Abstract
A high-affinity RNA aptamer (Kd = 50 nM) was efficiently identified by SELEX against a heteroaryl dihydropyrimidine structure, chosen as a representative drug-like molecule with no cross reactivity with mammalian or bacterial cells. This aptamer, its weaker-binding variants, and a known aptamer against theophylline were each embedded in a longer RNA sequence that was encapsidated inside a virus-like particle by a convenient expression technique. These nucleoprotein particles were shown by backscattering interferometry to bind to the small-molecule ligands with affinities similar to those of the free (non-encapsidated) aptamers. The system therefore comprises a general approach to the production and sequestration of functional RNA molecules, characterized by a convenient label-free analytical technique.
Keywords: RNA packaging, virus-like particles, aptamers, backscattering interferometry
DNA and RNA aptamers have been generated against a wide variety of targets, from proteins1 and small molecules2 to whole cells.3 Refinements in the aptamer binding of small molecules have provided especially high affinities,4,5 unique specificities,6 or the ability to recognize specific functional groups on a ligand.7 We wish to combine the capabilities of small molecule-binding aptamers with modified versions of their natural packaging and transport vehicles, virus capsid proteins. As our test case, we used virus-like particles (VLPs) assembled from the recombinant expression of the bacteriophage Qβ coat protein.8,9 These particles are stable to a variety of chemical and genetic modifications, allowing for decoration of the exterior with targeting, immunogenic, or labeling agents, leaving the interior available for packaging cargo molecules.9–11 The native virus uses a hairpin sequence in its (+)-ssRNA genome to bind to the coat protein interior,12,13 an interaction also employed by the closely related MS2 phage.14–16 As reported elsewhere, we have used this interaction to produce Qβ VLPs with encapsidated enzymes.17 Here we describe the selection of a high-affinity aptamer for a generic small molecule, the construction of Qβ VLP-packaged forms of that aptamer, analysis of the RNA packaged by the VLPs, and the characterization of the binding of these and related RNA molecules to their ligands.
Results and Discussion
Aptamer evolution and characterization
In order to provide a general packaging method, we wished to develop an RNA aptamer sequence to bind a single small molecule “handle” that we could append to any desired cargo. The ligand has to be chemically complex enough to provide high affinity binding interactions with oligonucleotides,18 but otherwise without biological effect. We chose the heteroaryldihydropyrimidine (HAP) structure 1 (Figure 1), a variant of a class of agents that misdirect the assembly of hepatitis B virus.19–21 Compound 1 is water soluble, nontoxic, and sufficiently dissimilar in structure to native biological molecules to minimize off-target binding. It features a 1,4-triazole linkage installed with copper-catalyzed azide-alkyne cycloaddition (CuAAC) chemistry,22–24 which enables convenient connection of the ligand to other molecules of interest.
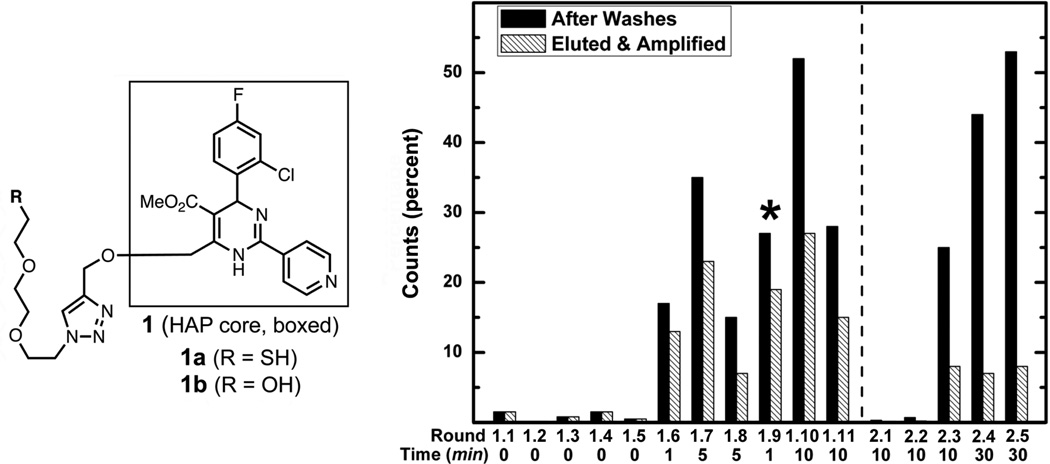
Figure 1.
(Left) Structure of the racemic HAP compounds used for selections and analyses. (Right) Percent of radioactive counts remaining on the HAP resin after buffer washes (solid bars) and the percent eluted, amplified, and carried into the following round (hashed bars). The difference between the bars for each round corresponds to the RNA eliminated from the pool with pre- or post-elution procedures. PCR hypermutagenesis was carried out before the round denoted with a star. The dotted line denotes re-design of the aptamer pool. After the fifth round, two pre-elution washes with solutions of 1b were performed at the incubation times given below the graph. Elution of material for amplification was done with 5.0 mM 1b for 30 minutes. The number of elution fractions retained in each round is specified in the Methods section
The selection of aptamer binders began with a naïve starting pool of RNA containing a 50-base random region flanked by primer binding regions. In the elution step, a long incubation time (30 min) with HAP 1b was used to allow recovery of RNA sequences with higher affinity. Once the pool was enriched in binding sequences, a brief “pre-elution” rinse with 1b was employed to remove lower-affinity (faster off-rate) members from the RNA population as has been done previously to promote the isolation of binders having especially high-affinities.25,26 The stringency of the selection was increased by increasing the incubation time of the washes. Hypermutagenic PCR was carried out once, after round 1.8, to generate diversity in the enriched pool.27 After eleven rounds of selection, the fraction of the input RNA retained by the column after extensive washes with buffer had dramatically increased (to 28%), of which a substantial fraction was eluted after two 10-minute pre-elution steps (Figure 1).
