Abstract
Most animals are diploid but haploid-only and male-haplo species have been described 1. Diploid genomes of complex organisms limit genetic approaches in biomedical model species such as in mice. To overcome this problem experimental induction of haploidy has been used in fish 2,3. In contrast, haploidy appears less compatible with development in mammals 4,5. Here we describe haploid mouse embryonic stem cells and show their application in forward genetic screening.
Experimentally induced haploid development in zebrafish has been utilized for genetic screening 2. Recently, haploid pluripotent cell lines from medaka fish have also been established 3. In contrast to fish, haploidy is not compatible with development in mammals. Although haploid cells have been observed in egg cylinder stage parthenogenetic mouse embryos 6 the majority of cells in surviving embryos become diploid. Previous attempts to establish pluripotent stem cell lines from haploid embryos have resulted in the isolation of parthenogenetic embryonic stem cells (ESCs) with a diploid karyotype 4. These studies reported the development of apparently normal haploid mouse blastocysts with a defined inner cell mass (ICM) 4,5. In order to investigate the haploid ICM, we cultured haploid mouse blastocysts in chemically defined medium with inhibitors of mitogen activated protein kinase kinase (MEK) and glycogen synthase kinase 3 (GSK3). This 2i medium 7 has previously been used for isolating ESCs from recalcitrant mouse strains 8 and rats 9 and may help to maintain certain characteristics of early mouse epiblast cells 10,11.
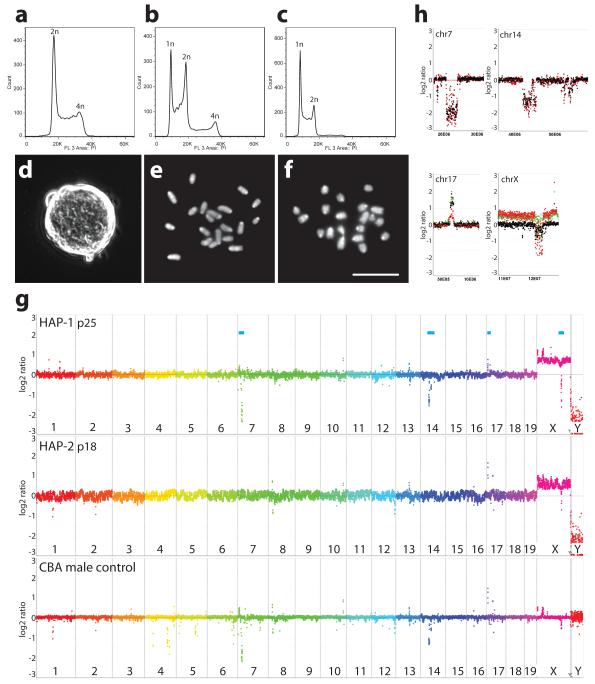
We generated haploid mouse embryos by activation of unfertilized oocytes isolated from superovulated B6CBAF1 hybrid female mice using strontium chloride. After culture in M16 medium 30 blastocysts (22 %) were obtained from 132 activated oocytes and used for ESC derivation. After removal of the zona and trophectoderm, ICMs were cultured in gelatinized 96 well dishes in 2i medium in the presence of LIF. 27 ESC lines were obtained (93 %). Individual ESC lines were expanded and their DNA content was analysed by flow analysis using diploid ESCs as controls (Fig. 1ab). In 6 ESC lines, at least 10 % of the cells had a haploid DNA content and the proportion of haploid cells could reach a conservatively estimated 60 % (Fig. 1b). Further enrichment was achieved by flow sorting of cells with a haploid DNA content after staining with HOECHST 33342 (Fig. 1c). This allowed expansion of haploid ESC lines for over 35 passages.
Figure 1. Derivation of haploid ESCs.
