostate tumors have utilized comparative genomic hybridization (CGH) arrays and first generation microarrays to identify regions of CNA loss and gain such as loss of 2q21-22, 5q13-21, 6q14-21, 8p21-23, 10q23-25, 13q14-22, 16q13-24, 18q12-23, and 21q22 as well as gain of 3q23-33, 7q21-33, 8q12-23, 17q24-25, and Xq11-23 (Sun et al., 2007; Ishkanian et al., 2009; Liu et al., 2006; Perner et al., 2006). These CNAs have highlighted important genes involved in prostate carcinogenesis such as PTEN at 10q23, MYC at 8q24, and the TMPRSS2:ERG gene fusion at 21q22 ( Ishkanian et al., 2009; Liu et al., 2006; Perner et al., 2006). The use of higher density single nucleotide polymorphism (SNP) arrays (≥1 million SNPs) with their increased genomic coverage allow for detecting smaller regions of CNA and improved delineation of region boundaries. More recently, to profile genomic alterations in prostate tumors, studies have integrated genomic approaches such as array CGH with high density SNP arrays (Liu et al., 2009); copy number analysis, gene expression, miRNA analysis, and exon sequencing (Taylor et al., 2010); or array CGH with next generation sequencing of cancer-related genes (Robbins et al., 2011). These integrated methods have suggested that metastatic prostate tumors originate from a common tumor progenitor cell (Liu et al., 2009), led to the identification of novel somatic mutations in genes such as MTOR, BRCA2, ARHGEF12, and CHD5 ( Robbins et al., 2011), and distinguished between low and high grade prostate cancers (Taylor et al., 2010).
The progression of prostate cancer is similarly characterized by the accumulation of genetic alterations as the tumor advances from a localized tumor to one that has metastasized. The identification of CNAs associated with clinicopathological characteristics may aid in elucidating key genes involved in prostate cancer development and progression. Moreover, this molecular characterization may ultimately provide insight into the development of new prognostic markers and therapeutic targets.
In this study, we characterized CNAs of 62 prostate cancer tumors, evaluating paired tumor and blood DNA samples using high resolution Illumina 1M SNP arrays, and examined the association between CNA and clinicopathological characteristics of prostate cancer. We used a pathway-based approach to focus our analyses and to improve the biological interpretability of our results. Specifically, for each tumor we identified individual genes affected by CNAs, and the pathways to which these genes contributed. We then evaluated the association between these pathways and clinicopathological characteristics of prostate cancer. Finally, we determined which specific genes may underlie the pathway associations.
METHODS
Study Subjects
Prostate cancer patients undergoing radical prostatectomy treatment at the Henry Ford Health System in Detroit, Michigan were studied. These men were recruited as part of an existing case-control study of prostate cancer (Rybicki et al., 2006). We selected 66 subjects for whom tumor tissue was available and that contained at least 70% cancer cells as determined by histological evaluation of Hematoxlyin & Eosin staining, and for which paired blood samples were available. The tumor tissue had a median content of cancer cells of 90%. As previously described, tumor DNA was extracted from freshly frozen prostate cancer tumors (Sanchez et al., 2009) and germline DNA was extracted from blood samples ( Rybicki et al., 2006). Diagnostic PSA levels, Gleason score, and tumor stage were obtained from the medical records; the latter two were from surgical samples. Family history was defined as a prostate cancer diagnosis in the biological father or full brother and collected via in person interview. Institutional Review Board approval was obtained for the study, and informed consent was acquired from all participants.
SNP Array Profiling
Two Illumina 1M-Duo bead arrays were genotyped for each patient (one on their tumor DNA sample, another on their germline DNA from blood) according to the manufacturer's specifications (Illumina, San Diego, CA). Briefly, approximately 750 ng of genomic DNA was whole genome amplified, fragmented, and hybridized to the BeadChips. Following allele specific primer annealing and single base extension, the arrays were imaged using the Beadstation Reader. All markers on the X and Y chromosomes as well as mitochondrial DNA markers were excluded.
For quality control, we first checked genotyping call rates for the 66 samples. Two samples had call rates < 90%, and the corresponding pairs were removed from analyses. Next, we searched for potential batch effects using principal components analysis (PCA), using the correlations of the total intensity value R across all markers between all possible pairs of samples. We focused on the first eigenvalue from the PCA, as it explained >88% of the total variation and subsequent eigenvalues each explained less than 1%. Two additional samples were clear outliers, with values along the first eigenvectors that were >8 standard deviations from the mean, and were removed. Thus, 62 samples were retained for the CNA analysis.
Copy Number Alterations
To distinguish somatic CNA losses and gains, we used the software BAFsegmentation (Staaf et al., 2008), which can account for tumor heterogeneity. Log2 ratios of the intensities of tumors relative to those from their own paired bloods (log2(tumor/blood)) were calculated using the Illumina Bead Studio Software (www.illumina.com). The segmentation algorithm calls gain/loss and copy number alteration using the tumors’ log2 ratios and mirror B allele frequencies (mBAF). mBAF is simply the reflection of the B allele frequency (BAF) with respect to 0.5, the value indicating allelic balance. For example, the mirror value of 0.4 is 0.6.
We filtered non-informative or noisy markers that could distort the mBAF segmentation results using the filtering schemes of Staaf et al. ( Staaf et al., 2008). First, markers that had the same homogenous genotypes in both tumor and its paired blood sample or without any genotype call were excluded. On average, this removed 71% of markers per chromosome. Second, markers with mBAF greater than a cutoff (0.97 for tumors and 0.95 for bloods) were excluded ( Staaf et al., 2008). Third, markers whose mBAF deviated more than two-fold from the average mBAF of its neighboring 20 markers were excluded. Finally, triplet filtering was performed on the remaining markers; this was based on the triplet sum, the absolute sum of the difference in mBAF between a given marker and its immediate neighbors, plus the markers distance from 0.5. Markers with triplet sums above 0.8 were considered outliers and were removed. In total, these filtering steps removed an additional 3% of markers per chromosome. On average, 26% of markers per chromosome passed through all filters and were retained for mBAF segmentation.
