Abstract
Cystatin A (gene: CSTA), is up-regulated in non-small-cell lung cancer(NSCLC) and dysplastic vs normal human bronchial epithelium. In the context that chronic obstructive pulmonary disease (COPD), a small airway epithelium (SAE) disorder, is independently associated with NSCLC(especially squamous cell carcinoma, SCC), but only occurs in a subset of smokers, we hypothesized that genetic variation, smoking and COPD modulate CSTA gene expression levels in SAE, with further up-regulation in SCC. Gene expression was assessed by microarray in SAE of 178 individuals [healthy nonsmokers (n=60), healthy smokers (n=82), and COPD smokers (n=36)], with corresponding large airway epithelium (LAE) data in a subset (n=52). Blood DNA was genotyped by SNP microarray. Twelve SNPs upstream of the CSTA gene were all significantly associated with CSTA SAE gene expression(p<0.04 to 5 × 10 −4). CSTA gene expression levels in SAE were higher in COPD smokers (28.4 ± 2.0) than healthy smokers (19.9 ± 1.4, p<10−3), who in turn had higher levels than nonsmokers(16.1 ± 1.1, p<0.04). CSTA LAE gene expression was also smoking-responsive (p<10−3). Using comparable publicly available NSCLC expression data, CSTA was up-regulated in SCC vs LAE (p<10−2) and down-regulated in adenocarcinoma vs SAE (p <10−7). All phenotypes were associated with significantly different proportional gene expression of CSTA to cathepsins. The data demonstrate that regulation of CSTA expression in human airway epithelium is influenced by genetic variability, smoking, and COPD, and is further up-regulated in SCC, all of which should be taken into account when considering the role of CSTA in NSCLC pathogenesis.
Keywords: cystatin, small airway epithelium, gene expression, genotype, COPD
Introduction
Cystatin A (CSTA; also referred to as stefin A, and acid cysteine proteinase inhibitor), a member of the class I or stefin subgroup of the 3 described cystatin families, is a 11kDa single chain intracellular cysteine protease inhibitor capable of inhibiting papain and cathepsins B, H and L, as well as the cysteine protease activity of the major house dust mite allergen Der p 1(1–3). Cystatin A has anti-apoptotic properties(4–6) and CSTA expression has been linked with neoplastic changes in squamous cell epithelium with CSTA levels paralleling the response to antitumor therapy(7–11). As with other members of the stefin family, CSTA has been proposed as a marker and prognostic indicator of lung cancer(11,12). In the lung, CSTA expression is up-regulated in dysplastic epithelium, and highly expressed in many lung cancers, especially squamous cell carcinoma(SCC) (7,10,12).
With this background, and based on the knowledge that the majority of lung cancers are derived from the small airway epithelium (SAE) of cigarette smokers(13–15), but that only a subset of smokers develop lung cancer(16,17), we hypothesized that the expression of CSTA in the SAE is modulated by genetic variation and up -regulated by smoking. Further, in the context that chronic obstructive pulmonary disease (COPD), a disease that starts in the small airways(13,18–21), is a risk factor for lung cancer independent of smoking (22–26), we asked whether CSTA is up-regulated in the SAE of smokers with COPD beyond that of smokers per se, and whether this up-regulation is also dependent on genetic variation. In view of the fact that both smoking and COPD are stronger risk factors for SCC than for adenocarcinoma of the lung, and that most SCCs are derived from large airways(15,27–30)we also examined CSTA expression in large airway epithelium (LAE) samples obtained in a subset of the study subjects, and compared our CSTA gene expression data to publicly available lung cancer microarray data derived using comparable methodologies (31), to see if CSTA is discordantly expressed in SCC and adenocarcinoma of the lung, relative to the SAE and LAE.
Materials and Methods
Study Subjects
Healthy nonsmokers, healthy smokers, and smokers with COPD were assessed in the Weill Cornell National Institutes of Health Clinical and Translational Sciences Center and Department of Genetic Medicine Clinical Research Facility using protocols approved by the Weill Cornell Medical College Institutional Review Board. Prior to inclusion in the study, each individual provided written informed consent. No abnormality was found in the nonsmokers (n=60) or the healthy smokers (n=82) following a standardized screening evaluation composed of a history, physical examination, complete blood count, serum chemistries, coagulation profile, liver function tests, urine studies, chest X-ray, EKG, and full lung function studies. The COPD smokers (n=36) were diagnosed in accordance with standard GOLD criteria(32). Blood was collected for genotyping on a random subset of individuals (nonsmokers, n=44; healthy smokers, n=48; COPD smokers, n=20). Fewer subjects with genotyped samples were available than subjects with SAE gene expression data because of a significantly longer temporal interest of our laboratory in the study of airway epithelium gene expression than in genomic studies. LAE samples were not given as high a priority as SAE samples when individuals were bronchoscoped, reflecting the focus of our lab on SAE gene expression profiles, and as a consequence, LAE samples were available for a random subset of individuals who underwent bronchoscopy (healthy nonsmokers, n=21; healthy smokers, n=31). Urine nicotine and cotinine were measured, in conjunction with serum carboxyhemoglobin, to verify the self-reported smoking status of each group. Detailed inclusion/exclusion criteria and characteristics of the study populations can be found in Supplementary Materials and Methods.
Collection of Airway Epithelium and Assessment of Gene Expression
Fiberoptic bronchoscopy was used to obtain small (and for a random subset, large) airway epithelial cells as previously described (33,34). Further details of the airway epithelial sampling, preparation of cDNA, hybridization of labelled cRNA to Affymetrix HG-U133 Plus 2.0 arrays and confirmation by TaqMan RT-PCR can be found in Supplementary Materials and Methods. No cell lines were used in the conduct of this research; all samples used were freshly collected and immediately processed for RNA or protein.
Correlation of Genotypes and Haplotypes with Gene Expression
Genomic DNA was extracted from stored blood samples using the Autogen FX robotic system in accordance with Autogen’s protocols (Autogen, Holliston, MA). Pre-printed bar-coded labels were affixed to sample containers to minimize sample mix-ups, and critical steps in sample processing were only undertaken when 2 technicians were present. The Affymetrix Human SNP Array 5.0 platform was used to assess genotype using the manufacturer’s protocols. The focus was on all 48 SNPs found on the SNP array whose chromosomal location was within 100,000 base pairs either side of the CSTA gene. To avoid artifactual association of genotype with expression caused by SNPs with known minor allele frequency> 5% situated within the target sequence of Affymetrix expression probe sets, the sequences of individual probes (obtained from NetAffx, Affymetrix, Santa Clara, CA) were cross-checked against the National Center for Biotechnology Information (NCBI) dbSNP build 129. Potential effects of copy number variation on associations of genotype with gene expression were assessed as outlined in Supplementary Materials and Methods.