Flow analysis of DNA after PI staining of (a) diploid control ESCs, (b) haploid ESC line HAP-1 at passage 7 (p7) and (c) HAP-1 (p11) after sorting at p7. (d) Colony morphology of haploid ESCs (HAP-1). (ef) Chromosome spreads of HAP-3 (e) and HAP-1 (f), (Scale bar = 10 μm). (g) CGH analysis of HAP-1 and HAP-2 ESCs and control male CBA kidney DNA. Relative copynumber is plotted at 200 kb resolution using a log2 scale. Genomic positions indicated by blue bars (top) are enlarged at 40 kb resolution in (h); CBA control (black), HAP-1 (red) and HAP-2 (green).
We further tested the requirements for deriving haploid mouse ESCs (Table 1). These experiments showed that removal of the trophectoderm by immunosurgery was not essential. Haploid ESCs could also be established using DMEM medium supplemented with Knockout Serum Replacement (KSR) and LIF showing that derivation without kinase inhibitors is possible (Table 1 and Suppl. Fig. 1). We further succeeded in isolating haploid ESCs from the 129Sv inbred mouse strain and two genetically modified mouse lines. In the latter several alleles had been bred to homozygosity and maintained on a mixed genetic background for several generations (Table 1, and Suppl. Fig. 2). In summary, we derived 25 haploid ESC lines in 7 independent experiments. Haploid ESC cultures could also be maintained on feeders in serum containing DMEM supplemented with LIF.
Table 1.
Derivation of haploid mouse ESC lines
| Exp. No |
derivation protocol | Names of ESC lines used in study |
genetic background |
Oocytes activated |
number of blastocysts |
ESC lines obtained |
ESC lines with haploid contribution (max. % haploid before sorting) |
|---|---|---|---|---|---|---|---|
| 1 | 2i / immunosurgery | HAP-1 to 6 † | B6CBAF1 | 132 | 30 | 27 | 6 (>60%) |
| 2 | 2i | HAP-7 | B6CBAF1 | 22 | 10 | 5 | 1 (>15%) |
| 3 | 2i | HTG-1 to 3 ‡ | mixed TG* | 50 | 32 | 3 | 3 (>90%) |
| 4 | KSR / immunosurgery | HAP-8 to 13 | B6CBAF1 | 273 | 48 | 22 | 6 (>10%) |
| 5 | 2i | H129B6-1 to 5 |
129B6F1 | 250 | 37 | 8 | 5 (>10%) 5 |
| 6 | 2i | HTX-1 | mixed TG** | 70 | 11 | 1 | 1 (>40%) |
| 7 | 2i | H129-1 to 3 | 129Sv | 140 | 13 | 10 | 3 (>60%) |
Contribution to chimeric mice was confirmed for the HAP-1 and HAP-2 haploid ESC lines.
Contribution to chimeric mice was confirmed for the HTG-1 haploid ESC line.
Derived from ROSA26nlsrtTA LC1 Xist2LOX homozygous female mice.
Derived from ROSA26nlsrtTA tetOPXist homozygous female mice30.
Haploid ESCs exhibited a typical mouse ESC colony morphology (Fig. 1d). Chromosome spreads showed 20 chromosomes corresponding to the haploid mouse chromosome set (Fig. 1ef). For further characterizing the genetic integrity we performed comparative genomic hybridization (CGH) of 4 haploid ESC lines and control DNA from the CBA strain and the mixed transgenic mouse line from which HTG-1 and HTG-2 ESCs were derived (Fig. 1gh and Suppl. Fig. 3 and 4). Copynumber variations (CNVs) that were detected in the genome of haploid ESC lines were also present in the strains of origin (Suppl. Table 1). Albeit some CNVs appeared haploid ESC specific, inspection of the actual signals (Suppl. Fig. 4) suggested that these CNVs were also present in the CBA or HTG control DNAs but not detected with the threshold applied. CNVs between the C57BL6 and CBA strain of mice were consistent with a previously reported analysis 12. Taken together these data show that haploid ESCs maintained an intact haploid genome without amplifications or losses.