For log ratio segmentation, no filtering of markers was done as they would not distort the segmentation results on log ratios. mBAF and log ratios were segmented in identical ways, using the same input parameters ( Staaf et al., 2008). To overlay the gain/loss status, we interpolated the CNA status of markers that were filtered. All excluded markers within CNA regions were classified as CNA, while those in non-CNA regions were classified as non-CNA. This interpolation is simply a direct transformation from CNAs in the segmentation output to marker-level CNA status.
CNA was defined as a segment with a median mBAF > 0.56 across four neighboring markers; loss and gain were defined as ≥4 marker segments having median log2 ratios < -0.15 or > 0.073, respectively, based on cutoffs described by Staaf et al. ( Staaf et al., 2008). Hence, copy-neutrals (i.e. loss of one of the two germline alleles with a concomitant duplication of the retained allele, preserving the diploid copy number but resulting in loss of heterozygosity) were defined as segments with an mBAF>0.56 and a log ratio between -0.15 to 0.073.
To localize specific chromosomal CNA regions, sub-regions were defined as the minimal common region of CNA (loss, copy-neutral or gain) shared by at least four tumors. By this definition, a single tumor with a long CNA region (e.g. loss of the complete p arm of chromosome 8) could contribute to multiple sub-regions. Specifically, a new sub-region was defined when the minimal common region between a group of four or more tumors changed. This would occur with the addition of a tumor with a CNA region directly adjacent to a previously defined sub-region or at the end of sub-region when a tumor CNA region contributes to a previously defined end of a common sub-region.
Statistical Analyses
Non-parametric tests were used to test for association between genome-wide percent CNA measures (i.e. percent gain, loss, copy-neutral, and loss and copy-neutral combined), and clinicopathological characteristics. For stage and grade, Kruskal-Wallis tests were used, and for diagnostic PSA, Spearman's rank correlations were calculated. To determine whether associations between CNA measures and clinicopathological characteristics differed by racial/ethnic group, one degree of freedom multiplicative interaction tests were calculated based on logistic (for stage and grade) and linear (diagnostic PSA) models.
To carry out the pathway-based analysis, the Gene Ontology (GO) hierarchy of Biological Process (BP) terms was used to provide an inclusive definition of biological pathways. To assess whether a pathway was “disrupted” in a single tumor, a list of CNA (gain, loss, and copy-neutral) affected genes in the given tumor was generated, and a Fisher's exact test was then used to determine if there was an enrichment beyond the expected proportion for each tumor (the proportion of annotated genes in the entire genome that corresponded to a GO BP term). Specifically, genes in CNA regions were mapped to the base pair boundary positions of each CNA region and gene positions were catalogued from the National Center for Biotechnology Information's RefSeq database (both using base-pair positions from the NCBI's build 36.3 of the human genome). Those genes either partially or completely located within all CNA regions made up the list of CNA affected genes for that specific tumor. For the Fisher's exact tests, a liberal p-value threshold of 0.1 was used to determine pathway disruption, and these tests were carried out using the DAVID Bioinformatics Resources (Huang et al., 2009).
BP pathway terms found to be disrupted in four or more tumors (n=136) were retained for reporting and further analysis. To determine the overlap of genes in this list of pathways, single-linkage agglomerative clustering was used to form clusters of pathway terms, where each member of the final cluster shared at least 80% of its genes with at least one other member of the cluster. This procedure produced 60 clusters, and the pathway BP term within each cluster with the highest number of disrupted tumors was used to represent that cluster for the purpose of determining the top ranking disrupted pathway terms (Table 4). The full table of terms, the number of tumors with affected genes over-represented for each term, and the mapping of all terms to clusters are provided in Supplemental Table 1.
Table 4.
Top ten pathways over-represented in genes affected by CNA in each tumor.
| GO Biological Process | # Tumors Affected (%) |
|---|---|
| Defense response to bacterium | 24 (38.7) |
| Regulation of apoptosis | 21 (33.9) |
| Response to organic substance | 19 (20.6) |
| Cell-cell signaling | 16 (25.8) |
| Protein complex assembly | 15 (24.2) |
| Response to oxidative stress | 14 (22.6) |
| Cell death | 14 (22.6) |
| Regulation of cell proliferation | 14 (22.6) |
| Positive regulation of developmental process | 14 (22.6) |
| Neurofilament cytoskeleton organization | 14 (22.6) |
Each of the 136 pathway terms was analyzed individually to determine if CNAs (Yes/No) were associated with stage, grade, and/or diagnostic PSA. The influence of pathway term disruption on stage (> 2C) and grade (Gleason grade <7 and 3+4 vs. 4+3 and >7) was analyzed using logistic regression. The influence of disruption on diagnostic PSA was determined using linear regression, after applying a log transformation to the PSA values. For each pathway term associated with one or more clinicopathological feature (p<0.05), regions of over-represented CNAs (> 5% tumors) containing genes involved in that pathway were tested for association with each of the clinicopathological features. All models were adjusted for age at diagnosis and race/ethnicity. All analyses were carried out using the R statistical programming language (http://www.R-project.org).
