Abstract
Purpose
To screen mutations in the FERM domain-containing 7 (FRMD7) gene in a Chinese family with X-linked idiopathic congenital nystagmus (ICN).
Methods
It has been reported that FRMD7 mutations account for approximately 47% of X-linked nystagmus in Chinese patients. We collected 5 ml of blood samples from members of a family with X-linked ICN and 100 normal controls. Mutations in FRMD7 were determined by sequencing PCR products.
Results
We identified a previously unreported 4 bp deletion in FRMD7 (c.1486–1489 del TTTT) in a Chinese family. The mutation co-segregated with the disease phenotype in patients and female carriers, while it was not detected in other relatives or in the 100 normal controls.
Conclusions
Our results expand the spectrum of FRMD7 mutations causing ICN, and further confirm the role of FRMD7 in the pathogenesis of ICN. Direct sequencing of FRMD7 could be used as a diagnostic testing of idiopathic congenital nystagmus.
Introduction
Idiopathic congenital nystagmus (ICN) is primarily a disorder of neurologic control system that stabilizes the eyes defined as conjugated, spontaneous, and involuntary ocular oscillations. The symptoms appear at birth or during the first three months of life. Many of the patients show X-linked inheritance. Three disease loci of X-linked ICN have been mapped to chromosome Xq26.2, Xp11.4 and Xp22.3 [1-3]. In 2006, Tarpey et al. [1] reported a new member of the FERM family (4.1 protein, ezrin, radixin, moesin) that mapped to chromosome Xq26.2, FERM domain-containing 7 (FRMD7), which was associated with X-linked ICN. Mutations associated with ICN include missense mutations, null mutations, deletions, and frame-shift mutations [1,4-14]. Mutations in FRMD7 are major causes of Chinese familial X-linked congenital nystagmus and account for approximately 47% of Chinese patients with the disorder [13].
FRMD7 contains 12 exons and encodes a protein with 714 amino acids. In this study, we present a previously unreported mutation in the 12th exon of FRMD7 in a Chinese family with ICN. We found a 4 bp deletion (c.1486–1489 del TTTT), resulting in a predicted truncation at amino acid residue 523. Our data expands on the spectrum of FRMD7 mutations causing ICN, and further confirm the role of FRMD7 in the pathogenesis of ICN.
Methods
Clinical data and 5 ml of blood samples were collected from a Chinese Han family with ICN. The Institutional Review Board approved the project and investigators followed the principles of the Declaration of Helskinki. Informed consent was obtained from each person.
Human genomic DNA was isolated from blood lymphocytes according to standard protocol (Roche Diagnostics Corporation, Shanghai, China), using the DNA Isolation Kits for Mammalian Blood according to the manufacturer’s instructions (Roche Diagnostics Corporation, Indianapolis, IN). PCR-amplification of FRMD7’s 12 exons and exon-intron boundaries was performed using a standard 40 μl PCR buffer system with primers listed in Table 1. DNA sequence analysis was determined by BigDye™ terminator cycle sequencing with an ABI-3130 Genetic Analyzer (ABI Corporation, Carlsbad, CA).
Table 1. Primers used to amplify the exons of FRMD7.