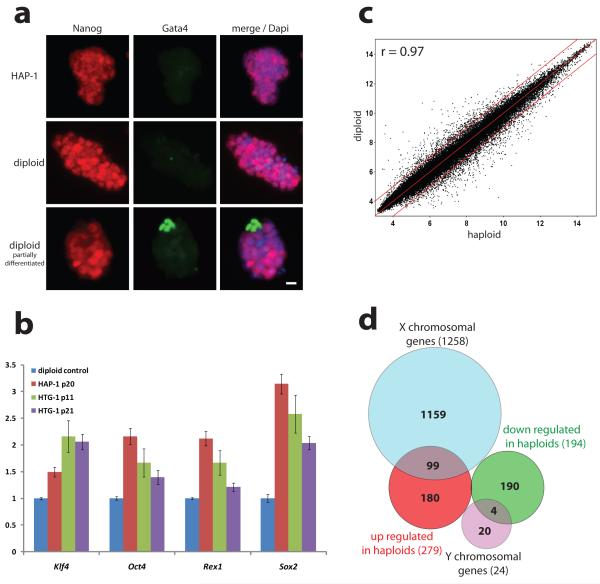
On the molecular level, haploid ESCs expressed pluripotency markers including Oct4, Rex1, Klf4, Sox2 and Nanog (Fig. 2ab). Genome-wide expression analysis showed a high correlation (Pearson correlation coefficient r=0.97 over all genes) between haploid ESCs and control diploid male ESCs (Fig. 2c and Suppl. Fig. 5). In haploid ESCs 279 and 194 genes were more than 2-fold up- or down-regulated (p<0.05), respectively (Suppl. Table 2). Among these, 99 X-linked genes were overexpressed and 4 Y-linked genes were lost in haploid ESCs consistent with different sex chromosome constitutions (Fig. 2d). Thus, haploid ESCs largely maintain a mouse ESC transcription profile.
Figure 2. Expression analysis of haploid ESCs.
(a) Immunofluorescence shows Nanog protein (red) in haploid (HAP-1) and diploid ESCs, and Gata4 (green) in differentiated cells (Scale bar = 10 μm). (b) Expression of pluripotency markers in haploid and diploid (set to 1) ESCs by real time PCR. Errorbars represent standard deviation (n=3). (c) Scatter plot showing log2 transformed average expression values from gene expression profiles of 3 haploid (HAP-1, HAP-2 and HTG-1) and three diploid J1 ESC lines for 45,001 probesets (r is the Paerson correlation coefficient; red lines indicate 2-fold up- and down-regulation). (d) Diagram of more than 2-fold up- and down-regulated genes in haploid ESCs.
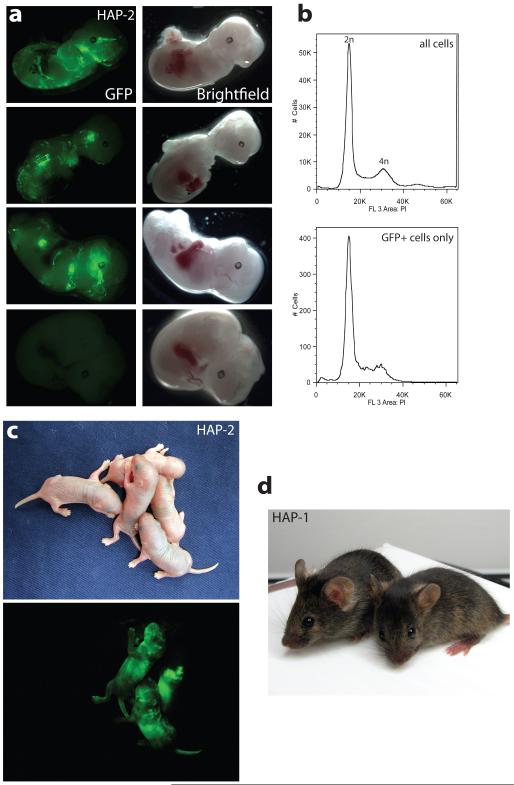
This prompted us to investigate the developmental potential of haploid ESCs. For this we introduced a piggyBac transposon vector for expressing green fluorescent protein (GFP) into HAP-2 ESCs. Flow sorting of cells for GFP fluorescence and DNA staining with Hoechst 33342 yielded a haploid ESC population that expressed GFP at high level showing that a haploid genome content was maintained during the transfection procedure (Suppl. Fig. 6). GFP marked haploid ESCs contributed substantially to chimeric embryos when injected into C57BL/6 blastocysts (Fig. 3a). The great majority of GFP positive cells extracted from chimeric embryos had a diploid DNA content (Fig. 3b) indicating that haploid ESCs contributed extensively to development after diploidization. We also obtained 2 male and 2 female live born chimeras with a substantial contribution from haploid ESCs (Fig. 3 c). These mice developed normally with apparent coat colour chimerism. Similar results were obtained with the HAP-1 and HTG-2 ESCs (Fig. 3d and Suppl. Fig. 7a). Furthermore, the diploid fraction of HAP-2 ESCs at passage 31 could be differentiated into Nestin positive cells following a neural in vitro differentiation protocol 13 (Suppl. Fig. 7b). Taken together these findings demonstrate that haploid ESCs maintain a wide differentiation potential.