RESULTS
Patient Characteristics
Table 1 presents characteristics of the 62 prostate cancer patients. The mean age was 60.8 years, 53% of the patients were European American and 47% were African American. Thirteen percent of the patients reported a positive family history of prostate cancer. Thirty-seven percent of the patients had a tumor stage >2C and 37% had a Gleason score of >7 or 4+3 (1°+2° Gleason grade). The mean PSA at diagnosis was 7.9 ng/ml.
Table 1.
Descriptive and clinicopathological characteristics of 62 prostate cancer (PCa) patients
| Total | |
|---|---|
| Number | 62 |
| Age; yrs* | 60.8 ± 5.5 |
| Race/Ethnicity; n (%) | |
| European American | 33 (53.2) |
| African American | 29 (46.8) |
| Positive Family History; n (%)† | 13 (23.2) |
| Stage; n (%) | |
| 2A | 1 (1.6) |
| 2C | 38 (61.3) |
| 3A | 12 (19.4) |
| 3B | 8 (12.9) |
| 4 | 3 (4.8) |
| Gleason grade; n (%) | |
| 6 | 17 (27.4) |
| 3 + 4 | 22 (35.5) |
| 4 + 3 | 8 (12.9) |
| 8 | 10 (16.1) |
| 9 | 5 (8.1) |
| PSA at diagnosis; ng/ml* | 7.9 ± 6.9 |
Mean ± SD
Numbers do not add to 62 due to missing data
Whole genome copy number alterations events
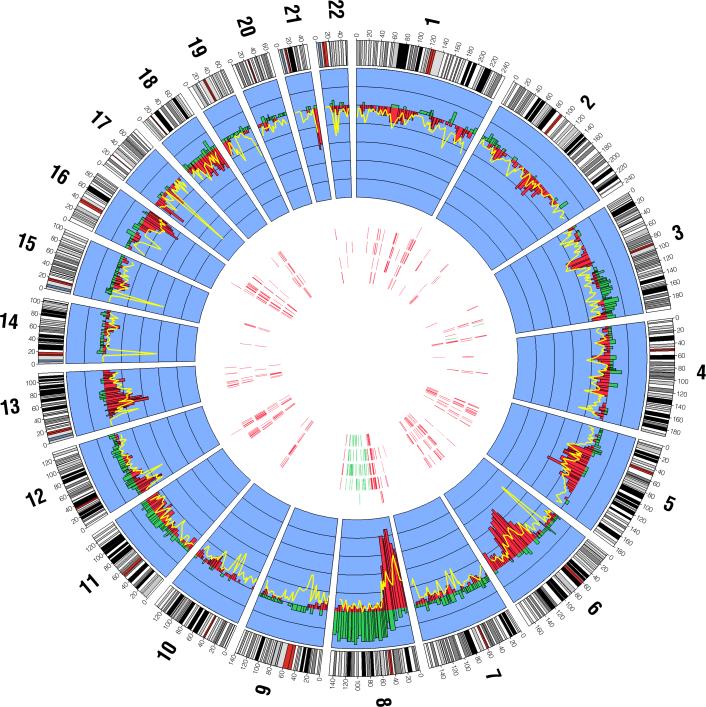
We identified a number of recurrent CNA events across the genome for the 62 prostate cancer tumors (Figure 1). Chromosome 8 displayed the most frequent gains and losses, averaging 8.8% and 12% of the entire chromosome across all samples, respectively. Common losses were also seen on chromosomes 6 (average = 6.5%), 13 (6.6%) and 18 (7.0%). Several frequent copy-neutral events were seen across the genome such as chromosome 13 (average = 4.9%). Next, we searched for regions of CNA by chromosomal arms (Supplemental Table 2). Chromosome 8q displayed CNAs with 302 regions of gains observed in 15 samples and 181 regions of loss of heterozygosity (LOH, defined as loss and copy-neutral) observed in 32 of the 62 tumors. Similarly, chromosome 8p harbored numerous regions of LOH (n=626 in 41 tumors). In addition, LOH was seen at chromosome 6q (37 samples and 403 regions), 13q (40 samples and 276 regions), and 16q (35 samples and 191 regions).
Figure 1.
The genome-wide distribution of CNA gains, losses, and copy-neutral events (loss of one allele and duplication of the second allele) among 62 prostate cancer cases. The inner blue band displays the proportion of CNA gains and losses for each cytological band as histograms across each chromosome. Gains are displayed in green and losses are displayed in red. Within the inner blue band, the distance between each black line corresponds to 0.1 units (i.e. 10%), and the black line where the gain and loss histograms radiate from corresponds to zero. The proportion of copy neutral events at each cytological band is displayed by a yellow line overlying the loss histograms. Within the interior portion of the figure, each of the five concentric bands corresponds to the top five GO biological process terms disrupted in the 62 tumors (arranged outward in: 1. defense response to bacterium, 2. regulation of apoptosis, 3. response to organic substance, 3. cell-cell signaling, 4. protein complex assembly). The vertical marks within each band correspond to the chromosome and base-pair locations of genes disrupted in each of the pathways in > 5% (4 or more) tumors, where the green marks indicates a gene that is predominantly gained and a red mark indicates a gene that is predominantly lost (either copy loss or copy neutral loss).
Overall, ≥ 50% of the prostate tumors had ≥ 4.55% of their autosomal genome affected by CNAs (Table 2). A single tumor had approximately 46% of its autosomal genome affected, while the next highest percentage was 15%. Within CNA categories, LOH was substantially more prevalent in comparison to allelic gains, with ≥50% of the tumors having ≥ 3.84% of their genomes affected by LOH versus ≥ 0.02% of the genome affected by gain. There were no significant differences between prostate tumors of European and African Americans in overall mean CNA or by CNA category (p > 0.14). Loss segments were longer than copy gain segments (median length: loss=835 kb versus gain=739.3 kb). In comparison, copy-neutral segments were the shortest length (median=187 kb).