Data Analysis and Statistics for SAE Expression and Genomic Data
Gene expression analyses on all samples was performed using the Microarray Suite 5.0 (MAS5) software. Publicly available data from Kuner et al (31) for 40 subjects with lung adenocarcinoma and 18 subjects with SCC of the lung were downloaded from the NCBI Web site, accession number GSE10245), using the gene expression data from the only CSTA probe set 204971_at on the Affymetrix HG-U133 Plus 2.0 chip, the same platform that was used on the SAE and LAE samples. The proportional relationship among CSTA and cathepsin gene expression was examined in all samples using the additional probe sets 200838_at (cathepsin B), 202295_s_at (cathepsin H) and 202087_s_at (cathepsin L). Gene expression data were normalized using GeneSpring version 7.2 software (Agilent Technologies) per chip and per gene across all samples. All microarray data have been deposited at the Gene Expression Omnibus (GEO) site (35; accession number GSE22047). Genomic data from the Affymetrix Human SNP 5.0 were assessed using the BRLMM-P Analysis Tool (BAT) 2.0 software (Affymetrix) to determine genotype for cis-SNPs in the vicinity of CSTA on chromosome 3. Associations between CSTA gene expression levels and genotype for the 48 SNPs located within 100 kb of the gene were performed using PLINK, with permutation testing used to control for the effect of genetic ancestry. Haplotype associations with expression were performed using PLINK and Haploview, with linear regression modeling(see Supplementary Materials and Methods).
Results
Association of CSTA Small Airway Epithelial Gene Expression with Genomic Variation
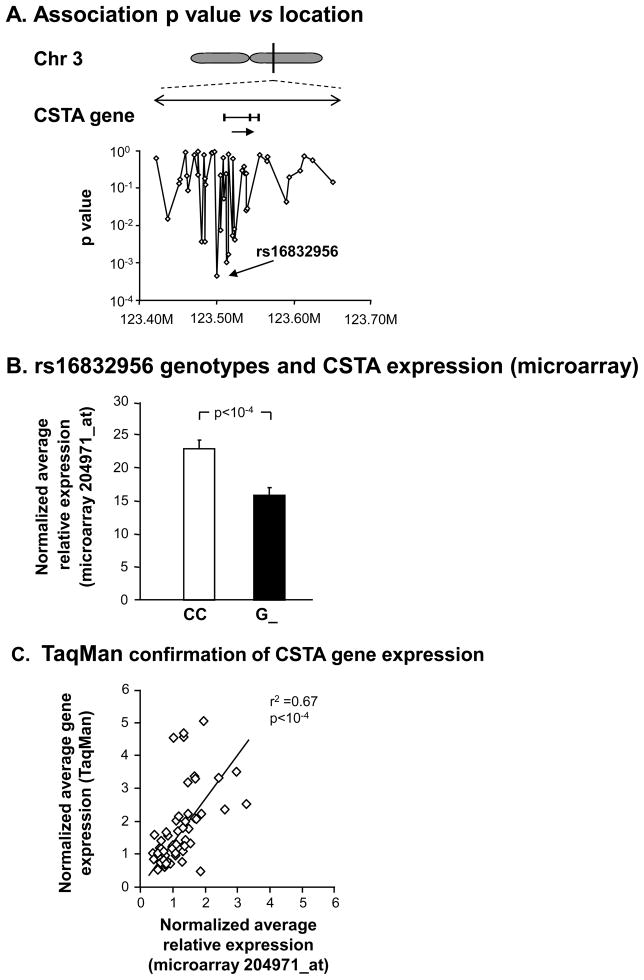
The study population demographic findings and characteristics of brushed SAE samples and microarray expression probe performance are detailed in Supplementary Results. Based on the hypothesis that genetic variability might modulate the level of CSTA expression in the SAE, we evaluated the correlation of CSTA gene expression with genotypes of cis-SNPs within 100 kbp either side of the CSTA gene using paired airway gene expression data and blood DNA SNP data from the subset of the study population for whom both gene expression and SNP data was available (healthy nonsmokers, n=44; healthy smokers, n=48; and COPD smokers, n=20). Ten SNPs located upstream of the CSTA gene, and 2 more within introns of the CSTA gene, displayed significant correlation of genotype with SAE expression levels of CSTA (Table I, Figure 1A). For the total combined group of subjects (healthy nonsmokers, healthy smokers and smokers with COPD), the most significant correlation of the levels of CSTA small airway gene expression was with the SNP rs16832956 (p< 5 × 10−4). Nine of the 12 SNPs identified had p values< 10−2in reference to the strength of their association with CSTA SAE gene expression. The 4 SNPs most upstream of CSTA were located within introns of the calcium-sensing receptor (CASR) gene, but none had any relationship of genotype with CASR gene expression levels (p>0.8, not shown).
Table I.
Association of CSTA Genotypes with CSTA Gene Expression in the Small Airway Epithelium1
| SNP identity3 | Chr. 3 location4 | Minor allele frequency5 | p value for association of genotype with expression level2 |
|||
|---|---|---|---|---|---|---|
| Assessed by microarray
| ||||||
| All subjects6 (n=112) | Healthy nonsmokers (n=44) | Healthy smokers (n=48) | COPD smokers (n=20) | |||
| rs1354162 | 123,436,767 | 0.040 | 0.02 | NS | NS | 4.6 × 10−3 |
| rs7652858 | 123,480,795 | 0.125 | 3.6 × 10−3 | NS | NS | NS |
| rs2134225 | 123,484,943 | 0.125 | 3.6 × 10−3 | NS | NS | NS |
| rs16832956 | 123,500,198 | 0.161 | 4.5 × 10−4 | NS | 3.4 × 10−3 | 0.010 |
| rs1402200 | 123,505,107 | 0.411 | 7.3 × 10−3 | NS | NS | NS |
| rs5008830 | 123,513,152 | 0.089 | 1.0 × 10−3 | 0.025 | 1.1 × 10−5 | 0.038 |
| rs2001548 | 123,515,479 | 0.101 | 1.7 × 10−3 | NS | 3.4 × 10−5 | 0.038 |
| rs4678180 | 123,520,487 | 0.455 | 5.2 × 10−3 | NS | NS | NS |
| rs9864290 | 123,522,752 | 0.451 | 7.8 × 10−3 | NS | NS | NS |
| rs6803098 | 123,523,300 | 0.453 | 4.1 × 10−3 | NS | NS | NS |
| rs9817571 | 123,538,670 | 0.099 | 0.03 | NS | 0.018 | NS |
| rs9842752 | 123,539,553 | 0.076 | 0.03 | NS | NS | NS |
CSTA gene expression levels in small airway epithelium as assessed by Affymetrix HGU133 Plus 2.0 microarray expression probe set 204971_at were correlated with Affymetrix Human SNP Array 5.0 cis-SNPs situated within 100 kbp of the CSTA gene location on chromosome 3.3q21.