Figure 3. Developmental potential of haploid ESCs.
(a) GFP marked haploid HAP-2 ESCs (p18) contribute to chimeric embryos at E12.5. 6 out of 9 embryos showed GFP contribution. A GFP negative embryo is shown as a control (below). (b) Representative flow analyses of DNA content of all cells (above) and GFP positive cells (below) extracted from a chimeric E12.5 embryo are shown. All 6 embryos gave similar results. (c) Live born chimeric mice were obtained from GFP marked HAP-2 ESCs. (d) Chimeric mice obtained from injection of HAP-1 ESCs into C57BL/6 blastocysts (black) show coat colour contribution from the ESCs (agouti).
To investigate the utility of haploid ESCs for genetic screening we performed a pilot screen for mismatch repair genes following a previously published strategy 14. For this, 5 × 106 haploid ESCs were co-transfected with a gene trap piggyBac transposon vector (Suppl. Fig. 8a) and a plasmid for expressing an optimized piggyBac transposase 15. Gene trap insertions were selected with puromycin. A pool of 1 × 107 cells was then cultured in the presence of 2-amino-6-mercaptopurine (6-TG) which is toxic to mismatch repair proficient cells. After 8 days 20 6-TG resistant colonies were isolated and the integration sites were mapped using Splinkerette PCR 16. Of 7 clones analysed we identified two independent insertions in Msh2 and one in Hprt (Suppl. Fig. 8b). Msh2 is a known mismatch repair gene and Hprt is required for converting 6-TG into a toxic metabolite 14. Thus, identification of mutations in autosomal genes was possible suggesting a potential for haploid ESCs in forward genetic screening in mammals.
The difficulty in obtaining haploid ESC lines in previous attempts might be explained by aberrant gene regulation such as aberrant dosage compensation and genomic imprinting. However, diploid ESCs from mouse and human parthenogenetic embryos have been established 17,18. Misregulation of X inactivation has been observed to some extent in haploid mouse embryos 5 and has also been shown to reduce the efficiency of producing cloned mice 19. Thus, it is conceivable that X inactivation is initiated aberrantly in haploid embryos during some ESC derivation procedures. Direct capture of naive pluripotent cells from ICM outgrowths as accentuated by the use of 2i conditions 11 could have contributed to the success of our study.
Previously, near-haploid cells have been observed in human tumours (for literature review see 20) and a near-haploid human tumour derived cell line has been described 21,22. These tumour cells carry genomic rearrangements and mutations that might stabilize the haploid genome. An interesting aspect of haploid ESCs is their developmental potential. We have observed rapid diploidization when haploid ESCs differentiate. The resulting diploid parthenogenetic cells can contribute to development 23. It is interesting to speculate whether differentiated haploid lineages can be generated perhaps through suppression of X inactivation and whether it is possible to derive haploid human ESCs.