Table 2.
Distribution of copy number alterations (CNAs) in prostate tumors
| Percentage of Autosomes Affected |
|||||
|---|---|---|---|---|---|
| CNA Category | Mean | Median | 25th-75th percentile | Minimum | Maximum |
| Total | 6.22 | 4.55 | 1.38-10.10 | 0.038 | 46.00 |
| Gain | 1.35 | 0.02 | <0.01-2.81 | 0 | 9.45 |
| LOH | 4.87 | 3.84 | 1.38-6.06 | 0.04 | 45.71 |
| Copy Loss | 2.68 | 1.53 | <0.01-4.50 | 0 | 12.65 |
| Copy Neutral Loss | 2.19 | 0.84 | 0.20-2.43 | 0.04 | 44.91 |
CNA and clinicopathological characteristics of prostate tumors
For most CNA categories, an increasing percentage of the genome affected by CNA was seen with increasing stage, grade, and diagnostic PSA levels (Table 3). In particular, higher stage and grade tumors had higher percentages of LOH in comparison to lower stage (p=0.012) and grade (p=0.025) tumors. For high (> 2c) and low stage tumors (< 2c), the median LOH percentages were 5.6 and 3.2, respectively. For high (≥7 and 4+3), medium (3+4), and low (<7) grade tumors, the median LOH percentages were 4.7, 4.0, and 1.9, respectively. Similarly, higher diagnostic PSA levels were associated with an increasing percentage of total CNA (rs=0.26, p=0.046) and LOH (rs=0.31, p-value=0.013).
Table 3.
Associations between genome-wide measures of CNAs and clinicopathological characteristics of prostate tumors
| Stage |
Grade |
PSA† | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CNA Category | >2c | ≤2c | >7 and 4+3 | <7 and 3+4 | ||||||||
| Median | IQR* | Median | IQR | P | Median | IQR | Median | IQR | P | Rho | P | |
| Total | 5.56 | 3.5-12.3 | 4.43 | 1.1-8.5 | 0.070 | 4.72 | 3.5-12.3 | 4.33 | 1.0-8.5 | 0.052 | 0.255 | 0.046 |
| Gain | 0.05 | 0.0-2.0 | 0.02 | 0.0-2.7 | 0.638 | 0.08 | 0-3.8 | 0.01 | 0.0-1.2 | 0.318 | 0.166 | 0.198 |
| LOH | 5.55 | 3.5-8.3 | 3.24 | 1.1-5.0 | 0.012 | 4.69 | 3.5-6.9 | 3.24 | 1.0-5.1 | 0.025 | 0.314 | 0.013 |
| Copy Loss | 2.88 | 1.0-6.0 | 0.49 | 0.0-3.8 | 0.053 | 2.88 | 0.8-5.4 | 0.49 | 0.0-3.8 | 0.019 | 0.181 | 0.160 |
| Copy Neutral Loss | 0.85 | 0.3-2.8 | 0.62 | 0.2-1.9 | 0.453 | 0.85 | 0.2-3.1 | 0.62 | 0.2-1.9 | 0.427 | 0.209 | 0.103 |
IQR=inter-quartile range
PSA=prostate specific antigen measurement at diagnosis, Spearmen correlation estimate
Additionally, we evaluated whether associations between CNAs and clinicopathological characteristics differed by racial/ethnic group. While there was some suggestion of a stronger positive association between genome-wide LOH and higher stage of disease in African Americans in comparison to European Americans (phet=0.068), this difference was not statistically significant. Furthermore, there was no evidence of heterogeneity by racial/ethnic group between CNAs, diagnostic PSA (phet ≥0.310) or grade (phet ≥0.268).
Pathway analysis of CNA of prostate tumors
To determine the biological involvement of CNA and any potential enrichment of biologically relevant pathways, we conducted gene ontology analysis and cataloged the top ten GOBP terms over-represented among the 62 prostate tumors (Table 4, Fisher's exact p-value<0.10). “Defense response to bacterium” was the most commonly over-represented pathway term (39% of tumors) and could be attributed to the large proportion (50%) of tumors with LOH on chromosome 8p23.1, which harbors the defensin gene cluster that is involved in host immune response. Among 25 genes within this pathway affected by LOH in more than 4 tumors, 22 (88%) were located within 8p23.1 (data not shown). For this pathway, outside of the defensin cluster, the second most common site affected by CNA was gain at 8q21.3, where nine tumors had a common gain. Of note, only a single gene (RIPK2) in the 8q21.3 region was implicated in the “defense response to bacterium” pathway and was the only gene in this pathway that showed gain in more than four tumors.
The second most common pathway term, “regulation of apoptosis” (34% of tumors), was not confined to one contiguous region of the genome but involved multiple regions. In this pathway, there were 174 genes that showed LOH events in more than four tumors (data not shown). Among these genes, 23 (13.2%) were located in a region extending from 8p23.3 to 8p11.21, where the largest number of tumors (n=30) were affected by LOH at 8p21.3, containing a cluster of the tumor necrosis factor receptor superfamily genes (TNFRSF10A, TNFRSF10B, and TNFRSF10D). A separate tumor necrosis factor superfamily cluster showing LOH in 12 tumors on chromosome 17p13.1 was also highlighted in this pathway, a region that also contained TP53. While this region overlapped with the defensin cluster described above, it is of note that none of the genes affected by LOH overlapped with those in the “regulation of apoptosis” pathway. On chromosome 1, an additional 23 (13.2%) genes showed LOH in more than 4 tumors, with the maximum of 6 of tumors LOH at 1q41 and implicating the TGFB2 gene. On chromosome 6, an additional 22 (12.6%) genes exhibited LOH in more than 4 tumors, with the largest number of tumors (21) showing LOH at 6q15, implicating the CASP8A2 gene. Chromosomes 5, 11, 13, and 17 all had more than 10 genes showing LOH in four or more tumors (15 (8.6%), 18 (10.3%), 10 (5.7%), and 10 (5.7%) genes respectively).