For analysis with the group of all subjects, p values represent Wald statistic following permutation analysis: after initial associations were identified, each association was further tested by performing 103 permutations within clusters of similar genetic ancestry, as defined by STRUCTURE, using PLINK software. For the subgroup analyses, t-tests are shown.
Only those SNPs with p values for association of genotype with expression of <0.05 are shown. SNPs are listed in ascending order of location on chromosome 3. 5′ to 3′, the first 4 SNPs listed are within introns of the upstream CASR gene, the next 6 are intergenic, and the last 2 listed SNPs are located within introns of CSTA.
Ch, chromosome. Locations refer to NCBI Human Genome Reference Genome Assembly, Build 36.3
Minor allele frequencies observed in the study population.
All subjects with SAE gene expression data and genotype data. Healthy nonsmokers (n=44), healthy smokers (n=48) and COPD smokers (n=20) combined. NS=not significant (p>0.05).
Figure 1.
Genetic modulation of CSTA SAE gene expression. A. Magnified view of the CSTA gene region, with direction of transcription (arrow). Below the gene, same scale, are the corresponding chromosomal locations of 48 cis-SNPs on the Affymetrix Human SNP Array 5.0 found within 100 kb either side of the gene. Shown are the correlations (Wald statistic) with microarray-assessed SAECSTA gene expression in 112 healthy nonsmokers, healthy smokers, and smokers with COPD. B. Microarray normalized average expression values of CSTA for genotypes of SNP rs16832956. Shown are data for all 112 genotyped subjects with CSTA SAE gene expression on the ordinate. G_=data for the combined genotypes GG and CG. C. TaqMan RT-PCR confirmation of microarray CSTA gene expression levels in SAE in a random subset of healthy nonsmokers (n=23), healthy smokers (n=28), and COPD smokers (n=13).
These associations persisted at this significance level after assessing 103 permutations within clusters of similar genetic ancestry suggesting that genotype rather than genetic ancestry was the cause of the observed effect. For example, G_ genotypes of rs16832956, the SNP with the strongest genetic association, had low CSTA expression, with a >1.4 ± 0.1-fold increase seen in CC genotypes (Figure 1B, p<1.6 × 10−4). The SAECSTA gene expression levels were confirmed by TaqMan real time RT-PCR (Figure 1C, r2=0.67, p<10−4).
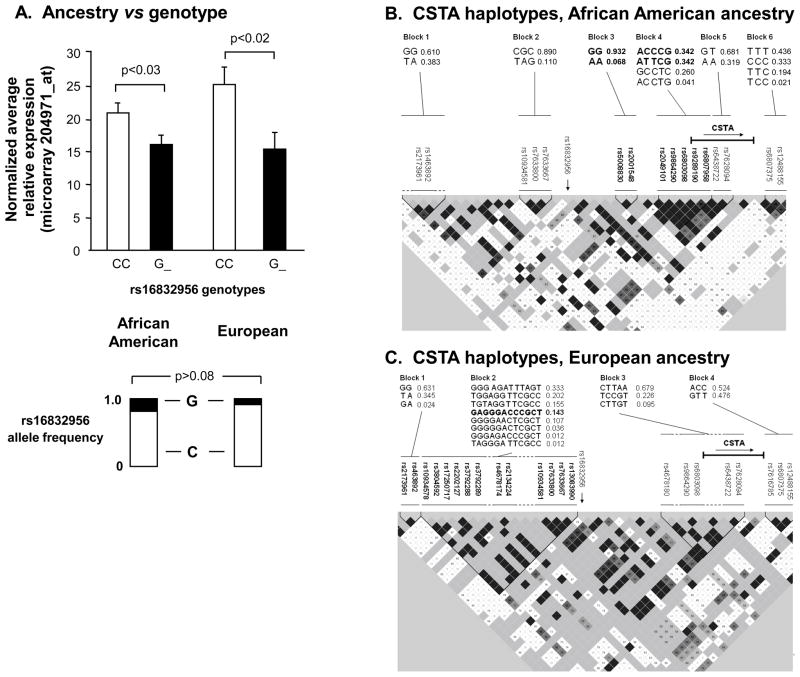
Influence of Ancestral Background on Genetic Associations with CSTA Small Airway Epithelium Gene Expression
Genetic ancestry could be a potential confounding variable limiting the generalizability of observations made in relation to the modulation of CSTA gene expression by genetic variability. However, the associations of rs16832956 genotypes with CSTA expression levels were similar among the 2major ancestral groupings (p< 0.03, both ancestral groups), with no significant difference in allele frequency among those of African American ancestry vs subjects of European ancestry (Figure 2A, G allele frequency 0.19 in African American ancestral group, G allele frequency 0.09 in European ancestral group, p>0.08). Haplotypes, comprised of SNPs surrounding the CSTA genomic location, were significantly associated with CSTA gene expression for specific ancestral subgroups examined separately(Figure 2B,C), with the strongest association seen in the group of individuals of European ancestry for haplotype GAGGGACCCGCT(Figure 2 C, p<0.008, Supplementary Results, Supplementary Table II).
Figure 2.
Effect of ancestry on genetic modulation of SAECSTA expression. A. CSTA expression based on genotypes of rs16832956 parsed by the 2 major ancestral groups, African American and European. Ordinate -normalized average relative CSTA gene expression. Allele frequencies as shown. B. Analysis of haplotypes derived from local CSTA cis-genotypes in subjects of African American genetic ancestry. Analyses performed on the subset of 56 subjects of African American ancestry with available genotype data. The genomic sequence and frequencies of the 6 haplotype blocks are shown in the upper portion of the figure, corresponding to the named SNPs indicated below. In the haplotype map, the darker the shading, the stronger the linkage disequilibrium (LD) between adjacent SNPs, and vice versa. The haplotypes in blocks 3 and 4 (highlighted in bold type, with the highlighted constituent SNPs shown beneath), were significantly associated with CSTA gene expression (see Supplementary Table II). The relative location and direction of the CSTA gene is indicated. The relative location of the SNP most strongly associated with CSTA gene expression, rs16832956, is indicated (vertical arrow). C. CSTA haplotype association data for 35 subjects of European genetic ancestry. The indicated haplotype in block 2, composed of the 12 SNPs shown in bold, was the only haplotype significantly associated with CSTA gene expression (see Supplementary Table II).