Methods summary
For the derivation of haploid ESCs mouse oocytes were activated in M16 medium as described 24. ESC culture in chemically defined 2i medium has been described previously 7,8. Cell sorting for DNA content was performed after staining with 15 μg/ml Hoechst 33342 (Invitrogen) on a MoFlo flow sorter (Beckman Coulter) selecting the haploid 1n peak. For analytic flow profiles cells were fixed in ethanol, RNase treated, and stained with propidium iodide (PI). For karyotype analysis cells were arrested in metaphase with demecolcine (Sigma). After incubation in hypotonic KCl buffer cells were fixed in methanol-acetic acid (3:1) and chromosome spreads were prepared and stained with DAPI. RNA was extracted using the RNeasy Kit (Quiagen). Transcription profiles were generated using Affymetrix GeneChip 430.2 arrays. Sample preparation, hybridization, and basic data analysis were performed by Imagenes (Berlin, Germany). Further analysis was performed using the Genespring GX software (Agilent). For CGH analysis genomic DNA was isolated from haploid ESC lines and hybridized to NimbleGen 3x720K whole-genome tiling arrays by Imagenes (Berlin, Germany) using C57BL/6 kidney DNA as a reference. For chimera experiments GFP labelled HAP-1 (p29), HAP-2 (p18) and HTG-2 (p23) ESCs were injected into C57BL/6 host blastocysts. Live born chimaeras were analysed for expression of GFP at postnatal day 2. Genetic screening was performed following a previously published strategy 25. In brief, HAP-1 ESCs were co-transfected with 2 μg piggyBac transposase expression vector 15 and 1μg piggyBac gene trap vector (Suppl. Fig. 8) using Lipofectamine 2000 (Invitrogen). Selection for transposon insertions was performed using 2 μg/ml puromycin for 8 days. 1×107 puromycin resistant ESCs were plated in two 15 cm dishes and mutations in mismatch repair genes were selected using 0.3 μg/ml 6-TG (Sigma). piggyBac integration sites in seven 6-TG resistant clones were mapped by Splinkerette PCR 16.
Methods
Derivation of haploid ESCs
Oocytes were isolated from superovulated females and activated in M16 medium using 5 mM strontium chloride and 2 mM EGTA as described 24. Embryos were subsequently cultured in M16 or KSOM medium microdrops covered by mineral oil. Under these conditions around 80 % of oocytes reached the 2-cell stage on the next morning. Thereafter development of preimplantation embryos was variable with a large number of embryos showing unequal sized blastomeres or unusual embryo morphology. Removal of the zona, immunosurgery for removal of the trophectoderm and ESC derivation was performed as described previously 7,8. ESCs were cultured in chemically defined 2i medium plus LIF as described 7,8 with minor modifications. 2i medium was supplemented with non essential amino acids and 0.35% BSA fraction V. Culture of ESCs on feeders was performed as previously described 26. Knockout serum replacement (KSR) was obtained from Invitrogen. Cell sorting for DNA content was performed after staining with 15 μg/ml Hoechst 33342 (Invitrogen) on a MoFlo flow sorter (Beckman Coulter). The haploid 1n peak was purified. Diploid cells did arise in cultures to various extents in all ESC lines. Periodic purification by flow sorting every four to five passages allowed us to maintain cultures containing a great majority of haploid ESCs in all cases. Analytic flow profiles of DNA content were recorded after fixation of the cells in ethanol, RNase digestion, and staining with propidium iodide (PI) on a Cyan analyser (Beckman Coulter). For karyotype analysis cells were arrested in metaphase using demecolcine (Sigma). After incubation in hypotonic KCl buffer cells were fixed in methanol-acetic acid (3:1) and chromosome spreads were prepared and stained with DAPI. Immunostaining was performed as described 27 using Nanog (Abcam; 1:100), Oct4 (Santa Cruz; 1:100), Nestin (Developmental Studies Hybridoma bank, Iowa City; 1:30) and Gata4 (Santa Cruz; 1:200) antibodies.