Next, to determine the effect of common pathways on clinicopathological characteristics, we restricted the pathways analyzed to those that were over represented in at least 10% of the tumors (n≥7) (Supplemental Table 1). Among them, those pathway terms significantly associated with worse outcomes (i.e. higher stage, grade, and diagnostic PSA levels) are presented in Table 5. The term “Regulation of apoptosis”, most commonly occurring in 21 tumors, was strongly associated with increasing tumor stage (p=0.004). In line with expected biological processes of prostate cancer, the most significant association with tumor grade was the term “Positive regulation of cell differentiation” (OR=5.73, 95%CI=1.44-22.78, p=0.013) and “Regulation of cellular component size” was significantly associated with tumor stage (OR=4.02, 95%CI=1.04-15.49, p=0.043). The most significant association was between the term “Reproductive cellular process” and diagnostic levels of PSA (p=3.84×10-4). While the most over-represented term (“Defense response to bacterium”; Table 4) was not significantly associated with these clinicopathological characteristics, one of its cluster members (“Response to wounding”) was associated with advanced stage (p=0.05), grade (p=0.041) and diagnostic PSA level (p=0.038). Similarly, the term “protein amino acid dephosphorylation” was associated with stage (p=0.013), grade (p=0.039), and diagnostic PSA level (p=0.004).
Table 5.
Common pathway terms (>10%) associated with stage, grade, and diagnostic PSA.*
| Stage |
Grade |
PSA |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Ontology Biological Process Term | N* | OR | 95% CI | P | OR | 95% CI | P | Beta | SE | P |
| Regulation of apoptosis | 21 | 6.76 | 1.82-25.15 | 0.004 | 1.61 | 0.52-5.04 | 0.410 | 0.31 | 0.164 | 0.067 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | 13 | 3.38 | 0.93-12.31 | 0.065 | 5.34 | 1.37-20.87 | 0.016 | 0.25 | 0.186 | 0.176 |
| Apoptosis | 12 | 1.87 | 0.51-6.85 | 0.347 | 4.36 | 1.13-16.83 | 0.033 | 0.14 | 0.193 | 0.477 |
| Positive regulation of cell differentiation | 12 | 2.27 | 0.63-8.19 | 0.209 | 5.73 | 1.44-22.78 | 0.013 | 0.57 | 0.179 | 0.002 |
| Regulation of cellular component size | 12 | 4.02 | 1.04-15.49 | 0.043 | 1.78 | 0.49-6.48 | 0.384 | 0.17 | 0.194 | 0.393 |
| Aromatic compound catabolic process | 12 | 4.91 | 1.22-19.78 | 0.025 | 2.90 | 0.76-11.03 | 0.118 | 0.19 | 0.198 | 0.347 |
| Response to wounding | 10 | 4.20 | 1.00-17.72 | 0.050 | 4.53 | 1.07-19.21 | 0.041 | 0.42 | 0.200 | 0.038 |
| M phase | 10 | 4.31 | 0.95-19.58 | 0.059 | 7.35 | 1.47-36.80 | 0.015 | 0.39 | 0.210 | 0.069 |
| Regulation of growth | 9 | 3.68 | 0.88-15.36 | 0.073 | 0.94 | 0.23-3.87 | 0.932 | 0.23 | 0.207 | 0.275 |
| Protein amino acid dephosphorylation | 9 | 10.02 | 1.62-61.94 | 0.013 | 5.63 | 1.09-29.15 | 0.039 | 0.64 | 0.211 | 0.004 |
| Metal ion transport | 9 | 1.21 | 0.27-5.49 | 0.802 | 5.27 | 1.04-26.78 | 0.045 | 0.27 | 0.228 | 0.242 |
| Transcription initiation | 8 | 5.11 | 0.91-28.83 | 0.065 | 1.41 | 0.27-7.26 | 0.681 | 0.42 | 0.236 | 0.081 |
| Steroid biosynthetic process | 8 | 7.42 | 1.37-40.31 | 0.020 | 1.72 | 0.38-7.75 | 0.483 | 0.37 | 0.222 | 0.102 |
| Reproductive developmental process | 7 | 2.56 | 0.48-13.72 | 0.271 | 1.42 | 0.27-7.61 | 0.680 | 0.22 | 0.250 | 0.381 |
| Reproductive cellular process | 7 | 3.89 | 0.76-19.87 | 0.103 | 4.26 | 0.83-21.74 | 0.081 | 0.80 | 0.213 | 3.84×10-4 |
| Cell division | 7 | 7.09 | 1.1-45.80 | 0.039 | 7.38 | 1.13-48.37 | 0.037 | 0.31 | 0.247 | 0.215 |
Adjusted for age and race. Bolded results indicate p-values < 0.05.