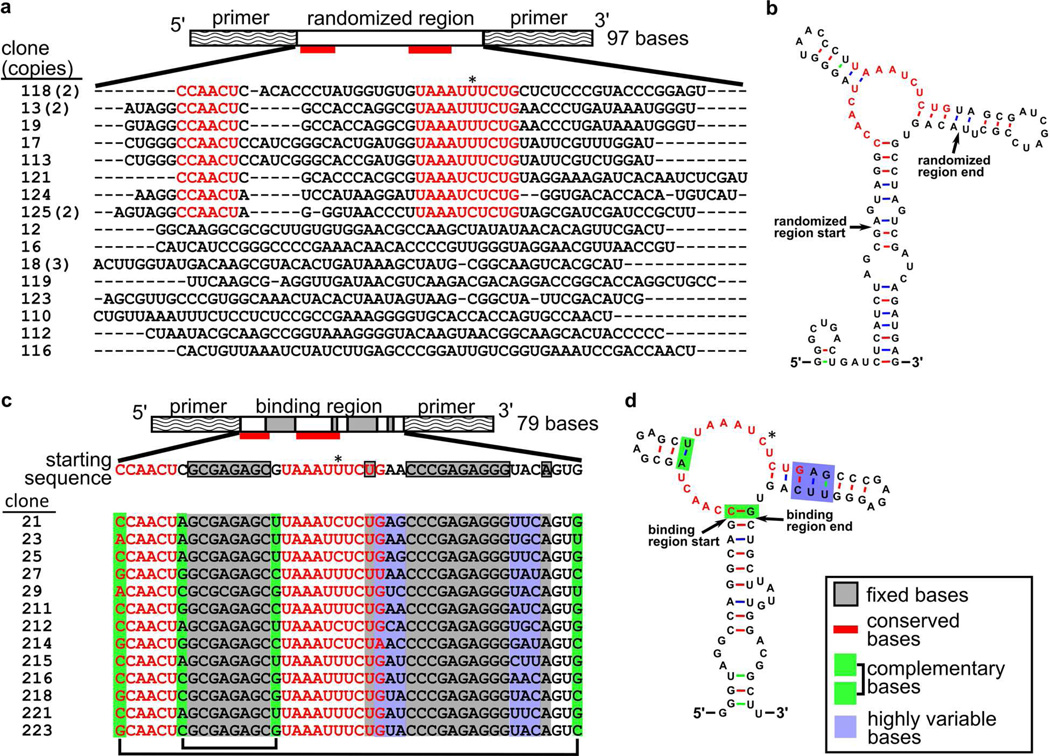
Members of the enriched pool were sequenced, and a common motif was found in the randomized regions of many of the clones (Figure 2a). The predicted secondary structures28 suggested that this central motif formed a bulge at the junction of two hairpins (Figure 2b and Figure S3). Sequences generated by in vitro transcription that contained this motif showed affinity for ligand 1b in preliminary binding experiments.
Figure 2.
Sequences generated after round 1.11 (a,b) and after round 2.5 (c,d) of selection. (a) Layout of the starting pool and sequences of the 50-base random region after round 1.11. The consensus sequence is indicated with red bars and red text; the pyrimidine (either U or C) position is starred. The number of clones with the given sequence is indicated in parentheses. (b) Predicted secondary structure of clone 125, a representative sequence containing the consensus motif. (c) Layout of the starting pool (entering round 2.1) and sequence of the starting pool and clones; (d) the predicted secondary structure of clone 21. The bases boxed in white were synthetically randomized at a 30% rate (10% for each base other than the one shown). Most of the consensus motif (marked in red) remained intact among clones with the exception of the starred pyrimidine position. Bases marked in gray in panel c were fixed; all other bases outside of the primer-binding regions were randomized. Bases marked in green varied but retained complementarity. Bases marked in blue varied among clones and did not always form a paired duplex.
We designed a second-generation pool of RNA sequences containing this putative binding motif in the hope of generating smaller, higher-affinity sequences (Figure 2c). The two predicted flanking hairpins were shortened, a shorter set of primers was used to flank the conserved motif, and several bases in the 3’ hairpin were randomized to test their participation in binding. After five additional rounds of selection (2.1 through 2.5), more than half of the input RNA remained bound to the column after washing with buffer. Two 30-minute pre-elution washing steps followed by a single application of 5 mM 1b eluted 8% of the input RNA, presumably of high affinity. Members of this enriched pool were sequenced and found to have significant similarity to the starting motif, suggesting that a local optimum in sequence space had been identified (Figure 2d).
The predicted28 lowest-energy secondary structure of one final sequence is shown in Figure 2d. The selected clones contained consistent CAACU and UAAAUYUC sequences (positions 16–20 and 32–39) that correspond to a bulge that likely binds the HAP ligand. Two pairs of randomized bases were found to be co-varying, suggesting a role in stapling the binding region together. Bases in the 3’ hairpin varied randomly and probably do not form contacts with the HAP structure. One sequence, designated aptamer 21, was chosen for further characterization.