Microarray analysis
RNA from biological triplicates of diploid ESCs and three independently derived haploid ESCs (HAP-1 p21, HAP-2 p24, HTG-1 p11) was extracted using the RNeasy kit (Quiagen). Gene expression analysis on Affymetrix GeneChip 430 2.0 arrays was performed by Imagenes Ges.m.b.H. (Berlin, Germany). Additional gene expression profiles of neural progenitors, mesodermal progenitors and mouse embryonic fibroblasts (MEF) were obtained from a previously published dataset (GEO accession number GSE12982 28). The data was analysed using Genespring GX software (Agilent Technologies). Data were normalized using the RMA algorithm. Lists showing differentially regulated genes (> 2 fold change; p < 0.05) are provided in Suppl. Table 2. p-values were established by an unpaired t test followed by FDR adjustment by the Benjamini Hochberg method. Hierarchical clustering was performed based on the Euclidean distances and complete linkage analysis. The relatedness of transcription profiles was determined by calculating the Pearson correlation coefficient (r). DNA samples for comparative genomic hybridization (CGH) experiments were extracted and sent to Imagenes Ges.m.b.H. (Berlin, Germany) for CGH analysis using NimbleGen 3x720K mouse whole-genome tiling arrays with an average probe spacing of 3.5 kb. Adult male C57BL/6 kidney DNA was used as a reference. A genomic overview of these analyses is presented in Fig.1g and Suppl. Fig. 3 at 200kb resolution and selected zoomed in regions at 40 kb resolution. The complete data set at 40 kb resolution is included in Suppl. Fig. 4.
Accession of datasets
Gene expression and CGH data sets can be accessed as the GEO reference series GSE30879 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30879). This series includes the GSE30744 (Expression analysis of haploid and diploid ES cells in 2i medium) and the GSE30749 (CGH analysis of haploid ES cells) data sets.
Quantitative gene expression analysis
RNA was extracted using the RNeasy kit (Quiagen) and converted into cDNA using the Quantitect reverse transcription kit (Quiagen). Real time PCR was performed on a StepOnePlus machine (Applied Biosystems) using the Fast Sybr green master mix (Applied Biosystems) and previously published primers 27. The ddCt method was used for quantification of gene expression. Expression levels were normalized to L32 ribosomal protein mRNA and values in diploid control ESCs were set to 1.
Embryo analysis
Haploid ESCs were co-transfected with a piggyBac vector carrying a CAG-GFP-IRES-hygro transgene and a piggyback transposase expression plasmid. Stable integrants were selected using 150 μg/ml Hygromycin for 7 days. The haploid fraction of HAP-1 (p29), HAP-2 (p18) and HTG-2 (p23) GFP positive cells were purified by flow sorting (Suppl. Fig. 6). GFP labelled ESCs were expanded and injected into C57BL/6 host blastocysts which were transferred to recipient females. Embryos were analysed at E9.5 and E12.5. Dissociation to single cells was performed by incubation in 0.25% Trypsin/EDTA for 15 min. Prior to PI staining cells were fixed in 4% PFA and permeabilized in PBS/0.25% Triton X-100. Live born chimeras were analysed at postnatal day 2 (P2) for expression of GFP using UV illumination. Images were obtained using a Canon Powershot S5 IS camera with a FHS/EF-3GY2 filter (BLS). All mouse experiments were conducted in accordance with institutional guidelines of the University of Cambridge. All necessary UK home office licenses were in place.
Gene Trap screen
The screen was performed based on a previously published protocol 25. 5×106 HAP-1 ESCs were co-transfected with 2 μg piggyBac transposase plasmid 15 and 1 μg piggyBac gene trap vector (Suppl. Fig. 8a) using Lipofectamine 2000. piggyBac insertions into expressed genes were selected with 2 μg/ml puromycin for 8 days. 1×107 ESCs corresponding to approximately 5,000 puromycin resistant colonies were then plated onto two 15cm dishes. Selection for mismatch deficient integrants was performed using 0.3μg/ml 6-TG (Sigma). 20 colonies were picked and piggyBac integration sites of seven clones were identified by Splinkerette PCR and mapped using iMapper 29.