| Exon | Forward primer | Reverse primer | Product length (bp) |
|---|---|---|---|
| 1 |
gctgagtttaagaaggctagagg |
atttgctattgttgtcccttgag |
563 |
| 2 |
aagggtaaatttgcagatgtagc |
acaaagagggaggacaaaaactag |
548 |
| 3 |
agggggcagattaaacgtag |
gcagtgccagaaaatgagata |
505 |
| 4 |
gaggggacggaagaggagagc |
ggcataacccccaagtggatac |
450 |
| 5 |
cccaaaaaggcatctgactg |
aggccatgctgtttctctctatc |
375 |
| 6,7 |
ccaaacacacacacccctatag |
cctatttctgtccccatctatcc |
851 |
| 8 |
Accccttcttgcttgcattc |
ggcaaaagaaaagacacaccatc |
440 |
| 9 |
ggagccaagtggaaaatcagaag |
cccatcttcctccctcctagttag |
480 |
| 10,11 |
gcgttctgagtagttgaggttgt |
gccagttctctccagtctataagg |
676 |
| 12,1 |
tctggaagtaggatggcattgag |
tgattggctctgggacctttta |
975 |
| 12,2 | ccccaattagagcagaggaaagg | gccaacccatactgtcaccattc | 962 |
Results
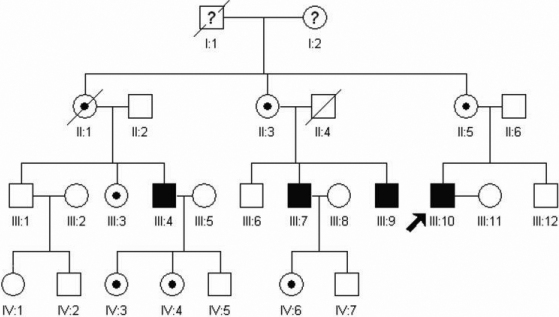
The family from Shandong Province, China, included 4 male patients and 5 female patients who were carriers (Figure 1). All patients in this family had various reduced visual acuity with a similar pattern of nystagmus.
Figure 1.
Pedigree of the Chinese family with ICN. The squares and circles represent males and females, respectively. The shaded symbols signify the affected individuals, the dotted circles represent female carriers, a diagonal line symbol indicates a deceased family member, and the arrow indicates the proband.
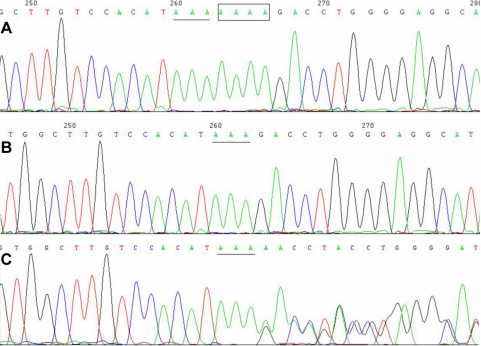
Sequencing of the 12 coding exons of FRMD7 in one of the patients revealed a deletion in exon 12 (c.1486–1489delTTTT: Figure 2), which resulted in frame-shift at codon 497 and a putative stop codon 26 amino acids downstream in the translated protein (p.F497fs26X). The mutation was confirmed, and was further extended to other family members. The mutation co-segregated with the disease phenotype in male patients and heterozygous female individuals, while it was not found in other unaffected relatives or in the 100 normal controls.
Figure 2.
Sequencing chromatograms. A: Reverse sequencing chromatograms of a normal individual, B: an affected male, and C: a female carrier, showing a 4 bp deletion.
Discussion
FRMD7 shows a strong sequence homology with 2 other FERM family members, FERM, RhoGEF and pleckstrin domain protein 1 (FARP1) and FARP2, which are known to promote the dendritic growth of spinal motor neuron subtypes and modulate the length of neurite branching in developing cortical neurons, respectively [15-17]. Recently, Pu et al. [18] reported that expression of FRMD7 in the fetal brain was mainly detected in the brainstem, which is associated with ocular motor control. Betts-Henderson et al. [19] found FRMD7 may play a role in multiple aspects of neuronal development.
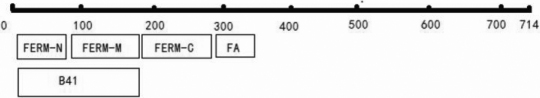
FRMD7 contains FERM-N, FERM-M, FERM-C, and FA structural domains in the NH2-terminus with conserved domains concentrated at the B41 and FERM-C domains (Figure 3). The B41 domain is located from residue 1 to 192, and the FERM-C domain is located from residue 186 to 279. Approximately 44 mutations causing congenital nystagmus have been reported in FRMD7, including 23 missense mutations, 8 splicing site mutations, 1 synonymous mutation, 4 nonsense mutations, 6 frame-shift mutations, and 2 deletions [1,4-14]. These mutations are mainly located in the FERM domain in FRMD7.