**N=number of tumors with over-representation of the pathway term at a p-value < 0.1.
To further investigate these associated pathway terms, we conducted a sub-region specific analysis to localize genes within these clinicopathogical-associated CNA regions. The term “Regulation of apoptosis” contained 109 LOH sub-regions (containing 174 genes) and 11 gain sub-regions (containing 20 genes) in genes underlying this pathway term that occurred in ≥5% of tumors. In total, 32 of the 109 LOH sub-regions were associated with tumor stage, and these 32 sub-regions contained 46 “Regulation of Apoptosis” genes. Of the 62 tumors, 23 had no “Regulation of Apoptosis” genes showing LOH in their genomes. Of the 39 with at least one of these genes affected by LOH, the median number of genes affected was 18, with an inter-quartile range from 10.5 to 22 genes. Of these 39 tumors with genes showing LOH, 32 had loss of at least one gene in this pathway as a result of LOH on chromosome 8p, the most common chromosomal region lost in prostate cancer. Furthermore, BNIP3L at 8p21, which was recently found to exhibit homozygous deletions in prostate tumors (Liu et al., 2008), demonstrated LOH in 5% of tumors and was significantly associated with increasing stage disease (p=0.007) (Table 6). Within the “Regulation of Apoptosis” pathway, the putative tumor suppressor gene, WWOX at 16q23, located in the second most common fragile site (FRA16D) in the human genome showed LOH in 17.7% of the tumors and was associated with higher stage of disease (p=0.047) (Table 6). In addition, LOH at CLN8 (p=0.006) and IDO1 (p=0.006)—which is also in the “Response to Wounding” pathway (below)—were most strongly associated with increasing stage (Table 6).
Table 6.
Genes in the “Regulation of Apoptosis” pathway and contained within sub-regions of LOH* associated with tumor stage.**
| CNA Frequency |
STAGE |
|||||
|---|---|---|---|---|---|---|
| Cytoband | GENE | LOH† | Gain | OR | 95% CI | P |
| 1p34 | PRDX1 | 4(1) | 0 | 6.77 | 0.61-75.42 | 0.120 |
| 1p13 | SORT1, ALX3 | 4(1) | 0 | -- | -- | -- |
| 2p22 | SOS1 | 5(2) | 1 | -- | -- | -- |
| 3q21 | KALRN, MBD4 | 4(1) | 2 | -- | -- | -- |
| 5q11 | IL31RA, MAP3K1 | 10(5) | 0 | 5.39 | 1.16-24.94 | 0.031 |
| 5q13 | TAF9, NAIP | 11(3) | 0 | 7.46 | 1.59-35.09 | 0.011 |
| 5q13 | F2R | 10(3) | 0 | 8.41 | 1.59-44.54 | 0.012 |
| 5q14 | JMY, RASGRF2 | 9(2) | 0 | 5.61 | 1.11-28.26 | 0.037 |
| 5q31 | IL4 | 4(1) | 0 | -- | -- | -- |
| 5q31 | PPP2CA | 5(2) | 0 | 11.76 | 1.09-127.21 | 0.042 |
| 8p23 | CLN8 | 21(2) | 0 | 5.36 | 1.62-17.73 | 0.006 |
| 8p23 | CTSB | 26(6) | 0 | 4.94 | 1.47-16.57 | 0.010 |
| 8p22 | DLC1 | 28(6) | 0 | 4.67 | 1.44-15.10 | 0.010 |
| 8p21 | PPP3CC | 30(7) | 0 | 4.72 | 1.44-15.42 | 0.010 |
| 8p21 | TNFRSF10B, TNFRSF10D, TNFRSF10A | 30(5) | 0 | 4.83 | 1.46-15.98 | 0.010 |
| 8p21 | NKX2-6, NEFL, GNRH1, BNIP3L | 29(5) | 0 | 5.09 | 1.56-16.63 | 0.007 |
| 8p21 | TRIM35 | 27(6) | 1 | 4.82 | 1.46-15.90 | 0.010 |
| 8p21 | CLU | 28(6) | 1 | 3.99 | 1.25-12.78 | 0.020 |
| 8p12 | BAG4 | 18(5) | 5 | 4.75 | 1.35-16.67 | 0.015 |
| 8p11 | ADAM9 | 18(4) | 4 | 4.21 | 1.24-14.35 | 0.021 |
| 8p11 | IDO1 | 21(7) | 2 | 5.32 | 1.62-17.48 | 0.006 |
| 8p11 | SFRP1 | 20(7) | 2 | 4.16 | 1.29-13.37 | 0.017 |
| 8p11 | IKBKB, POLB | 17(5) | 2 | 3.51 | 1.05-11.70 | 0.041 |
| 12p13 | CD27, ING4 | 6(3) | 1 | 1.95 | 0.33-11.68 | 0.464 |
| 13q12 | HMGB1 | 4(2) | 0 | 6.88 | 0.59-80.32 | 0.124 |
| 13q13 | BRCA2 | 4(2) | 0 | 6.87 | 0.59-80.15 | 0.124 |
| 13q22-q31 | EDNRB, POU4F1 | 6(3) | 0 | 2.00 | 0.31-12.81 | 0.465 |
| 13q33 | ERCC5 | 5(2) | 0 | 4.99 | 0.65-38.26 | 0.122 |
| 16q23 | BCAR1, CFDP1 | 11(2) | 0 | 4.20 | 1.01-17.57 | 0.049 |
| 16q23 | WWOX | 11(3) | 0 | 4.30 | 1.02-18.18 | 0.047 |
| 16q23 | CDH13 | 15(4) | 0 | 4.05 | 1.06-15.50 | 0.041 |
| 17p13 | ALOX15B | 10(1) | 0 | 2.06 | 0.50-8.60 | 0.319 |
A total of 109 sub-regions with four or more tumors with LOH were tested for association within the “regulation of apoptosis” pathway.
Adjusted for age and race. Bolded results indicate p-values < 0.05.
LOH: the total number of tumors with LOH for each sub-region is listed before the parentheses, and the number of those tumors with copy neutral LOH are listed in within the parentheses.