References used only in the Methods section
- 26.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 27.Leeb M, et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–76. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong J, Zhu F, Stalker J, Adams DJ. iMapper: a web application for the automated analysis and mapping of insertional mutagenesis sequence data against Ensembl genomes. Bioinformatics. 2008;24:2923–5. doi: 10.1093/bioinformatics/btn541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–77. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary Material
Acknowledgements
We thank Austin Smith and Jenny Nichols for critical discussion, Kenneth Jones for advice on cell culture, Sabine Dietmann for bioinformatics support, and Rachael Walker for cell sorting. We are further indebted to Bill Mansfield and Charles-Etienne Dumeau for their expert help and the BSU team at the centre for maintaining the mouse colony. This work was supported by a Wellcome Trust Senior Research Fellowship to AW (grant reference 087530/Z/08/A) and an EMBO Long Term Fellowship to ML.
Footnotes
Author contributions ML performed the experiments, analysed the data and wrote the manuscript. AW performed some experiments, wrote the paper and supervised the study.
Competing interests The authors declare that a patent application covering haploid embryonic stem cells has been filed.
References
- 1.Otto SP, Jarne P. Evolution. Haploids--hapless or happening? Science. 2001;292:2441–3. doi: 10.1126/science.1062890. [DOI] [PubMed] [Google Scholar]
- 2.Wiellette E, et al. Combined haploid and insertional mutation screen in the zebrafish. Genesis. 2004;40:231–40. doi: 10.1002/gene.20090. [DOI] [PubMed] [Google Scholar]
- 3.Yi M, Hong N, Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430–3. doi: 10.1126/science.1175151. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman MH, Robertson EJ, Handyside AH, Evans MJ. Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol. 1983;73:249–61. [PubMed] [Google Scholar]
- 5.Latham KE, Akutsu H, Patel B, Yanagimachi R. Comparison of gene expression during preimplantation development between diploid and haploid mouse embryos. Biol Reprod. 2002;67:386–92. doi: 10.1095/biolreprod67.2.386. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman MH. Chromosome analysis of early postimplantation presumptive haploid parthenogenetic mouse embryos. J Embryol Exp Morphol. 1978;45:85–91. [PubMed] [Google Scholar]
- 7.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nichols J, et al. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat Med. 2009;15:814–8. doi: 10.1038/nm.1996. [DOI] [PubMed] [Google Scholar]
- 9.Buehr M, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–98. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–22. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols J, Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- 12.Cutler G, Marshall LA, Chin N, Baribault H, Kassner PD. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 2007;17:1743–54. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard SM, Benchoua A, Lowell S. Neural stem cells, neurons, and glia. Methods Enzymol. 2006;418:151–69. doi: 10.1016/S0076-6879(06)18010-6. [DOI] [PubMed] [Google Scholar]
- 14.Li MA, Pettitt SJ, Yusa K, Bradley A. Genome-wide forward genetic screens in mouse ES cells. Methods Enzymol. 2010;477:217–42. doi: 10.1016/S0076-6879(10)77012-9. [DOI] [PubMed] [Google Scholar]
- 15.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkers H, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–9. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 17.Mai Q, et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 2007;17:1008–19. doi: 10.1038/cr.2007.102. [DOI] [PubMed] [Google Scholar]
- 18.Revazova ES, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–49. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 19.Inoue K, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–9. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 20.Sukov WR, et al. Nearly identical near-haploid karyotype in a peritoneal mesothelioma and a retroperitoneal malignant peripheral nerve sheath tumor. Cancer Genet Cytogenet. 2010;202:123–8. doi: 10.1016/j.cancergencyto.2010.07.120. [DOI] [PubMed] [Google Scholar]
- 21.Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp Cell Res. 1999;252:273–80. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 22.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–5. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 23.Jiang H, et al. Activation of paternally expressed imprinted genes in newly derived germline-competent mouse parthenogenetic embryonic stem cell lines. Cell Res. 2007;17:792–803. doi: 10.1038/cr.2007.70. [DOI] [PubMed] [Google Scholar]
- 24.Kishigami S, Wakayama T. Efficient strontium-induced activation of mouse oocytes in standard culture media by chelating calcium. J Reprod Dev. 2007;53:1207–15. doi: 10.1262/jrd.19067. [DOI] [PubMed] [Google Scholar]
- 25.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–5. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.