Figure 3.
Graphic structure of FRMD7.
We are reporting the third frame-shift mutation in Chinese people. The presence of the mutation in all patients and carriers, and its absence in unaffected individuals and the 100 unrelated controls, support that the identified mutation causes the pathogenesis of ICN. The mutation identified in this study was found in the 12th exon, resulting in a truncated FRMD7 with FERM domain, while the COOH-terminus of FRMD7 protein was deleted. In the Pu et al. [18] study, a nonsense mutation type (COOH-terminally truncated protein) exhibited a different subcellular localization pattern from the wild type, which suggests that the COOH-terminus of FRMD7 may play a key role in the subcellular localization of FRMD7.
Conclusion
Our results expand the spectrum of FRMD7 mutations causing ICN and also confirm the role of FRMD7 in the pathogenesis of ICN. Direct sequencing of FRMD7 can be used as a method in gene diagnosis of idiopathic congenital nystagmus.
Acknowledgments
We are grateful to the patients and their family members for their cooperation in this study. This study was supported by the National Natural Science Foundation (No. 30950007). The study was approved by the Ethics Committee of the Peking University 3rd Hospital and conformed to the Declaration of Helsinki. Great thanks for the comments from Mark Tso, Professor, Wilmer Eye Institute, Johns Hopkins University. Many thanks should also be given to Tina-Marie Gauthier, Ophthalmic Medical Editor (tinamgauthier@earthlink.net), for her editorial assistance.
References
- 1.Tarpey P, Thomas S, Sarvananthan N, Mallya U, Lisgo S, Talbot CJ, Roberts EO, Awan M, Surendran M, McLean RJ, Reinecke RD, Langmann A. Mutations in FRMD7, a newly identified member of the FERM family, cause X-linked idiopathic congenital nystagmus. Nat Genet. 2006;38:1242–4. doi: 10.1038/ng1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabot A, Rozet JM, Gerber S, Perrault I, Ducroq D, Smahi A, Souied E, Munnich A, Kaplan J. A gene for X-linked idiopathic congenital nystagmus (NYS1) maps to chromosome Xp11.4-p11.3. Am J Hum Genet. 1999;64:1141–6. doi: 10.1086/302324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu JY, Ren X, Yang X, Guo T, Yao Q, Li L, Dai X, Zhang M, Wang L, Liu M, Wang QK. Identification of a novel GPR143 mutation in a large Chinese family with congenital nystagmus as the most prominent and consistent manifestation. J Hum Genet. 2007;52:565–70. doi: 10.1007/s10038-007-0152-3. [DOI] [PubMed] [Google Scholar]
- 4.Li N, Wang L, Cui L, Zhang L, Dai S, Li H, Chen X, Zhu L, Hejtmancik JF, Zhao K. Five novel mutations of the FRMD7 gene in Chinese families with X-linked infantile nystagmus. Mol Vis. 2008;14:733–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Li N, Wang X, Wang Y, Wang L, Han R, Liu Y, Zhao K. Investigation of the gene mutations in two Chinese families with Xlinked infantile nystagmus. Mol Vis. 2011;17:461–8. [PMC free article] [PubMed] [Google Scholar]
- 6.Fingert JH, Roos B, Eyestone ME, Pham JD, Mellot ML, Stone E. Novel intragenic FRMD7 deletion in a pedigree with congenital X-linked nystagmus. Ophthalmic Genet. 2010;31:77–80. doi: 10.3109/13816810903584989. [DOI] [PubMed] [Google Scholar]