Effect of Smoking Status and COPD on CSTA Gene Expression in the SAE
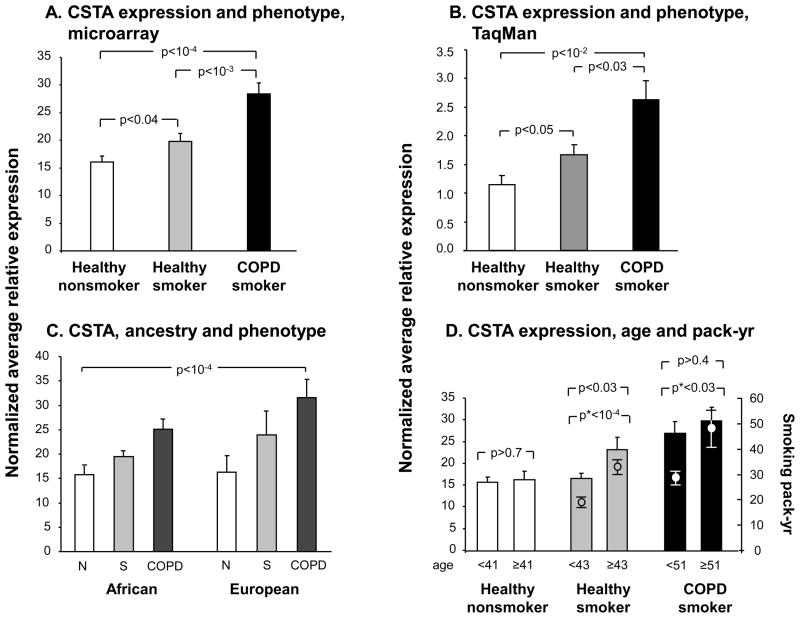
Gene expression levels of CSTA in SAE were significantly higher in the group of healthy smokers (n=82) compared to healthy nonsmokers (n=60, p<0.04, pairwise Student’s t-test), but were even more up-regulated in smokers with COPD (n=36) compared to the healthy smokers (Figure 3A, p <10−3, pairwise Student’s t-test; p<10−4by analysis of variance for all 3 groups). The association of CSTA expression with phenotype was confirmed by RT-PCR, and was explained neither by genetic ancestry nor by age, though there was evidence of a possible dose-response relationship (Figure 3B–D, Supplementary Results). The effect of genotype on SAE CSTA gene expression seen in the total genotyped study population, was also observed when healthy nonsmokers, healthy smokers and COPD smokers were examined separately (Supplementary Results, Supplementary Figures 1A,B).
Figure 3.
CSTA gene expression levels in SAE of healthy nonsmokers, healthy smokers and smokers with COPD. A. Average relative mRNA expression values of CSTA (healthy nonsmokers, n=60; healthy smokers, n=82; and COPD smokers, n=36) as detected by microarray. B. TaqMan RT-PCR confirmation of the association of small airway epithelium CSTA gene expression levels with phenotype in a subset of healthy nonsmokers (n=23), healthy smokers (n=28), and smokers with COPD (n=13). C. Average relative expression level of CSTA for the same subjects as in A, with genetic ancestry on the abscissa. Two factor ANOVA: p <10−4 for phenotype; p>0.09 for the factor “ancestry”; interaction among phenotype and ancestry p>0.5. D. Average relative expression level of CSTA in SAE as a function of age and smoking history. Left ordinate: CSTA gene expression, bar plots; right ordinate: smoking history of the 2 groups of smokers (in pack-yr), dot and whisker plots and asterisk p values. Each of the phenotypic groups are divided by mean age (of that group). Error bars, standard error. p values are from pairwise student t-tests, except where otherwise indicated.
Western Analysis of Cystatin A Protein Expression
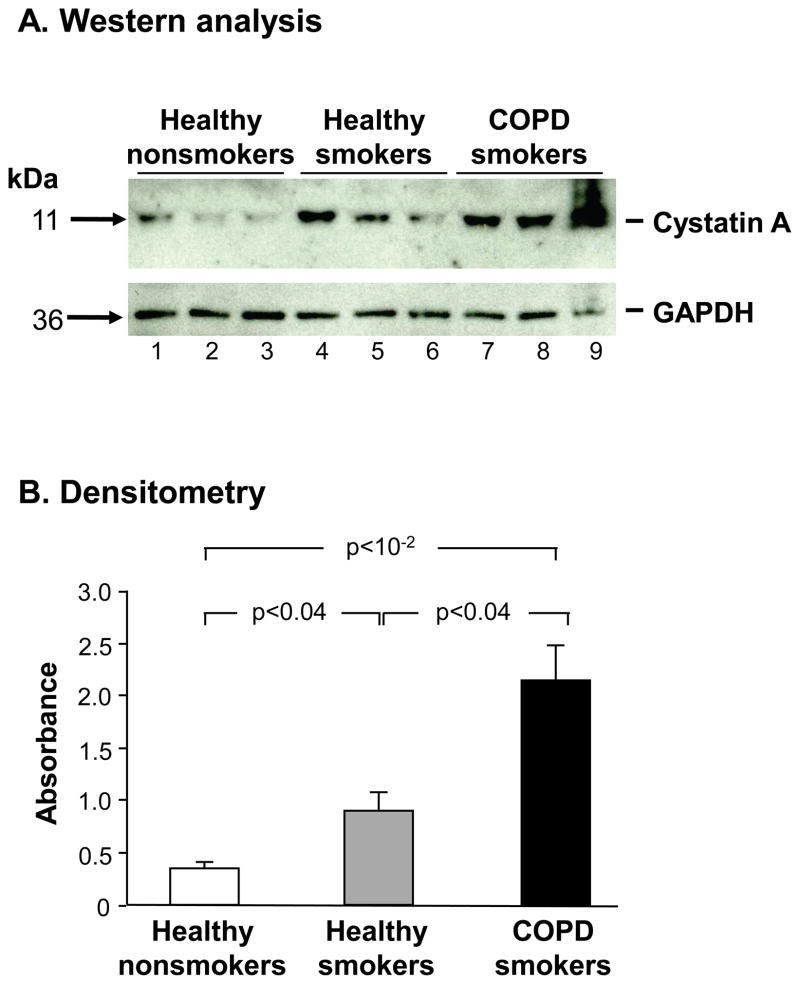
Whole-cell lysates of small airway epithelium from healthy nonsmokers, healthy smokers and smokers with COPD were quantitatively assessed for cystatin A expression using Western analysis. Western analysis revealed increased cystatin A protein expression in healthy smokers compared to nonsmokers, with even higher expression in smokers with COPD compared to the healthy smokers (Figure 4A), which was confirmed quantitatively by densitometry (Figure 4B, p<0.04 by pairwise comparisons by Student’s t-test; p<10−2by analysis of variance for all 3 phenotypic groups).