Measurements of binding of HAP aptamers
To test binding specificity, we generated a random sequence of similar length, flanked by the fixed primer regions of aptamer 21, as a negative control (21-R) and a sequence containing three point mutations within the conserved motif that were expected to diminish binding (21-E) (Table 1). The three HAP aptamers were evaluated by surface plasmon resonance (SPR), performed by flowing RNA over an SPR chip functionalized with a mixture of HAP molecule 1b and a short PEG molecule to inhibit nonspecific adsorption (Figure S6). The aptamer 21 sequence showed a Kads of approximately 50 nM (Table 2), a binding constant that compares favorably to other aptamer-small molecule systems (Table S5).18 As expected, sequence 21-E was found to have a weaker Kads of approximately 223 nM, demonstrating the importance of the conserved motif for binding, and the negative control RNA (21-R) engaged in no discernable binding.
Table 1.
Sequences of the theophylline-binding aptamer ThA and HAP-binding aptamers 21, 21-E, and 21-R, with bases that differ from aptamer 21 marked in red.
| Aptamer | Sequence |
|---|---|
| 21 | |
| 21-E | |
| 21-R | |
| ThA |
Table 2.
Surface adsorption and dissociation constants for HAP aptamers and the theophylline aptamer. Mutant 21-R gave no signal and was used as the reference sample in BSI experiments in free or packaged form as appropriate, so no Kd is given.
| Aptamer | Small molecule |
1/Kads (SPR) | Kd (BSI) | Notes |
|---|---|---|---|---|
| 21 | 1b | 49.9 ± 9.5 nM | 36.1 ± 23.6 pM | |
| 21-E | 1b | 223 ± 92 nM | 350 ± 240 pM | |
| 21-R | 1b | no binding b | no binding b | |
| Qβ@21(s) a | 1b | n.a. | 30.0 ± 3.5 pM | |
| Qβ@21(d) a | 1b | n.a. | 24.0 ± 2.4 pM | |
| Qβ@21-E(s) a | 1b | n.a. | 3.4 ± 1.3 nM | |
| Qβ@21-E(d) a | 1b | n.a. | 2.6 ± 1.1 nM | |
| Qβ@21-R(s) a | 1b | n.a. | no binding b | |
| Qβ@21-R(d) a | 1b | n.a. | no binding b | |
| ThA | Theophylline | n.d. | 1.32 ± 0.53 nM | literature: 0.21 – 0.86 µM c–g |
| ThA | Caffeine | n.d. | no binding b | literature: ~3.5 mM h |
The @ symbol denotes an aptamer packaged inside the Qβ virus-like particle shell; values of Kd are calculated per particle. (s) indicates the use of the single-pasmid method, and (d) notes the use of a dual-plasmid system.
No signal was observed at the highest ligand concentration tested.
0.4 µM (eq. filtration).44
0.21 µM (eq. filtration).45
0.30 µM (UV-vis).46
0.86 ± 0.12 µM (ITC).47
0.3 ± 0.1 µM (fluorescence).40
Affinity determination was not performed for this aptamer sequence and caffeine, so the value listed is for a similar theophylline aptamer with the same binding site sequence (TCT8-4).6
(n.a. = not applicable; n.d. = not determined.)
Encapsidation and analysis of HAP aptamers in Qβ VLPs
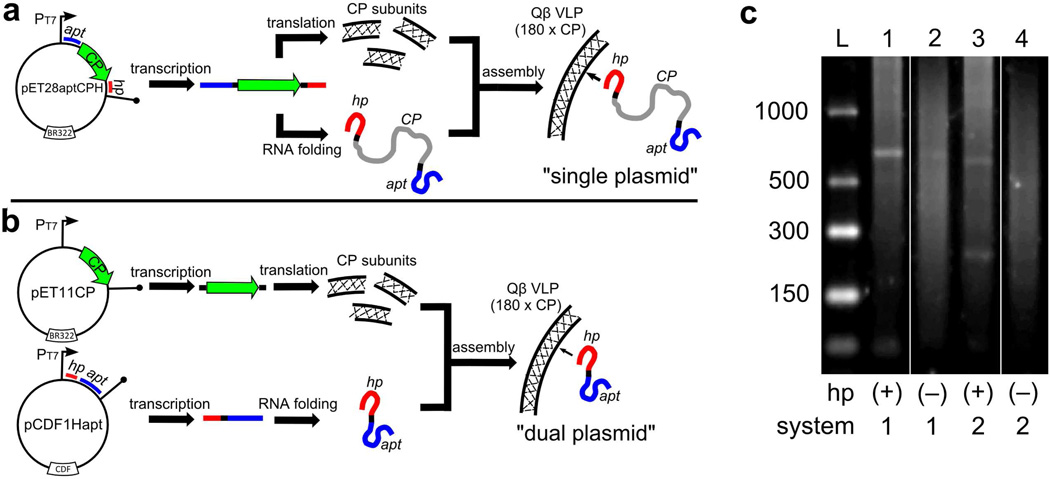
Qβ VLPs encapsidating aptamers 21, 21-E, and 21-R were produced in E. coli with “single-plasmid” and “dual-plasmid” expression systems, differing in the organization of the genomic information. The former, an approach similar to that used for the expression and packaging of heterologous mRNA in MS2 VLPs,29 involves placing the Qβ hairpin and aptamer 21 at opposite ends of the Qβ coat protein gene (plasmid pET28apt21CPH, Figure 3a), giving rise to a relatively long (658 nt) mRNA sequence. In the latter approach, we placed the coat protein and aptamer/hairpin sequences on separate plasmids (pET11CP and pCDF1Hapt21, respectively) with different origins of replication and different antibiotic resistance markers; the aptamer of interest was therefore encoded into a much smaller (228 nt) polynucleotide (Figure 3b). The dual-plasmid method is more modular, since coat protein and aptamer variants possessing different structural or functional properties can be separately introduced. We also generated constructs containing aptamer 21 but lacking the Qβ hairpin (pET28apt21CP and pCDF1apt21).