- 7.Shiels A, Bennett TM, Prince JB, Tychsen L. X-linked idiopathic infantile nystagmus associated with a missense mutation in FRMD7. Mol Vis. 2007;13:2233–41. [PubMed] [Google Scholar]
- 8.Zhang B, Liu Z, Zhao G, Xie X, Yin X, Hu Z, Xu S, Li Q, Song F, Tian J, Luo W, Ding M, Yin J, Xia K, Xia J. Novel mutations of the FRMD7 gene in X-linked congenital motor nystagmus. Mol Vis. 2007;13:1674–9. [PubMed] [Google Scholar]
- 9.Zhang Q, Xiao X, Li S, Guo X. FRMD7 mutations in Chinese families with X-linked congenital motor nystagmus. Mol Vis. 2007;13:1375–8. [PubMed] [Google Scholar]
- 10.Schorderet DF, Tiab L, Gaillard MC, Lorenz B, Klainguti G, Kerrison JB, Traboulsi EI, Munier FL. Novel mutations in FRMD7 in X-linked congenital nystagmus. Mutation in brief #963. Online. Hum Mutat. 2007;28:525. doi: 10.1002/humu.9492. [DOI] [PubMed] [Google Scholar]
- 11.He X, Gu F, Wang Y, Yan J, Zhang M, Huang S, Ma X. A novel mutation in FRMD7 causing X-linked idiopathic congenital nystagmus in a large family. Mol Vis. 2008;14:56–60. [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan Y, Vargel I, Kansu T, Akin B, Rohmann E, Kamaci S, Uz E, Ozcelik T, Wollnik B, Akarsu NA. Skewed X inactivation in an X linked nystagmus family resulted from a novel, p.R229G, missense mutation in the FRMD7 gene. Br J Ophthalmol. 2008;92:135–41. doi: 10.1136/bjo.2007.128157. [DOI] [PubMed] [Google Scholar]
- 13.He X, Gu F, Wang Z, Wang C, Tong Y, Wang Y, Yang J, Liu W, Zhang M, Ma X. A novel frameshift mutation in FRMD7 causing X-linked idiopathic congenital nystagmus. Genet Test. 2008;12:607–13. doi: 10.1089/gte.2008.0070. [DOI] [PubMed] [Google Scholar]
- 14.Self JE, Shawkat F, Malpas CT, Thomas NS, Harris CM, Hodgkins PR, Chen X, Trump D, Lotery AJ. Allelic variation of the FRMD7 gene in congenital idiopathic nystagmus. Arch Ophthalmol. 2007;125:1255–63. doi: 10.1001/archopht.125.9.1255. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang B, Su YS, Sockanathan S. FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 2009;61:359–72. doi: 10.1016/j.neuron.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo T, Yamashita T, Yamaguchi A, Sumimoto H, Hosokawa K, Tohyama M. A novel FERM domain including guanine nucleotide exchange factor is involved in Rac signaling and regulates neurite remodeling. J Neurosci. 2002;22:8504–13. doi: 10.1523/JNEUROSCI.22-19-08504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyofuku T, Yoshida J, Sugimoto T, Zhang H, Kumanogoh A, Hori M, Kikutani H. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat Neurosci. 2005;8:1712–9. doi: 10.1038/nn1596. [DOI] [PubMed] [Google Scholar]
- 18.Pu J, Li Y, Liu Z, Yan Y, Tian J, Chen S, Zhang B. Expression and localization of FRMD7 in human fetal brain, and a role for F-actin. Mol Vis. 2011;17:591–7. [PMC free article] [PubMed] [Google Scholar]
- 19.Betts-Henderson J, Bartesaghi S, Crosier M, Lindsay S, Chen HL, Salomoni P, Gottlob I, Nicotera P. The nystagmus-associated FRMD7 gene regulates neuronal outgrowth and development. Hum Mol Genet. 2010;19:342–51. doi: 10.1093/hmg/ddp500. [DOI] [PubMed] [Google Scholar]