Figure 4.
Western analysis of SAECSTA protein expression in nonsmokers, healthy smokers and smokers with COPD. A. Western analysis. Upper panel. SAECSTA protein expression in healthy nonsmokers (lanes 1–3), healthy smokers (lanes 4–6) and smokers with COPD (lanes 7–9). Lower panel: protein loading control antibody (GAPDH). B. Quantification of the data in panel A by densitometry normalized to GAPDH. P values, pairwise student t-test. Error bars, standard error.
CSTA Gene Expression in LAE, SCC and Adenocarcinoma
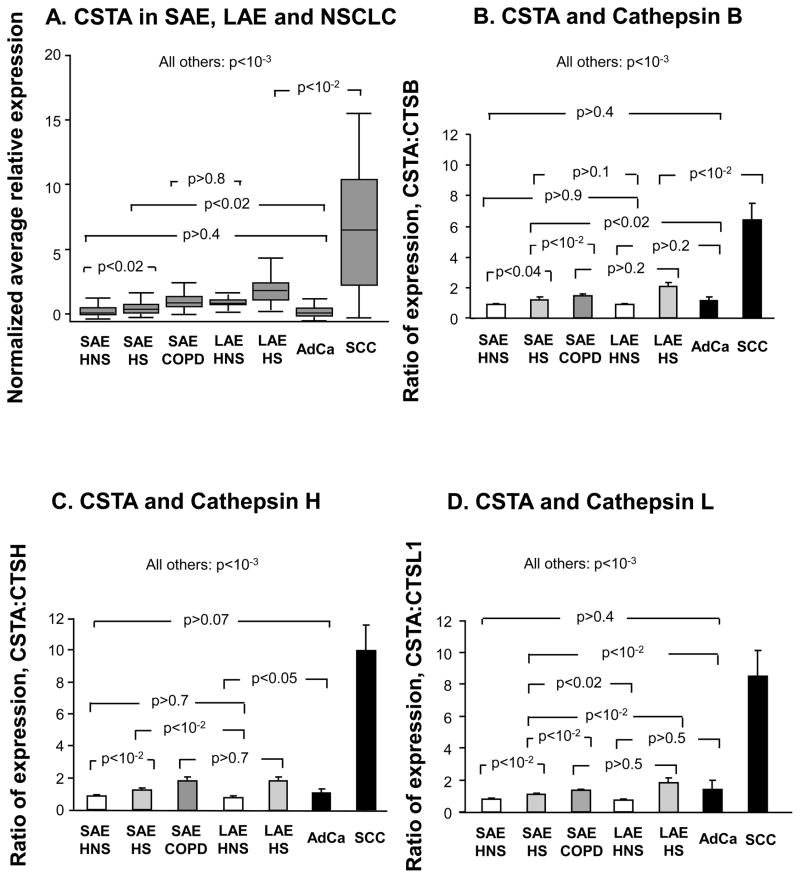
Most lung cancers, especially adenocarcinoma, are derived from the SAE, and yet it is well established that smoking and COPD are more strongly linked to SCC than to adenocarcinoma (14,28,30). SCCs tend to arise in the more central airways (15). A comparison of CSTA expression was made among SAE, LAE and NSCLC specimens (see Supplementary Results for details of the study population demographics and characteristics of brushed LAE samples). CSTA expression levels in LAE of healthy nonsmokers were similar to the up-regulated levels seen in the SAE of COPD smokers. LAECSTA expression was smoke -responsive (Figure 5A, p<10−3)and but further up-regulated in SCC (Figure 5A, p<10−2, all comparisons). In contrast, adenocarcinoma CSTA gene expression levels were significantly down-regulated compared to all but the SAE of healthy nonsmokers (Figure 5A, Supplementary Results).
Figure 5.
Comparison of CSTA gene expression in SAE, LAE, and in lung cancer. A. Normalized average microarray CSTA gene expression levels are shown on the ordinate as box and whisker plots with median, interquartile range and range, for each of the groups depicted on the abscissa. B. Ratio of microarray-determined gene expression level of CSTA to cathepsin B within individual subjects, for each of the 7 indicated phenotypic groups, including data from non-small cell lung cancer cells. C. The ratio of microarray -derived SAE, LAE and tumor CSTA levels to corresponding cathepsin H gene expression levels. D. Shown on the ordinate is the same analysis as in B and C but with cathepsin L as the denominator. Error bars in B–D represent standard error. P values are from ANOVA. HNS, healthy nonsmoker. HS, healthy smoker. COPD, smokers with chronic obstructive pulmonary disease. SAE, small airway epithelium. AdCa, adenocarcinoma of the lung. SCC, squamous cell carcinoma of the lung.
In view of the marked up-regulation of CSTA observed in SCC subjects, a comparison was made of the ratio of CSTA gene expression to expression levels of the 3 known cathepsin targets (B, H and L) of cystatin A with in individuals. These expression ratios differed significantly among most phenotypic and tissue groupings, except for adenocarcinoma vs nonsmoker SAE (Supplementary Results, Figure 5B–D).
Discussion
The cytosolic cysteine protease inhibitor cystatin A, coded for by the CSTA gene, has attracted interest with a number of reports relating expression of CSTA protein levels to neoplastic states in a variety of tissues including those of epithelial origin(7–12,36). Immunohistochemical data have suggested that CSTA is expressed at higher levels in dysplastic human bronchial epithelium, in SCC in particular, and less often in adenocarcinoma of the lung, compared to normal bronchial epithelium (11,12). In light of the fact that COPD, a disease that arises in the small airways, is a risk factor for lung cancer independent of the risk attributable to smoking, but only occurs in a minority of smokers(16–20,22–27,29), we asked: does genetic variation modulate CSTA gene expression levels in the human SAE; is the expression of CSTA in SAE influenced by both smoking and by COPD; and since smoking and COPD each have a propensity toward SCC rather than adenocarcinoma (28–30), is CSTA expression differentially influenced by SCC vs adenocarcinoma of the lung relative to SAE and LAE expression levels in the non-cancerous samples ?