Figure 3.
Generation of Qβ-packaged aptamers. (a) A single-plasmid approach, which produces RNA sequences encoding a fusion of the Qβ hairpin, coat protein, and aptamer. (b) A dual-plasmid approach, in which the coat protein and hairpin-labeled aptamer sequences are expressed from two separate plasmids. (c) Agarose gel electrophoresis of RNA extracted from the indicated VLPs. The lanes were loaded with the following samples (expected length of packaged RNA in parentheses and boxed): (1) pET28apt21CPH (658 nt), (2) pET28apt21CP (626 nt) (3) pET11CP + pCDF1Hapt21 (228 nt), (4) pET11CP + pCDF1apt21 (188 nt). On the left is single-stranded RNA ladder. “hp” indicates the presence or absence of the Qβ hairpin, “system” denotes production by the single (1) or dual (2) plasmid methods. A band corresponding to the coat protein transcript (587 nt) is also visible in lanes 3 and 4.
The VLPs obtained from these systems were indistinguishable in size and protein content from standard Qβ VLPs by size-exclusion chromatography, dynamic light scattering, and gel electrophoresis analysis (Figure S11). RT-PCR of RNA extracted from the VLPs confirmed that the particles packaged their expected RNA payloads (Figure S12), but other RNA was encapsidated as well. Treatment of the particles with RNase produced no detectable change in the packaged RNA nor in its binding properties (Figure S13), showing that the capsid shell is impermeable to the enzyme.
RNA could be isolated from VLPs after chemical denaturation of the protein, gel electrophoresis of which showed discrete bands corresponding to the predicted RNA transcript lengths (Figure 3c). However, a broad distribution of other RNA molecules was also observed, suggesting degradation of transcripts in the E. coli cytosol and/or packaging of cellular mRNA.30,31 The transcripts lacking hairpin were packaged less efficiently than the transcripts containing hairpin (lane 1 vs. 2; lane 3 vs. 4).
The RNA isolated from pET28apt21CPH particles was also subjected to Next Generation sequencing.32,33 Briefly, the extracted RNA was fragmented and ligated to adaptor sequences, then subjected to reverse transcription and PCR to amplify the complementary DNA. DNA products containing an insert of approximately 40 bases between the adaptors were separated by gel electrophoresis, loaded onto the flowcell, and subjected to bridge amplification to generate clonal clusters for sequencing. Making the assumption that no strong amplification bias occurred to skew the results, approximately 14% of the fragments read in this analysis were derived from the aptamer-CP-hairpin sequence expressed under the T7 promotor, 35% of the fragments were from the rest of the plasmid carrying that sequence, and 31% were from sequences matching portions of the E. coli genome. Since the transcription terminator is not perfect, we expect the entire plasmid to be transcribed a fraction of the time under accelerated T7 promotion, and its size should make it well-represented among the fragments identified in the sequencing experiment, even if it is packaged relatively infrequently. These results are consistent with directed packaging by the Qβ hairpin, but also confirm that other RNA molecules associate with the positively-charged interior surface of the capsid protein. They do not allow us to estimate the number of aptamers packaged per particle, because the RNA extraction and processing steps produce extensive cleavage (Figure 3C) and may well skew the populations of the detected sequences. However, it is very likely that the number of packaged aptamers is small, probably fewer than 5 per particle.
Because capsid-packaged aptamers are sequestered from direct contact with their environment, it is impossible to use SPR, and fluorophore labeling was judged to be likely to distort interactions with oligonucleotide binding agents. We therefore turned to the technique of backscattering interferometry (BSI),34,35 as the label-free analytical technique most applicable to the situation. While BSI has been successfully applied to antibody-antigen,35,36 protein-protein,34,37 and protein-ligand interactions,34,38,39 aptamer-ligand binding has not previously been examined. BSI detects changes in bulk refractive index caused by binding interactions, presumably due to changes in hydration and dipole moment of the binding partners. Since the structure and conformational flexibility of some aptamers varies significantly in the presence and absence of ligand,40–43 we anticipated that BSI would be well suited to this application.
BSI analyses of aptamer samples equilibrated with increasing concentrations of ligand 1b generated single-site binding curves that gave Kd values shown in Table 2 (data shown in Supplementary Information). The absolute magnitudes of the binding constants measured by this technique were approximately 1000-fold stronger than the values obtained by SPR: 40 pM vs. 50 nM for aptamer 21, and 350 pM vs. 220 nM for 21-E. Similarly, BSI analysis of the binding of the known aptamer ThA to theophylline was also found to give values more than two orders of magnitude stronger than measurements obtained with other methods (Table 2). Caffeine, known to interact far more weakly with ThA than theophylline, did not show detectable binding in the concentration range studied by BSI.