The over-represented term “Response to Wounding” and “Protein Amino Acid Dephosphorylation” in CNA regions that were associated with stage, grade, and diagnostic PSA, contained 77 sub-regions with 115 genes and 23 sub-regions and 24 genes, respectively. Interestingly, GATM within the LOH region at 15q22, occurring in 4.8% of tumors, was associated with higher stage (p=0.043), higher grade (p=0.046), and higher diagnostic PSA (p=0.01) (Supplemental Table 3). For the term “Response to Wounding”, the most significant association was observed for IL5, IL13 at 5q31 (LOH in 4.8% of tumors) and diagnostic PSA (p=2.59×10-6). For the term “Protein Amino Acid Dephosphorylation” (Supplemental Table 4), PPP2CA within a LOH sub-region at 5q31 was most strongly associated with diagnostic PSA (p=1.48×10-4).
DISCUSSION
In the present study, we examined 62 paired blood and tumor prostate cancer samples, using the Illumina 1M SNP array, to characterize CNAs across the genome and to investigate the impact of CNAs on clinicopathological characteristics of prostate cancer. Our analysis identified common CNA events at >10% frequency on chromosome arms 6q, 8p, 8q, 12p, 13q, 16q, and 18q. In addition, we found that CNA events were associated with prostate cancer stage, grade, and diagnostic PSA. Within distinct subregions of chromosomal arms with frequent CNAs, we identified through gene ontology analyses that the “Regulation of Apoptosis” pathway was associated with stage of disease and the “Response to Wounding” and “Protein Amino Acid Dephosphorylation” pathways were associated with stage, grade, and diagnostic PSA.
Previous characterization of prostate tumors have identified chromosomal alterations such as 8p loss, 8q gain, 10q loss, and 16q gain (Sun et al., 2007; Ishkanian et al., 2009; Liu et al., 2008; El Gammal et al., 2010; Torring et al., 2007) We observed similar CNAs among prostate tumors in our study, and these regions harbor known tumor suppressors and oncogenes such as MYC at 8q24 gain (Jenkins et al., 1997), PTEN at 10q23 loss ( Jenkins et al., 1997), and ATBF1 at 16q22 loss (Sun et al., 2005). Genomic alterations at chromosome 8 have been seen consistently across numerous prostate tumor studies. A meta-analysis of 41 array CGH studies of prostate cancer reported 8p loss and 8q gain were the most common regions of genomic instability, occurring in a third and half of all tumors, respectively ( Sun et al., 2007). We observed 8p LOH and gain in 66% and 19% of tumors, respectively and 8q LOH and gain in 24% and 56% of tumors, respectively. These findings support previous observations of the frequent genomic alterations in prostate tumors at the 8p and 8q loci.
Previous CNA studies of clinicopathological characteristics of prostate cancer have implicated numerous chromosomes including 1p, 1q, 2p, 3p, 4q, 6p, 8p, 8q, 9q, 11q, 12p, 12q, 15q, 17q, and 22q ( Sun et al., 2007; Torring et al., 2007). In the current study, we detected similar results for a majority of the previously reported chromosome arms for the following clinicopathological characteristics: stage (1p, 2p, 6p, 8p,11q, 17q), grade (2p, 3p, 8p, 15q), and PSA (1p, 3p, 8p, 12p, 17q, 22q). However, we found no associations with prostate cancer clinicopathological features at 1q, 4q, 8q, 9q, and 12q. The discrepancy in these findings may be due to heterogeneity in patient samples and tumor content as normal cell contamination may mask the detection of LOH as well as variation in robustness and quantification of CNAs by array CGH, microsatellite, and SNP array methods. In addition, array CGH methods are unable to detect copy-neutral events and therefore there is no analogue to our LOH as well as copy-neutral categories.
The over-representation of the regulation of apoptosis pathway within CNA regions and its strong association with stage of disease reinforces the importance of cellular proliferation and apoptosis in prostate cancer. Within the apoptosis pathway, several previously associated prostate cancer genes such as WWOX and DLC-1 were found significantly associated with stage of disease. WWOX, WW domain-containing oxidoreductase, is a putative tumor suppressor gene previously reported to be down-regulated in prostate cancer cell lines and tumor tissue, which is consistent with the effects of LOH at this locus on increase stage of disease (Qin et al., 2006). DLC-1, deleted in liver cancer 1, a regulator of Rho family of small GTPases, is frequently deleted or down-regulated in several forms of cancer (Yuan et al., 1998). Decreased expression of DLC-1 in prostate cancer has been correlated with aberrant promoter hypermethylation and deacetylation (Guan et al., 2006). Taken together the transcriptional silencing of DLC-1 with stage-associated LOH at this locus supports the involvement of DLC-1 in the pathogenesis of prostate cancer.
Our study suggests new pathways and genes that may be important in clinicopathological characteristics of prostate cancer. For example, the “Response to Wounding” pathway was associated with higher stage, grade, and diagnostic PSA, with 16% of tumors showing LOH sub-regions containing genes involved in this pathway category. In particular, glycine amidinotransferase (GATM) that encodes for the mitochondrial enzyme involved in creatine biosynthesis, an important protein for energy supply, exhibited LOH associated with all three clinicopathological characteristics, suggesting the importance of the biosynthesis of creatine in aggressive prostate cancer. In addition, LOH occurring at the loci for the anti-inflammatory cytokines, IL-5 (interleukin-5) and IL-13 (interleukin-13), demonstrated the strongest association with diagnostic PSA levels, emphasizing the involvement of inflammation or other aspects of the immune systems in prostate cancer. Alternatively, LOH at these loci may lead to detection bias as it is possible that men with LOH at anti-inflammatory cytokines may be more likely to have intraprostatic inflammation, resulting in higher PSA levels and subsequent biopses and diagnoses of prostate cancer, which may lead to a spurious association. Furthermore, the protein amino acid dephosphorylation pathway was found to be over-represented within regions LOH and associated with increasing stage, grade, and diagnostic PSA. Within this pathway, the PP2AC locus associated with higher stage and diagnostic PSA encodes for the catalytic subunit of protein phosphatase 2a (PP2A), which has been reported to be downregulated in androgen-independent prostate cancer cells in comparison to androgen dependent cells (Singh et al., 2008). This is consistent with the involvement of LOH at this locus and advanced clinicopathological characteristics.