Using fiberoptic bronchoscopy, pure populations of small airway epithelium were obtained from 60 carefully phenotyped healthy nonsmokers, 82 healthy smokers, and 36 smokers with COPD, and for the majority, both SAECSTA gene expression and corresponding blood CSTA genotypes were assessed by microarray. The data demonstrate that CSTA gene expression levels in the human SAE are modulated by 12 SNPs with in the vicinity of the gene, a genetic association that is not confounded by genetic ancestry. There is an association of local cis-haplotypes with CSTA gene expression for 4 haplotypes in the case of subjects of African American ancestry and 1 haplotype in those of European ancestry. In addition, healthy smokers have higher CSTA gene expression levels than healthy nonsmokers, with even higher levels observed in smokers with COPD compared to the healthy smokers independent of pack-yr, albeit with a suggestive evidence of a dose-response relationship for smokers. The microarray expression was confirmed at the transcript level by quantitative TaqMan PCR, and at the protein level by Western analysis. The genetic modulation of SAECSTA gene expression was a consistent finding when the 3 phenotypic groups were examined separately. Finally, tumor tissues from individuals with SCC of the lung have the highest relative levels of CSTA gene expression, but tumor tissue from adenocarcinoma subjects have substantially lower levels of CSTA than in any of the other tissues and phenotypes examined (except for being similar to healthy nonsmokers). Interestingly, the phenotypic states examined (healthy, COPD, lung cancer) are each associated with significantly different proportional gene expression of CSTA to cathepsins B, H and L. These observations are in keeping with the concept that the CSTA gene plays a role in the evolution of the bronchial epithelium of peripheral and central airways from normal to disease, and suggests a complex interplay among genetics, smoking, COPD and lung cancer histologic subtype in relation to CSTA gene expression. Cystatin A appears to have a more plausible connection to the specific evolution of healthy smoker and COPD smoker airway epithelium into SCC rather than into adenocarcinoma, in keeping with the known stronger relationship of the separate risk factors of smoking and COPD with SCC than with adenocarcinoma of the lung(27–30).
Small Airway Epithelium, Smoke-induced Lung Disease and Genetics
There is now a large body of evidence pointing to the small airway epithelium (defined as bronchi of ≥6 generations, <2 mm in diameter) as the earliest site of pathological involvement in COPD, the primary site of airflow limitation in this disorder, and the site of development of most non-small cell lung cancers(13–15,18–21). Consistent with these observations in the major smoke-induced lung diseases, morphologic changes are found in small airways of asymptomatic smokers with normal lung function (37,38). COPD is a relevant phenotype to study in the progression from healthy airway epithelium to lung cancer, an airway epithelial-derived disease, because COPD is itself an independent risk factor for the development of lung cancer, with a contribution to lung cancer risk that is separate from the risk attributable to cigarette smoking (22–26). Since only 15 to 20% of smokers will develop COPD, together with evidence of familial clustering in COPD and non-small cell lung cancer and twin concordance for the risk for COPD (15–17,22,39–41), it is likely that genetic variation influences disease risk. The fact that SCC is more common than adenocarcinoma in smokers and in COPD subjects also raises the question of the role of the LAE in the pathogenesis of these tumor subtypes. Although the current study was not designed to focus on LAE gene expression/genotype relationships, it is interesting to note that the smoke-responsive nature of CSTA expression in SAE was also observed in LAE. However, the levels of CSTA in healthy smoker LAE are still considerably less than in SCC subjects and taken together with the smoke-responsiveness, are therefore less likely to reflect mere constitutive expression of CSTA in a LAE-derived tumor. Although the observations of genetic modulation of CSTA gene expression and the up-regulation of such expression in SAE from COPD smokers compared to healthy smokers is intriguing, the present study was not designed to address the question of whether or not CSTA expression is genetically associated with COPD. Testing this hypothesis would require the genotyping of a large number of subjects in order to have sufficient statistical power to reach a definitive conclusion.
Although the data shows some evidence for a dose-response relationship between intensity of smoke-exposure and CSTA gene expression, disease also influences the expression levels, as evidenced by the greater relative increase in CSTA levels in COPD and SCC vs healthy smokers compared to the smaller increase in healthy smokers vs nonsmokers, even though the latter comparison would have the greater difference in smoking exposure. Also, reciprocal effects of disease state and pack-yr on CSTA gene expression in SAE were observed in the healthy smoker and COPD smoker age-defined subgroups.
Analysis of a published gene expression dataset that used microarray technology to study lung cancer tissues did not show a significant effect of smoking status on CSTA gene expression (42). Others have noted that cystatin A levels are higher in early-detected lung cancers compared to lung cancers presenting later in the disease course, despite higher pack-yr values in the latter (43). These disparate observations highlight the need for further study to clarify if there is an effect of smoking-intensity on CSTA gene expression in lung airway epithelium and cancers derived from the airway epithelium. The observations suggest that CSTA expression is not uniformly smoking-responsive and is likely to be specific to cell populations. Controlling for the influence of age while examining pack-yr consumption is challenging and would likely require large datasets, because more often than not, individuals with higher pack-yr histories will also be older. From the data in the present study, it seems more likely that it is the intensity of smoking rather than age that exerts the greater influence on CSTA expression levels, as evidenced by the concordance among the magnitudes of pack-yr difference and the difference in SAE CSTA levels in comparing the 2 smoking phenotypic groups (healthy smokers and COPD smokers) parsed by median age.
Cystatin A in Lung Disease
In the normal human airway epithelium, cystatin A is detected in the basal cells by immunohistochemistry, with more extensive staining in preneoplastic bronchial epithelium (12). Strong staining for cystatin A has also been observed in lung cancer tissue, especially SCC but also in adenocarcinoma and large cell carcinoma (11,12). For SCC, cystatin A expression was less marked in less well differentiated tumors, in which case lower cystatin A expression was also associated with tumor recurrence (12). Quantitative up-regulation of cystatin A protein has been noted in human lung cancer tissues vs control lung using ELISA, and univariate survival analyses showed that individuals with higher levels of cystatin A had a better survival probability, suggesting that cystatin A is up-regulated in lung tumors to counteract potentially harmful tumor-associated activity(11,12). The present study confirms the previously published immunohistochemical data showing an up-regulation of CSTA in SCC vs normal airway epithelium, but also reveals a down-regulation of CSTA in adenocarcinoma relative to levels in the healthy state, challenging data from another cohort that suggested an up-regulation of cystatin A in some (but not in all) bronchogenic adenocarcinomas compared to normal airway epithelium (12).