We do not yet understand why BSI reports tighter binding, and experiments to probe the possible causes are underway. This type of discrepancy has been observed previously for other molecules, but to a much lesser extent.38 It should be noted that the BSI measurement is done in solution whereas SPR is performed on a surface, one giving an equilibrium dissociation value (Kd) and the other an adsorption isotherm (1/Kads).38,48 However, in all cases BSI accurately reports the relative differences in affinities for comparisons of different receptor molecules binding the same ligand, or different ligands binding the same receptor. Thus, the binding affinities of aptamers 21, 21-E, and 21-R retain their relative ordering when analyzed by both SPR and BSI, and the difference in binding of caffeine vs. theophylline by ThA was correctly assessed by BSI.
Importantly, binding of ligands to the VLP-packaged aptamers was clearly observed by BSI, with results summarized in Table 2. Assuming that the number of packaged aptamers per particle is small, the the affinities of aptamers 21 and 21E for 1b were in the same approximate range in their packaged vs. free forms. In both cases the randomized aptamer 21-R showed no binding by BSI. No differences were observed in any comparison between the Qβ-packaged aptamers using single- vs. dual-plasmid methods, showing that the length of the RNA in which the aptamer is embedded has little effect on binding, both sequences being substantially longer than the aptamer itself. Calculated predictions of secondary structure for both transcripts28 showed the proposed active structure (Figure 2D) to be well represented among the lowest-energy possible alignments.
Conclusions
A variation of the SELEX method developed by Davis and Szostak, featuring stringent selection by pre-washing with the ligand of interest,49 produced a consensus high-affinity sequence for a “generic” drug-like small molecule after only two phases of selection. The encapsidation of this aptamer, as well as a known aptamer for theophylline, inside Qβ capsids by assembly during the production of the particle in E. coli imbued the particles with the ability to bind the cognate ligand tightly. The label-free solution phase method of backscattering interferometry was also shown for the first time to be suitable for the characterization of RNA aptamers both outside and inside the VLP shell. While the absolute values obtained differed substantially from those determined by other methods, BSI provided accurate relative binding affinities in a convenient and rapid manner, requiring very small amounts of RNA (10–100 pmoles).
Combined with our previous use of a peptide-binding aptamer to assist protein packaging,17 these results show that RNA aptamer sequences are functional when embedded in polynucleotides that are spontaneously assembled with coat protein subunits, and therefore can be used as general genetically-encoded binding agents inside virus-like particles. The capsid protects the aptamers from degradation by nucleases while allowing small-molecule ligand entry and providing sufficient room for the conformational changes in the aptamer required for binding. It is therefore anticipated that the full range of RNA function can be brought to engineered protein nanoparticles in this manner.
Experimental Section
HAP molecules were synthesized with procedures similar to those described previously.19 Detailed synthetic schemes and characterization data are provided in the Supplementary Information. All RNA samples were generated by run-off transcription and purified on TBE-urea polyacrylamide gels, extracted with a crush-and-soak technique, desalted, and concentrated. A table of DNA primers and detailed experimental procedures may also be found in the Supplementary Information.
Selection procedure
Thiol-functionalized HAP 1a was immobilized on epoxy-Sepharose (GE Healthcare) in water/DMF in the presence of sodium hydroxide. Unreacted epoxy groups were capped with ethanolamine, and the resin was washed extensively and stored at 4 °C in water containing 0.02% sodium azide. A second batch of resin, capped only with ethanolamine, was prepared for counterselection. The selection buffer contained 150 mM NaCl, 5 mM MgCl2, and 50 mM HEPES, pH 7.4 at 20 °C. Taq DNA polymerase (Roche) was used for PCR amplification, SuperScript III reverse transcriptase (Invitrogen) was used to generate cDNA. T7 RNA polymerase (USB Corporation) was used in the presence of α-32P-CTP to generate radiolabeled run-off transcripts.
At the start of each round of selection, the RNA in folding buffer was heated to 80 °C for 3 min, then allowed to cool to room temperature over 15 min. The RNA was first applied to the counterselection resin, the flow-through of which was applied to the HAP-labeled resin. The HAP-labeled resin was washed with 15 column volumes of folding buffer and subjected to successive 30 min incubations with a high concentration (5 mM) solution of HAP 1b. Aliquots of the wash and elution fractions were subjected to scintillation counting. The elution fractions were pooled and subjected to RT-PCR, followed by run-off transcription, to generate the pool used for the subsequent round of selection.
In some rounds, two “pre-elution” rinses with HAP 1b were performed before the elution step.26,49 The length of the pre-elution incubations was increased during the course of the selection as indicated in Figure 1. A solution of 5 mM 1b was used for the pre-elution rinses, except in round 1.6, in which 1.3 mM 1b was used. RNA molecules that preferentially bound to soluble ligand 1b over resin-immobilized ligand were selected by decreasing the number of fractions used to elute the products, from four elution fractions in rounds 1.1 through 1.5, to two elution fractions in rounds 1.6 though 2.4, to one fraction in round 2.5. Before round 1.9, hypermutagenic PCR was performed by the method of Wain-Hobson and coworkers,27 with a target error rate of 10% per base.
Generation of starting pools
Approximately 3 × 1014 DNA template molecules were used for transcription of the initial pool, and ~6 × 1014 DNA template molecules were used for transcription of the pre-organized pool in the second phase of selection. The DNA templates had the following sequences, with the T7 promoter underlined. The initial, naive pool contained a 50-base random region50 was: TCTAACACGACTCACTATAGGGCTGACTGATCTCATCTAGCG(N50)-ACAGTGCCTAGTCGATCAGATGAG. The antisense strand of this sequence was generated with PCR. The pre-organized pool for the second phase of selection, with bases marked in bold randomized at 30% per position was: AAGCCGTCCACATAAGCAGCACTGTACCCTCTCGGGTTCAGAAATTTACGCTCTCGCGAGTTGGCTGCCTGGCCTACCCTATAGTGAGTCGTATTAGA. This antisense strand was annealed with a primer (AGTAATACGACTCACTATAGGGTAGGCC) to perform transcription.