Although this is one of the largest CNA studies of prostate cancer aggressiveness using a high resolution SNP array, our small sample size limits the implications of our findings. An important strength of this study is the use of blood DNA as the source of germline DNA, mitigating the issue of field effects that can been seen with the use of normal adjacent tissue as a source germline DNA. Furthermore, by utilizing the high density SNP array in paired tumor and normal samples, we are able to assess the contribution of both copy-neutral loss of heterozygosity in addition to hemizygous loss of heterozygosity.
In summary, our data demonstrate CNA events as important contributors to differences in clinicopathological characteristics of prostate cancer. To validate the role of these CNA events in prostate cancer aggressiveness, further high resolution characterization of larger patient populations having detailed clinical, pathology, and outcome information is needed. If replicated, CNAs may ultimately guide decision making in distinguishing between aggressive and non-aggressive prostate cancer and the likelihood of progression. Moreover, by identifying the specific pathways and genes within these unstable genomic regions, we may further expand our understanding of the underlying biological processes that drive prostate cancer development and progression.
Supplementary Material
Acknowledgements
The Illumina 1M SNP genotyping arrays were processed by the Cleveland Clinic Genomics Core (http://www.lerner.ccf.org/services/gc/).
Financial Support: This work was supported by the National Institute of Health grants (CA88164, CA94211, CA98683, CA112355, and ES011126).
References
- 1.El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Köllermann J, Sauter G, Simon R, Wilczak W, Schwarz J, Bokemeyer C, Brümmendorf TH, Izbicki JR, Yekebas E, Fisch M, Huland H, Graefen M, Schlomm T. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 2.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res. 2006;12:1412–9. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 3.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishkanian AS, Mallof CA, Ho J, Meng A, Albert M, Syed A, van der Kwast T, Milosevic M, Yoshimoto M, Squire JA, Lam WL, Bristow RG. High-resolution array CGH identifies novel regions of genomic alteration in intermediate-risk prostate cancer. Prostate. 2009;69:1091–100. doi: 10.1002/pros.20959. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–31. [PubMed] [Google Scholar]
- 6.Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, Turner AR, Kim JW, Meyers DA, Zheng SL, Isaacs WB, Xu J. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer. 2006;45:1018–32. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Xie CC, Zhu Y, Li T, Sun J, Cheng Y, Ewing CM, Dalrymple S, Turner AR, Sun J, Isaacs JT, Chang BL, Zheng SL, Isaacs WB, Xu J. Homozygous deletions and recurrent amplifications implicate new genes involved in prostate cancer. Neoplasia. 2008;10:897–907. doi: 10.1593/neo.08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, Nelson WG, Yegnasubramanian S, Luo J, Wang Y, Xu J, Isaacs WB, Visakorpi T, Bova GS. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 10.Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S, Croce CM, Morrison CD, Klein RD, Huebner K. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66:6477–81. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 11.Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, Beckstrom-Sternberg J, Barrett M, Long J, Chinnaiyan A, Lowey J, Suh E, Pearson JV, Craig DW, Agus DB, Pienta KJ, Carpten JD. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybicki BA, Neslund-Dudas C, Nock NL, Schultz LR, Eklund L, Rosbolt J, Bock CH, Monaghan KG. Prostate cancer risk from occupational exposure to polycyclic aromatic hydrocarbons interacting with the GSTP1 Ile105Val polymorphism. Cancer Detect Prev. 2006;30:412–22. doi: 10.1016/j.cdp.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez JA, Krumroy L, Plummer S, Aung P, Merkulova A, Skacel M, DeJulius KL, Manilich E, Church JM, Casey G, Kalady MF. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg. 2009;96:1196–204. doi: 10.1002/bjs.6683. [DOI] [PubMed] [Google Scholar]
- 14.Singh AP, Bafna S, Chaudhary K, Venkatraman G, Smith L, Eudy JD, Johansson SL, Lin MF, Batra SK. Genome-wide expression profiling reveals transcriptomic variation and perturbed gene networks in androgen-dependent and androgen-independent prostate cancer cells. Cancer Lett. 2008;259:28–38. doi: 10.1016/j.canlet.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staaf J, Lindgren D, Vallon-Christersson J, Isaksson A, Göransson H, Juliusson G, Rosenquist R, Höglund M, Borg A, Ringnér M. Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome SNP arrays. Genome Biol. 2008;9:R136. doi: 10.1186/gb-2008-9-9-r136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Frierson HF, Chen C, Li C, Ran Q, Otto KB, Cantarel BL, Vessella RL, Gao AC, Petros J, Miura Y, Simons JW, Dong JT. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat Genet. 2005;37:407–12. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, Xu J. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 18.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torring N, Borre M, Sorensen KD, Andersen CL, Wiuf C, Orntoft TF. Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50K SNP mapping array. Br J Cancer. 2007;96:499–506. doi: 10.1038/sj.bjc.6603476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.