Whereas all of the functions of cystatin A are not clear, other than its known cysteine protease inhibitory effects, there is some evidence that it inhibits apoptosis in the presence of stimuli such as UVB radiation and viruses(4–6). Although deficiency states have been described arising from mutations in the genes that code for cystatins B and C, resulting in phenotypes of progressive myoclonous epilepsy and hereditary cystatin C amyloid angiopathy respectively, to date no functional mutations have been reported for CSTA in humans(44,45).
The putative role of cystatin A as a tumor suppressor is supported by observations in other tissues, including breast, prostate and esophageal tumors, with evidence that cystatin A can inhibit tumor cell growth, angiogenesis, invasion and metastasis(9,36,46). The observations in the present study that CSTA is down-regulated in adenocarcinoma of the lung with evidence of less-well opposed cysteine protease activity vs other smoke-exposed phenotypes, are compatible with a tumor suppressor role in adenocarcinoma, but with a different function in the progression of some smokers to COPD and SCC, perhaps reflecting an “excess” of cystatin relative to its cognate proteases, the cathepsins. In contrast with these observations, a homozygous mutant mouse model involving chromosomal deletion of csta, the murine homolog of human CSTA, together with 3 other genes, showed no phenotypic abnormality, the animals were not overtly susceptible to spontaneous or irradiation-induced tumor formation, and had evidence of compensatory gene expression of genes phylogenetically related to csta (47).
Based on reports that CSTA is up-regulated in dysplastic airway epithelium and in lung cancer, the findings of the present study, which shows that the regulation of CSTA expression in SAE is influenced by genetic variability, smoking, and COPD, and that CSTA is differentially expressed by lung cancer histological subtypes, suggests that each of these factors should be controlled for when considering the use of CSTA as a marker related to the pathogenesis of lung cancer. The progressive rise in CSTA expression observed in disease states (i.e., COPD and SCC) does not appear to be offset by a corresponding rise in cognate cathepsin levels in the same tissue from any given individual, an observation that may have implications for disease pathogenesis if intracellular cysteine protease-antiprotease homeostasis is relevant, particularly in genetically predisposed smokers.
Supplementary Material
Acknowledgments
We thank N Mohamed and T Benios for help in preparing this manuscript. These studies were supported, in part, by R01 HL074326; P50 HL084936 and UL1-RR024996.
Footnotes
These studies were supported, in part, by R01 HL074326; P50 HL084936 and UL1-RR024996
References
- 1.Barrett AJ. The cystatins: a diverse superfamily of cysteine peptidase inhibitors. Biomed Biochim Acta. 1986;45:1363–74. [PubMed] [Google Scholar]
- 2.Takai T, Kato T, Hatanaka H, et al. Modulation of allergenicity of major house dust mite allergens Der f 1 and Der p 1 by interaction with an endogenous ligand. J Immunol. 2009;183:7958–65. doi: 10.4049/jimmunol.0713276. [DOI] [PubMed] [Google Scholar]
- 3.Vray B, Hartmann S, Hoebeke J. Immunomodulatory properties of cystatins. Cell Mol Life Sci. 2002;59:1503–12. doi: 10.1007/s00018-002-8525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorklund HV, Johansson TR, Rinne A. Rhabdovirus-induced apoptosis in a fish cell line is inhibited by a human endogenous acid cysteine proteinase inhibitor. J Virol. 1997;71:5658–62. doi: 10.1128/jvi.71.7.5658-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones B, Roberts PJ, Faubion WA, Kominami E, Gores GJ. Cystatin A expression reduces bile salt-induced apoptosis in a rat hepatoma cell line. Am J Physiol. 1998;275:G723–G730. doi: 10.1152/ajpgi.1998.275.4.G723. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi H, Komatsu N, Ibe M, Ishida-Yamamoto A, Hashimoto Y, Iizuka H. Cystatin A suppresses ultraviolet B-induced apoptosis of keratinocytes. J Dermatol Sci. 2007;46:179–87. doi: 10.1016/j.jdermsci.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Calkins CC, Sloane BF. Mammalian cysteine protease inhibitors: biochemical properties and possible roles in tumor progression. Biol Chem Hoppe Seyler. 1995;376:71–80. [PubMed] [Google Scholar]
- 8.Korolenko TA, Poteryaeva ON, Falameeva OV, Levina OA. Cystein proteinase inhibitor stefin A as an indicator of efficiency of tumor treatment in mice. Bull Exp Biol Med. 2003;136:46–8. doi: 10.1023/a:1026084712399. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Ding F, Zhang L, et al. Overexpression of stefin A in human esophageal squamous cell carcinoma cells inhibits tumor cell growth, angiogenesis, invasion, and metastasis. Clin Cancer Res. 2005;11:8753–62. doi: 10.1158/1078-0432.CCR-05-0597. [DOI] [PubMed] [Google Scholar]
- 10.Rinne A, Jarvinen M, Rasanen O, Dorn A. Demonstration of an epidermal SH-protease inhibitor in normal epithelium and in some human neoplasms--an immunological study (author’s transl) Acta Histochem Suppl. 1980;22:325–9. [PubMed] [Google Scholar]
- 11.Werle B, Schanzenbacher U, Lah TT, et al. Cystatins in non-small cell lung cancer: tissue levels, localization and relation to prognosis. Oncol Rep. 2006;16:647–55. [PubMed] [Google Scholar]
- 12.Leinonen T, Pirinen R, Bohm J, et al. Biological and prognostic role of acid cysteine proteinase inhibitor (ACPI, cystatin A) in non-small-cell lung cancer. J Clin Pathol. 2007;60:515–9. doi: 10.1136/jcp.2006.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petty RD, Nicolson MC, Kerr KM, Collie-Duguid E, Murray GI. Gene expression profiling in non-small cell lung cancer: from molecular mechanisms to clinical application. Clin Cancer Res. 2004;10:3237–48. doi: 10.1158/1078-0432.CCR-03-0503. [DOI] [PubMed] [Google Scholar]
- 14.Rosado-de-Christenson ML, Templeton PA, Moran CA. Bronchogenic carcinoma: radiologic-pathologic correlation. Radiographics. 1994;14:429–46. doi: 10.1148/radiographics.14.2.8190965. [DOI] [PubMed] [Google Scholar]
- 15.Toh CK. The changing epidemiology of lung cancer. Methods Mol Biol. 2009;472:397–411. doi: 10.1007/978-1-60327-492-0_19. [DOI] [PubMed] [Google Scholar]