Production of RNA for SPR and BSI
RNA for SPR and BSI studies was produced by transcription with T7 RNA polymerase, followed by PAGE purification. The HAP aptamer sequences (Table 1) were inserted into the pCDF-1b vector (Novagen) using standard cloning techniques. The DNA templates were amplified using Phusion polymerase (Finnzymes) with primers HA1 (AAGCCGTCCACATAAGCAG) and HA2 (TCTAATACGACTCACTATAGGGTAGGCCAGGCAG), the latter of which adds the T7 promoter sequence. The antisense strand ThAtemp-comp (GACGCTGCCAAGGGCCTTTCGGCTGGTATCGCCTATAGTGAGTCGTATTAGA) was annealed with ThAtrans-F (TCTAATACGACTCACTATAGGC) prior to transcription to generate the theophylline aptamer.
Production of Qβ VLPs
Two methods were employed for generating packaged aptamers. In a “ dual-plasmid” system, E. coli BL21(DE3) were co-transformed with one plasmid (pET11CP) encoding the Qβ coat protein and another (pCDF-1b) encoding the aptamer sequence of interest. These two plasmids contain different antibiotic resistance genes and origins of replication, so their copy numbers are stably maintained. In the “single-plasmid” system, expression was performed with a single plasmid (pET28aptCPH) containing the sequence for both the coat protein and aptamer. In each case, the RNA sequence that the native Qβ genome uses to associate with the interior coat protein surface was installed on the RNA containing the aptamer, to promote its encapsidation. This system is explained in detail elsewhere,17 as well as in the Supplementary Information. Particles were expressed and purified as previously described.51 After purification on 10–40% continuous sucrose gradients and ultracentrifugation, VLPs were characterized by size-exclusion chromatography (Superose 6) and protein size analysis (Agilent Bioanalyzer Protein 80 chip).
Next Generation Sequencing
1µg of RNA was isolated from the Qβ VLPs as described in Supporting Information and was prepared for Next Generation Sequencing using a slightly modified version of the Illumina protocol,52 in which 12 cycles of PCR were performed instead of 15 and standard Truseq adapters and barcoded primers were used for sample identification. The sample was then spiked into a phi X control lane of a HiSeq v2 single-read flowcell at approximately 20%, and 40 base sequencing of the insert product and 7 base barcode sequencing was carried out using an Illumina GAIIx system and standard HiSeq sequencing reagents. Reads belonging to the RNA sample were identified based on the barcode sequence located in the primer region using the standard Illumina Truseq barcoding protocols. The separated barcoded reads (8.8 million) were aligned in CLC Bio Workbench (www.clcbio.com) to the pET28aptCPH plasmid sequence and to the E. coli genome. All insert and vector reads were in the predicted direction of transcription relative to the vector transcript start site.
Surface plasmon resonance
The gold surface of a SpotReady™ 25 chip (GWC Technologies) was coated with aminooctanethiol to generate a monolayer. The bifunctional LC-SMCC linker (Thermo Fisher) was used to functionalize the monolayer with a 3:1 molar ratio of HAP-thiol 1a and a PEG-thiol. Chips were stored in fresh PBS and used within 2 weeks. For data acquisition, a functionalized chip was mounted into an SPRImager®II (GWC Technologies), and folding buffer was introduced into the flow cell. The CCD camera alignment was adjusted to provide recommended spot intensities (~80% of maximal signal). Various concentrations of RNA aptamers were flowed over the chip at a constant flow rate (~0.2mL/min), and the change in spot intensity upon binding was recorded over the course of 15 minutes. Bound RNA was removed from the chip surface between measurements by extensive rinsing with 10X PBS and re-equilibration in folding buffer. Complete removal was denoted by full recovery of spot intensities. Spot intensity changes were plotted against RNA concentrations and the resulting curves fit to a Langmuir adsorption isotherm to obtain surface adsorption constants (Kads) for each RNA aptamer-ligand pair.
Backscattering interferometry
RNA aptamer-ligand binding was examined by incubating a fixed amount of RNA with varying concentrations of a ligand for 8 hours in the dark at 4 °C. This long incubation period was not necessary, but was used for experimental convenience. Microfluidic channels were pre-treated by incubation with a solution of concentrated sulfuric acid for 5 min followed by extensive rinsing in 18.2 MΩ-cm water. For each sample, a solution of “negative control” (nonbinding) aptamer and ligand was injected into the channel and the BSI signal measured for 15 seconds. The channel was rinsed and an equilibrated solution of “unknown” aptamer and ligand was injected into the channel. This procedure was then repeated iteratively for mixtures containing increasing concentrations of ligand. The binding signal was calculated as the observed difference in phase between the nonbinding aptamer-ligand solution and the “unknown” (undetermined ligand affinity) aptamer-ligand solution. This corrected binding signal was plotted versus concentration to produce a saturation binding curve and fitted to a square hyperbolic function to calculate the solution equilibrium binding affinities (Kd). Aptamer 21-R was used as the nonbinding aptamer for experiments performed on aptamers 21 or 21-E; aptamer 21 was the nonbinding control for the theophylline aptamer.
Supplementary Material
Acknowledgments
This work was supported by The Skaggs Institute for Chemical Biology and the National Institutes of Health (RR021886, AI067417, EB003537). MMB was supported by the National Institute of Allergy and Infectious Diseases (T32AI007354). We thank Dr. Steven Head and Ms. Lana Schaffer for assistance with Next Generation sequencing, Prof. Adam Zlotnick for introducing us to HAP molecules, Dr. Sreenivas Punna for the synthesis of the initial HAP compound, and Dr. Phoebe Glazer and Prof. David Goodin for use of the SPR instrument.
Footnotes
Supporting Information Available. Details of all experiments, including syntheses, RNA production, analytical procedures, and data processing. Also included is a complete table of aptamer-small molecule interactions reported in the literature. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Shamah SM, Healy JM, Cload ST. Complex Target SELEX. Accounts Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 4.Carothers JM, Oestreich SC, Davis JH, Szostak JW. Informational Complexity and Functional Activity of RNA Structures. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic Characterization of an Engineered Tetracycline-Binding Riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenison RD, Gill SC, Pardi A, Polisky B. High-Resolution Molecular Discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 7.Sazani PL, Larralde R, Szostak JW. A Small Aptamer with Strong and Specific Recognition of the Triphosphate of ATP. J. Am. Chem. Soc. 2004;126:8370–8371. doi: 10.1021/ja049171k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozlovska TM, Cielens I, Dreilinna D, Dislers A, Baumanis V, Ose V, Pumpens P. Recombinant RNA Phage Qβ Capsid Particles Synthesized and Self-Assembled in Escherichia Coli. Gene. 1993;137:133–137. doi: 10.1016/0378-1119(93)90261-z. [DOI] [PubMed] [Google Scholar]
- 9.Strable E, Prasuhn DE, Jr, Udit AK, Brown S, Link AJ, Ngo JT, Lander G, Quispe J, Potter CS, Carragher B, et al. Unnatural Amino Acid Incorporation into Virus-Like Particles. Bioconjugate Chem. 2008;19:866–875. doi: 10.1021/bc700390r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udit AK, Everett C, Gale AJ, Kyle JR, Ozkan M, Finn MG. Heparin Antagonism by Polyvalent Display of Cationic Motifs on Virus-Like Particles. ChemBioChem. 2009;10:503–510. doi: 10.1002/cbic.200800493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee D, Liu A, Voss N, Schmid S, Finn MG. Multivalent Display and Receptor-Mediated Endocytosis of Transferrin on Virus-Like Particles. ChemBioChem. 2010;11:1273–1279. doi: 10.1002/cbic.201000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witherell GW, Uhlenbeck OC. Specific RNA Binding by Qβ Coat Protein. Biochemistry. 1989;28:71–76. doi: 10.1021/bi00427a011. [DOI] [PubMed] [Google Scholar]
- 13.Lim F, Spingola M, Peabody DS. The RNA-Binding Site of Bacteriophage Qβ Coat Protein. J. Biol. Chem. 1996;271:31830–31845. doi: 10.1074/jbc.271.50.31839. [DOI] [PubMed] [Google Scholar]
- 14.Pickett GG, Peabody DS. Encapsidation of Heterologous RNAs by Bacteriophage MS2 Coat Protein. Nucleic Acids Res. 1993;21:4621–4626. doi: 10.1093/nar/21.19.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, DuBois DB. Armored RNA Technology for Production of Ribonuclease-Resistant Viral RNA Controls and Standards. J. Clin. Microbiol. 1998;36:3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, Brown WL, Stockley PG. Cell-Specific Delivery of Bacteriophage-Encapsidated Ricin A Chain. Bioconjugate Chem. 1995;6:587–595. doi: 10.1021/bc00035a013. [DOI] [PubMed] [Google Scholar]
- 17.Fiedler JD, Brown SD, Lau J, Finn MG. RNA-Directed Packaging of Enzymes within Virus-Like Particles. Angew. Chem. Int. Ed. 2010;49:9648–9651. doi: 10.1002/anie.201005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carothers JM, Goler JA, Kapoor Y, Lara L, Keasling JD. Selecting RNA Aptamers for Synthetic Biology: Investigating Magnesium Dependence and Predicting Binding Affinity. Nucleic Acids Res. 2010;38:2736–2747. doi: 10.1093/nar/gkq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick AA. Heteroaryldihydropyrimidine Activates and Can Misdirect Hepatitis B Virus Capsid Assembly. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourne CR, Finn MG, Zlotnick A. Global Structural Changes in Hepatitis B Virus Capsids Induced by the Assembly Effector HAP1. J. Virol. 2006;80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. Small-Molecule Effectors of Hepatitis B Virus Capsid Assembly Give Insight into Virus Life Cycle. J. Virol. 2008;82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J. Org. Chem. 2002;67:3057–3062. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 23.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Hong V, Presolski SI, Ma C, Finn MG. Analysis and Optimization of Copper-Catalyzed Azide-Alkyne Cycloaddition for Bioconjugation. Angew. Chem. Int. Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis JH, Szostak JW. Isolation of High-Affinity GTP Aptamers from Partially Structured RNA Libraries. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11616–11621. doi: 10.1073/pnas.182095699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger A, Burgstaller P, Von der EH, Roeder A, Famulok M. RNA Aptamers That Bind L-Arginine with Sub-Micromolar Dissociation Constants and High Enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartanian JP, Henry M, Wain-Hobson S. Hypermutagenic Pcr Involving All Four Transitions and a Sizeable Proportion of Transversions. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legendre D, Fastrez J. Production in Saccharomyces Cerevisiae of MS2 Virus-Like Particles Packaging Functional Heterologous mRNAs. J. Biotechnol. 2005;117:183–194. doi: 10.1016/j.jbiotec.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 30.LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutants of the Bacteriophage MS2 Coat Protein that Alter Its Cooperative Binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 31.Bundy BC, Franciszkowicz MJ, Swartz JR. Escherichia Coli-Based Cell-Free Synthesis of Virus-Like Particles. Biotechnol. Bioeng. 2008;100:28–37. doi: 10.1002/bit.21716. [DOI] [PubMed] [Google Scholar]