- 16.Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J. Developing COPD: a 25 year follow up study of the general population. Thorax. 2006;61:935–9. doi: 10.1136/thx.2006.062802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rennard SI, Vestbo J. COPD: the dangerous underestimate of 15% Lancet. 2006;367:1216–9. doi: 10.1016/S0140-6736(06)68516-4. [DOI] [PubMed] [Google Scholar]
- 18.Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J. 2003;22:672–88. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- 19.Celli BR, MacNee W. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23:932–46. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- 20.Hogg JC, Chu F, Utokaparch S, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–53. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 21.Kim V, Rogers TJ, Criner GJ. New concepts in the pathobiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2008;5:478–85. doi: 10.1513/pats.200802-014ET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen BH, Diamond EL, Graves CG, et al. Lancet. Vol. 2. 1977. A common familial component in lung cancer and chronic obstructive pulmonary disease; pp. 523–6. [DOI] [PubMed] [Google Scholar]
- 23.Kassem J, Crystal RG. Chronic obstructive pulmonary disease and lung cancer. In: Schwab M, editor. Encyclopedia of Cancer. Germany: Springer-Verlag; 2008. [DOI] [Google Scholar]
- 24.Koshiol J, Rotunno M, Consonni D, et al. Chronic obstructive pulmonary disease and altered risk of lung cancer in a population-based case-control study. PLoS One. 2009;4:e7380. doi: 10.1371/journal.pone.0007380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tockman MS. Other host factors and lung cancer susceptibility. In: Samet JM, editor. Epidemiology of Lung Cancer (Lung Biology in Health and Disease) New York: Informa Healthcare; 1994. pp. 397–412. [Google Scholar]
- 26.Turner MC, Chen Y, Krewski D, Calle EE, Thun MJ. Chronic obstructive pulmonary disease is associated with lung cancer mortality in a prospective study of never smokers. Am J Respir Crit Care Med. 2007;176:285–90. doi: 10.1164/rccm.200612-1792OC. [DOI] [PubMed] [Google Scholar]
- 27.Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1438. [PMC free article] [PubMed] [Google Scholar]
- 28.Khuder SA. Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer. 2001;31:139–48. doi: 10.1016/s0169-5002(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 29.Purdue MP, Gold L, Jarvholm B, Alavanja MC, Ward MH, Vermeulen R. Impaired lung function and lung cancer incidence in a cohort of Swedish construction workers. Thorax. 2007;62:51–6. doi: 10.1136/thx.2006.064196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasswa-Kintu S, Gan WQ, Man SF, Pare PD, Sin DD. Relationship between reduced forced expiratory volume in one second and the risk of lung cancer: a systematic review and meta-analysis. Thorax. 2005;60:570–5. doi: 10.1136/thx.2004.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuner R, Muley T, Meister M, et al. Global gene expression analysis reveals specific patterns of cell junctions in non -small cell lung cancer subtypes. Lung Cancer. 2009;63:32–8. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 32.Rabe KF, Hurd S, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–55. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 33.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 34.Hackett NR, Heguy A, Harvey BG, et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29:331–43. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 35. [last accessed January 4, 2011];Gene Expression Omnibus (GEO) site. http://www.ncbi.nlm.nih.gov/geo.
- 36.Parker BS, Ciocca DR, Bidwell BN, et al. Primary tumour expression of the cysteine cathepsin inhibitor Stefin A inhibits distant metastasis in breast cancer. J Pathol. 2008;214:337–46. doi: 10.1002/path.2265. [DOI] [PubMed] [Google Scholar]
- 37.Niewoehner DE, Kleinerman J, Rice DB. Pathologic changes in the peripheral airways of young cigarette smokers. N Engl J Med. 1974;291:755–8. doi: 10.1056/NEJM197410102911503. [DOI] [PubMed] [Google Scholar]
- 38.Willemse BW, ten Hacken NH, Rutgers B, Postma DS, Timens W. Association of current smoking with airway inflammation in chronic obstructive pulmonary disease and asymptomatic smokers. Respir Res. 2005;6:38. doi: 10.1186/1465-9921-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brody JS, Spira A. State of the art. Chronic obstructive pulmonary disease, inflammation, and lung cancer. Proc Am Thorac Soc. 2006;3:535–7. doi: 10.1513/pats.200603-089MS. [DOI] [PubMed] [Google Scholar]
- 40.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 41.Webster PM, Lorimer EG, Man SF, Woolf CR, Zamel N. Pulmonary function in identical twins: comparison of nonsmokers and smokers. Am Rev Respir Dis. 1979;119:223–8. doi: 10.1164/arrd.1979.119.2.223. [DOI] [PubMed] [Google Scholar]
- 42.Chitale D, Gong Y, Taylor BS, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28:2773–83. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bianchi F, Hu J, Pelosi G, et al. Lung cancers detected by screening with spiral computed tomography have a malignant phenotype when analyzed by cDNA microarray. Clin Cancer Res. 2004;10:6023–8. doi: 10.1158/1078-0432.CCR-04-0619. [DOI] [PubMed] [Google Scholar]
- 44.Palsdottir A, Abrahamson M, Thorsteinsson L, et al. Mutation in cystatin C gene causes hereditary brain haemorrhage. Lancet. 1988;2:603–4. doi: 10.1016/s0140-6736(88)90641-1. [DOI] [PubMed] [Google Scholar]
- 45.Pennacchio LA, Lehesjoki AE, Stone NE, et al. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1) Science. 1996;271:1731–4. doi: 10.1126/science.271.5256.1731. [DOI] [PubMed] [Google Scholar]
- 46.Mirtti T, Alanen K, Kallajoki M, Rinne A, Soderstrom KO. Expression of cystatins, high molecular weight cytokeratin, and proliferation markers in prostatic adenocarcinoma and hyperplasia. Prostate. 2003;54:290–8. doi: 10.1002/pros.10196. [DOI] [PubMed] [Google Scholar]
- 47.Bilodeau M, MacRae T, Gaboury L, et al. Analysis of blood stem cell activity and cystatin gene expression in a mouse model presenting a chromosomal deletion encompassing Csta and Stfa2l1. PLoS One. 2009;4:e7500. doi: 10.1371/journal.pone.0007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.