- 32.Schuster SC. Next-Generation Sequencing Transforms Today's Biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Gerstein M, Snyder M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD, Sorensen HS. Free-Solution, Label-Free Molecular Interactions Studied by Back-Scattering Interferometry. Science. 2007;317:1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 35.Baksh MM, Kussrow AK, Mileni M, Finn MG, Bornhop DJ. Label-Free Quantification of Membrane-Ligand Interactions Using Backscattering Interferometry. Nat. Biotechnol. 2011;29:357–360. doi: 10.1038/nbt.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markov DA, Swinney K, Bornhop DJ. Label-Free Molecular Interaction Determinations with Nanoscale Interferometry. J. Am. Chem. Soc. 2004;126:16659–16664. doi: 10.1021/ja047820m. [DOI] [PubMed] [Google Scholar]
- 37.Latham JC, Stein RA, Bornhop DJ, Mchaourab HS. Free-Solution Label-Free Detection of α-Crystallin Chaperone Interactions by Back-Scattering Interferometry. Anal. Chem. 2009;81:1865–1871. doi: 10.1021/ac802327h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kussrow A, Kaltgrad E, Wolfenden ML, Cloninger MJ, Finn MG, Bornhop DJ. Measurement of Monovalent and Polyvalent Carbohydrate-Lectin Binding by Back-Scattering Interferometry. Anal. Chem. 2009;81:4889–4897. doi: 10.1021/ac900569c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kussrow AK, Enders CS, Castro AR, Cox DL, Ballard RC, Bornhop DJ. The Potential of Backscattering Interferometry as an in vitro Clinical Diagnostic Tool for the Serological Diagnosis of Infectious Disease. Analyst. 2010 doi: 10.1039/c0an00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jucker FM, Phillips RM, McCallum SA, Pardi A. Role of a Heterogeneous Free State in the Formation of a Specific RNA–Theophylline Complex. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 41.Duchardt-Ferner E, Weigand JE, Ohlenschlager O, Schtnidtke SR, Suess B, Wohnert J. Highly Modular Structure and Ligand Binding by Conformational Capture in a Minimalistic Riboswitch. Angew. Chem. Int. Ed. 2010;49:6216–6219. doi: 10.1002/anie.201001339. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking Structural Motifs Mediate Molecular Discrimination by a Theophylline-Binding RNA. Nat. Struct. Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 43.Flinders J, DeFina SC, Brackett DM, Baugh C, Wilson C, Dieckmann T. Recognition of Planar and Nonplanar Ligands in the Malachite Green–RNA Aptamer Complex. ChemBioChem. 2004;5:62–72. doi: 10.1002/cbic.200300701. [DOI] [PubMed] [Google Scholar]
- 44.Zimmermann GR, Shields TP, Jenison RD, Wick CL, Pardi A. A Semiconserved Residue Inhibits Complex Formation by Stabilizing Interactions in the Free State of a Theophylline-Binding RNA. Biochemistry. 1998;37:9186–9192. doi: 10.1021/bi980082s. [DOI] [PubMed] [Google Scholar]
- 45.Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A. Molecular Interactions and Metal Binding in the Theophylline-Binding Core of an RNA Aptamer. RNA. 2000;6:659–667. doi: 10.1017/s1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson PC, Mecozzi S. Unusually Short RNA Sequences: Design of a 13-Mer RNA That Selectively Binds and Recognizes Theophylline. J. Am. Chem. Soc. 2005;127:5290–5291. doi: 10.1021/ja0432463. [DOI] [PubMed] [Google Scholar]
- 47.Lee SW, Zhao L, Pardi A, Xia TB. Ultrafast Dynamics Show That the Theophylline and 3-Methylxanthine Aptamers Employ a Conformational Capture Mechanism for Binding Their Ligands. Biochemistry. 2010;49:2943–2951. doi: 10.1021/bi100106c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Y, Wolf LK, Georgiadis RM. Secondary Structure Effects on DNA Hybridization Kinetics: A Solution Versus Surface Comparison. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen DH, DeFina SC, Fink WH, Dieckmann T. Binding to an RNA Aptamer Changes the Charge Distribution and Conformation of Malachite Green. J. Am. Chem. Soc. 2002;124:15081–15084. doi: 10.1021/ja027635d. [DOI] [PubMed] [Google Scholar]
- 50.Unrau PJ, Bartel DP. RNA-Catalysed Nucleotide Synthesis. Nature. 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- 51.Brown SD, Fiedler JD, Finn MG. Assembly of Hybrid Bacteriophage Qβ Virus-Like Particles. Biochemistry. 2009;48:11155–11157. doi: 10.1021/bi901306p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive Comparative Analysis of Strand-Specific RNA Sequencing Methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. available from the Illumina web site at: https://icom.illumina.com/download/summary/K1tU5zFy8E6swZfRoxla7Q. See also a protocol titled "Directional mRNA-Seq Sample Preparation Guide,"
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.